Methodological Validation and Inter-Laboratory Comparison of Microneutralization Assay for Detecting Anti-AAV9 Neutralizing Antibody in Human
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and rAAV-EGFP-2A-Gluc Viruses
2.2. Serum or Plasma Samples
2.3. Optimizing Key Variables for Anti-AAV9 MN Assay Development
2.4. AAV9 MN Assay Protocol
2.5. Quality Control
2.6. Study Design, Method Transfer and Validation
2.7. Statistical Analysis
3. Results
3.1. Anti-AAV9 MN Assay Establishment
3.2. Key Parameters Validation
3.2.1. TCP Assay
3.2.2. Assay Precision
3.2.3. Sensitivity and Hook Effect
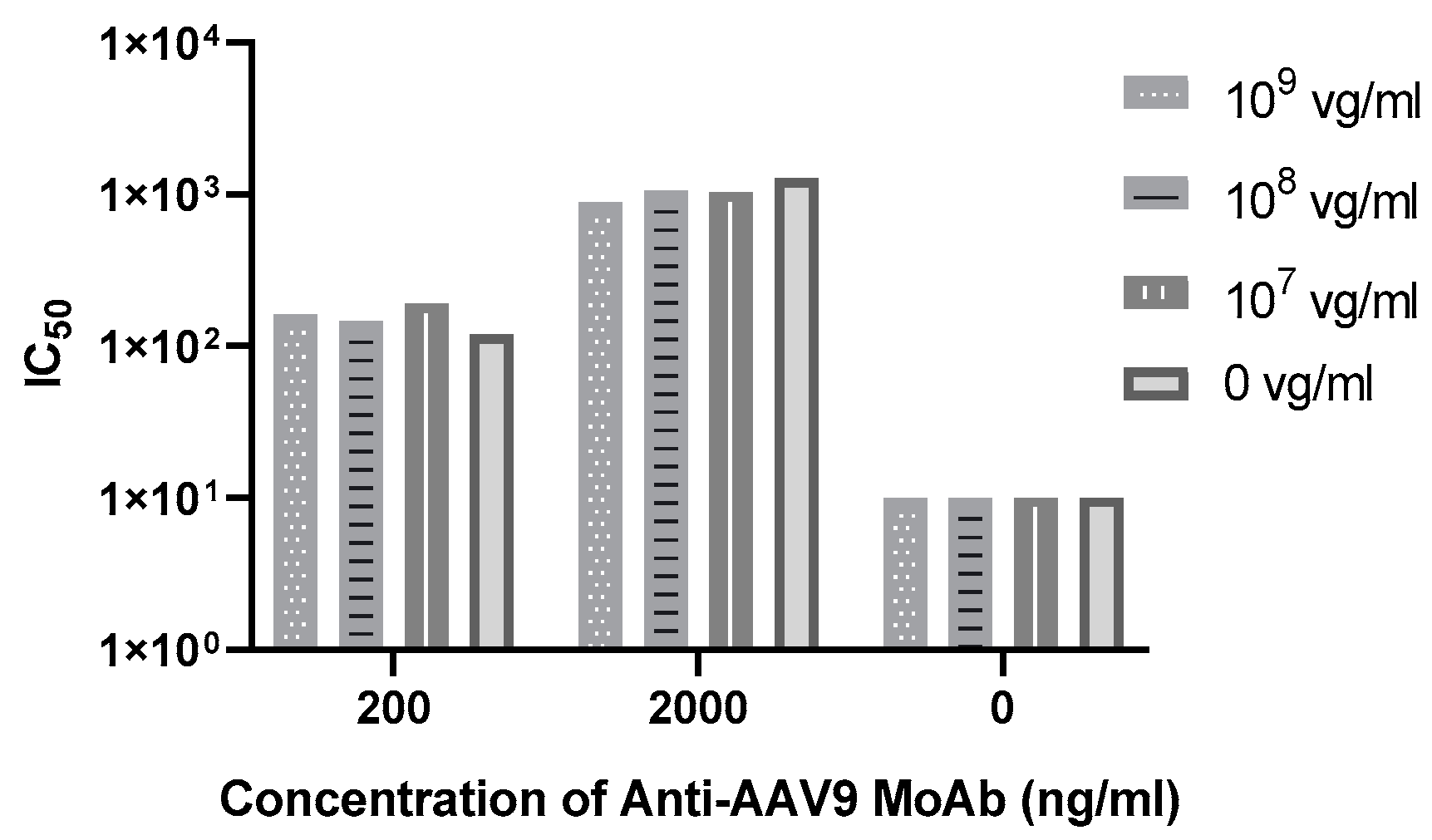
3.2.4. Drug Tolerance
3.2.5. Specificity
3.2.6. Robustness and Stability
3.2.7. System Suitability
3.2.8. Assay Repeatability
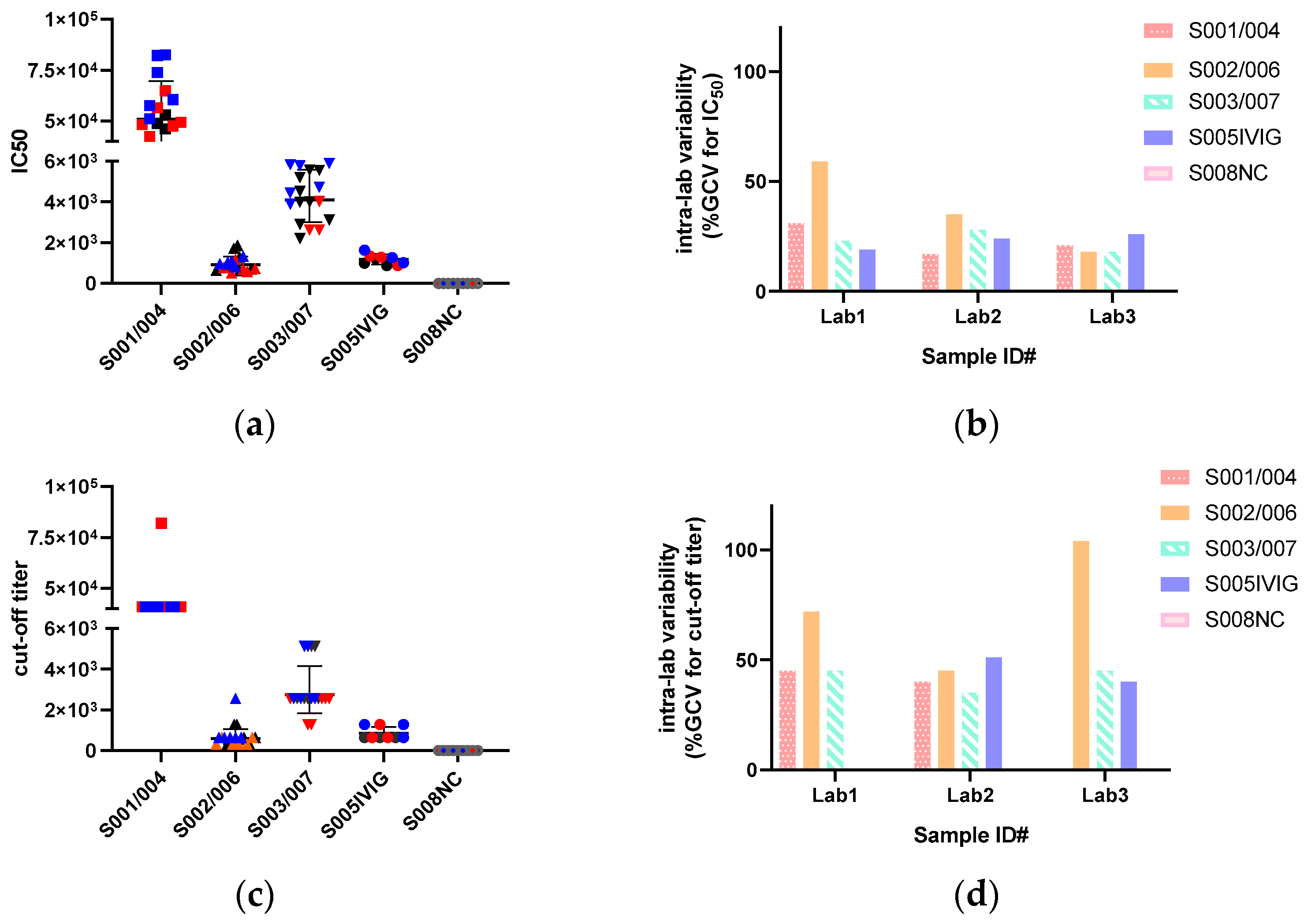
3.3. IC50 Are Less Variable than Cut-Off Titers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kruzik, A.; Fetahagic, D.; Hartlieb, B.; Dorn, S.; Koppensteiner, H.; Horling, F.M.; Scheiflinger, F.; Reipert, B.M.; de la Rosa, M. Prevalence of Anti-Adeno-Associated Virus Immune Responses in International Cohorts of Healthy Donors. Mol. Ther. Methods Clin. Dev. 2019, 14, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Louis Jeune, V.; Joergensen, J.A.; Hajjar, R.J.; Weber, T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods 2013, 24, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Aronson, S.J.; Veron, P.; Collaud, F.; Hubert, A.; Delahais, V.; Honnet, G.; de Knegt, R.J.; Junge, N.; Baumann, U.; Di Giorgio, A.; et al. Prevalence and Relevance of Pre-Existing Anti-Adeno-Associated Virus Immunity in the Context of Gene Therapy for Crigler-Najjar Syndrome. Hum. Gene Ther. 2019, 30, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.; Levy, D.I.; Petropoulos, C.J.; Bashirians, G.; Winburn, I.; Mahn, M.; Somanathan, S.; Cheng, S.H.; Byrne, B.J. Binding and neutralizing anti-AAV antibodies: Detection and implications for rAAV-mediated gene therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2023, 31, 616–630. [Google Scholar] [CrossRef]
- van der Marel, S.; Comijn, E.M.; Verspaget, H.W.; van Deventer, S.; van den Brink, G.R.; Petry, H.; Hommes, D.W.; Ferreira, V. Neutralizing antibodies against adeno-associated viruses in inflammatory bowel disease patients: Implications for gene therapy. Inflamm. Bowel Dis. 2011, 17, 2436–2442. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, C.; Dong, Z.; Zhu, X.; Zheng, X.; Liu, Z.; Zhou, J.; Yu, S.; Wu, X.; Dong, X. Using an In Vivo Mouse Model to Determine the Exclusion Criteria of Preexisting Anti-AAV9 Neutralizing Antibody Titer of Pompe Disease Patients in Clinical Trials. Viruses 2024, 16, 400. [Google Scholar] [CrossRef]
- West, C.; Federspiel, J.D.; Rogers, K.; Khatri, A.; Rao-Dayton, S.; Ocana, M.F.; Lim, S.; D’Antona, A.M.; Casinghino, S.; Somanathan, S. Complement Activation by Adeno-Associated Virus-Neutralizing Antibody Complexes. Hum. Gene Ther. 2023, 34, 554–566. [Google Scholar] [CrossRef]
- Meadows, A.S.; Pineda, R.J.; Goodchild, L.; Bobo, T.A.; Fu, H. Threshold for Pre-existing Antibody Levels Limiting Transduction Efficiency of Systemic rAAV9 Gene Delivery: Relevance for Translation. Mol. Ther. Methods Clin. Dev. 2019, 13, 453–462. [Google Scholar] [CrossRef]
- Al-Zaidy, S.A.; Mendell, J.R. From Clinical Trials to Clinical Practice: Practical Considerations for Gene Replacement Therapy in SMA Type 1. Pediatr. Neurol. 2019, 100, 3–11. [Google Scholar] [CrossRef]
- ICH. M10 Bioanalytical Method Validation and Study Sample Analysis. 2021. Available online: https://www.cde.org.cn/ichWeb/guideIch/toGuideIch/4/1 (accessed on 15 June 2024).
- Boutin, S.; Monteilhet, V.; Veron, P.; Leborgne, C.; Benveniste, O.; Montus, M.F.; Masurier, C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010, 21, 704–712. [Google Scholar] [CrossRef]
- Dai, Y.; Dong, H.; Gleason, C.; Mora, J.; Kolaitis, G.; Balasubramanian, N.; Surapaneni, S.; Kozhich, A.; Jawa, V. Comparison of Pre-existing Anti-AAV8 Total Antibody Screening and Confirmatory Assays with a Cell-Based Neutralizing Assay in Normal Human Serum. AAPS J. 2023, 25, 35. [Google Scholar] [CrossRef]
- Calcedo, R.; Vandenberghe, L.H.; Gao, G.; Lin, J.; Wilson, J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009, 199, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Petry, H.; Brooks, A.; Orme, A.; Wang, P.; Liu, P.; Xie, J.; Kretschmer, P.; Qian, H.S.; Hermiston, T.W.; Harkins, R.N. Effect of viral dose on neutralizing antibody response and transgene expression after AAV1 vector re-administration in mice. Gene Ther. 2008, 15, 54–60. [Google Scholar] [CrossRef]
- Falese, L.; Sandza, K.; Yates, B.; Triffault, S.; Gangar, S.; Long, B.; Tsuruda, L.; Carter, B.; Vettermann, C.; Zoog, S.J.; et al. Strategy to detect pre-existing immunity to AAV gene therapy. Gene Ther. 2017, 24, 768–778. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, W.; Zhang, H.; Wang, Y.; Zhao, J.; Song, A.; Xie, H.; Zhao, C.; Gao, D.; Wang, Y. Neutralizing antibodies against AAV2, AAV5B and AAV8 in healthy and HIV-1-infected subjects in China: Implications for gene therapy using AAV vectors. Gene Ther. 2014, 21, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Connolly, A.M.; Lehman, K.J.; Griffin, D.A.; Khan, S.Z.; Dharia, S.D.; Quintana-Gallardo, L.; Rodino-Klapac, L.R. Testing preexisting antibodies prior to AAV gene transfer therapy: Rationale, lessons and future considerations. Mol. Ther. Methods Clin. Dev. 2022, 25, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Liu, S.; Ou, L. rAAV immunogenicity, toxicity, and durability in 255 clinical trials: A meta-analysis. Front. Immunol. 2022, 13, 1001263. [Google Scholar] [CrossRef]
- Zincarelli, C.; Soltys, S.; Rengo, G.; Rabinowitz, J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 1073–1080. [Google Scholar] [CrossRef]
- Dong, X.; Tian, W.; Yuan, Z.; Tan, S.; Wu, X. the novel recombinant AAV5/5 vevtor and the packaging system. Chin. J. Biotechnol. 2010, 26, 679–686. [Google Scholar]
- Yu, Z.; Zhou, S.; Luo, N.; Ho, C.Y.; Chen, M.; Chen, H. TPP Combined with DGUC as an Economic and Universal Process for Large-Scale Purification of AAV Vectors. Mol. Ther. Methods Clin. Dev. 2020, 17, 34–48. [Google Scholar] [CrossRef]
- Shen, M.; Dong, X.; Tsong, Y. Statistical evaluation of several methods for cut-point determination of immunogenicity screening assay. J. Biopharm. Stat. 2015, 25, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Devanarayan, V.; Finco, D.; Gunn, G.R., 3rd; Kirshner, S.; Richards, S.; Rup, B.; Song, A.; Subramanyam, M. Recommendations for the validation of cell-based assays used for the detection of neutralizing antibody immune responses elicited against biological therapeutics. J. Pharm. Biomed. Anal. 2011, 55, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.; Devanarayan, V.; Amaravadi, L.; Barrett, Y.C.; Bowsher, R.; Finco-Kent, D.; Fiscella, M.; Gorovits, B.; Kirschner, S.; Moxness, M.; et al. Recommendations for the validation of immunoassays used for detection of host antibodies against biotechnology products. J. Pharm. Biomed. Anal. 2008, 48, 1267–1281. [Google Scholar] [CrossRef]
- Wood, J.; Stephenson, I.; Heath, A. Report of a WHO collaborative study to assess the suitability of a candidate International Standard for antibody to influenza H5N1 virus, Version 17 July 2008. In Proceedings of the Expert Committee on Biological Standardization, Geneva, Switzerland, 13–17 October 2008. [Google Scholar]
- Cao, L.; Ledeboer, A.; Pan, Y.; Lu, Y.; Meyer, K. Clinical enrollment assay to detect preexisting neutralizing antibodies to AAV6 with demonstrated transgene expression in gene therapy trials. Gene Ther. 2023, 30, 150–159. [Google Scholar] [CrossRef]
- Butala-Flores, E.; Nguyen, T.; Selvan, N.; Armstrong, L.; Miller, M.; Kamen, L.; Lester, T.; Wernyj, R.; Khanna, R.; McNally, J.; et al. Validation of Anti-Adeno Associated Virus Serotype rh10 (AAVrh.10) Total and Neutralizing Antibody Immunogenicity Assays. Pharm. Res. 2023, 40, 2383–2397. [Google Scholar] [CrossRef]
- Sierra-Delgado, J.A.; Likhite, S.; Bautista, P.K.; Gómez-Ochoa, S.A.; Echeverría, L.E.; Guío, E.; Vargas, C.; Serrano, N.C.; Meyer, K.C.; Rincon, M.Y. Prevalence of Neutralizing Antibodies against Adeno-Associated Virus Serotypes 1, 2, and 9 in Non-Injected Latin American Patients with Heart Failure-ANVIAS Study. Int. J. Mol. Sci. 2023, 24, 5579. [Google Scholar] [CrossRef]
- Wei, C.; Li, D.; Zhang, M.; Zhao, Y.; Liu, Y.; Fan, Y.; Wang, L.; Liu, J.; Chang, X.; Jiang, Y.; et al. Prevalence of Adeno-Associated Virus-9-Neutralizing Antibody in Chinese Patients with Duchenne Muscular Dystrophy. Hum. Gene Ther. 2024, 35, 26–35. [Google Scholar] [CrossRef]
- Gardner, M.R.; Mendes, D.E.; Muniz, C.P.; Martinez-Navio, J.M.; Fuchs, S.P.; Gao, G.; Desrosiers, R.C. High concordance of ELISA and neutralization assays allows for the detection of antibodies to individual AAV serotypes. Mol. Ther. Methods Clin. Dev. 2022, 24, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Ye, J.; Leng, M.; Gan, C.; Tang, N.; Li, W.; Valencia, C.A.; Dong, B.; Chow, H.Y. Enhanced sensitivity of neutralizing antibody detection for different AAV serotypes using HeLa cells with overexpressed AAVR. Front. Pharmacol. 2023, 14, 1188290. [Google Scholar] [CrossRef]
- Haar, J.; Blazevic, D.; Strobel, B.; Kreuz, S.; Michelfelder, S. MSD-based assays facilitate a rapid and quantitative serostatus profiling for the presence of anti-AAV antibodies. Mol. Ther. Methods Clin. Dev. 2022, 25, 360–369. [Google Scholar] [CrossRef]
- Kavita, U.; Dai, Y.; Salvador, L.; Miller, W.; Adam, L.P.; Levesque, P.C.; Zhang, Y.J.; Ji, Q.C.; Pillutla, R.C. Development of a Chemiluminescent ELISA Method for the Detection of Total Anti-Adeno Associated Virus Serotype 9 (AAV9) Antibodies. Hum. Gene Ther. Methods 2018, 29, 237–250. [Google Scholar] [CrossRef] [PubMed]
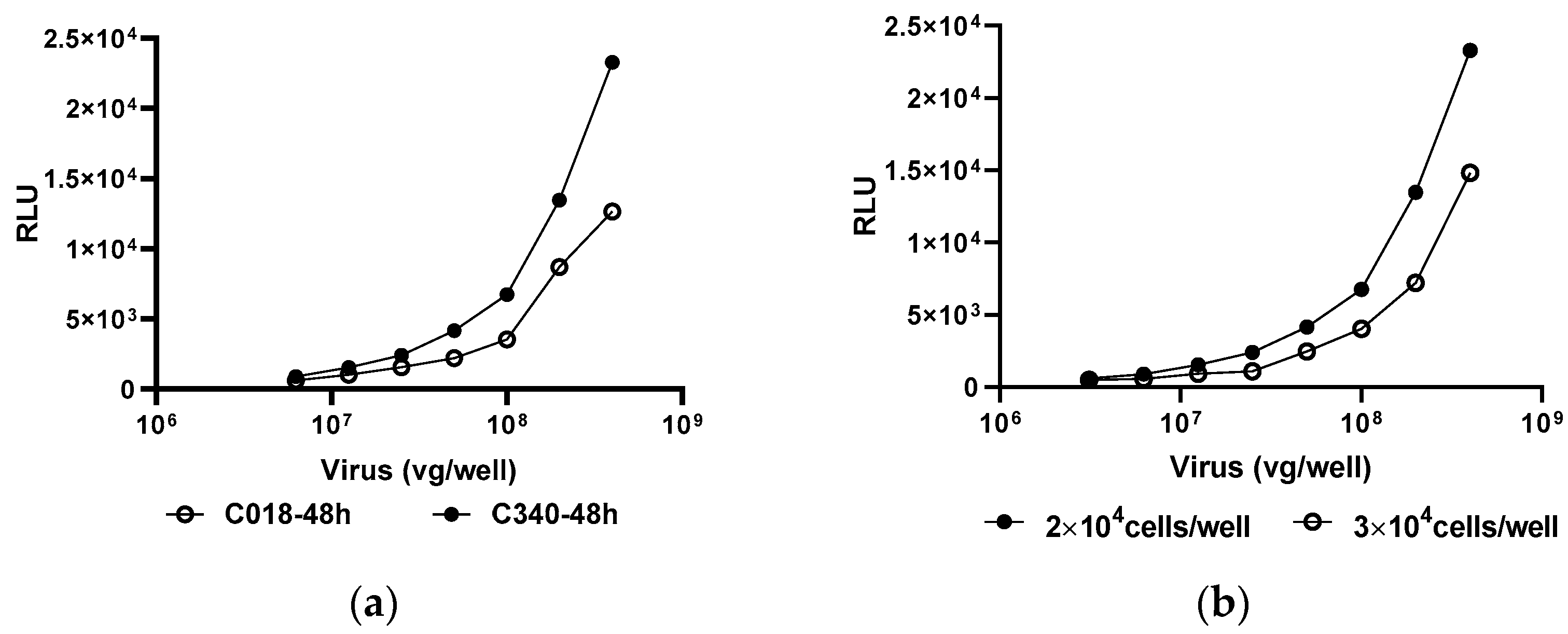


| RA1 | RA2 | RA3 | RA4 | RA5 | RA6 | CV | |
|---|---|---|---|---|---|---|---|
| IC50 (ng/mL) | 7.35 | 7.8 | 6.29 | 6.45 | 7.69 | 6.29 | 10% |
| 95%CI of IC50 | 6.47~8.35 | 6.07~10.76 | 5.82~6.81 | 4.22~10.55 | 5.82~10.29 | 5.82~6.81 | - |
| R2 | 0.99 | 0.97 | 0.99 | 0.94 | 0.97 | 0.99 | - |
| Parameters | Lab 1 | Lab 2 | Lab 3 |
|---|---|---|---|
| TCP determination | 1:21 | - | 1:15 |
| Intra-assay and inter-assay precision (%GCV, fold change in titer) | |||
| NC | 0–58%, 1–2; 39%, 3 | 0–38%, 1–2; 19%, 2 | 11–31%, 1–3; 19%, 2 |
| LPC | 7–35%, 1–2; 37%, 3 | 10–35%, 1–2; 41%, 3 | 8–26%, 1–2; 22%, 2 |
| MPC | 9–36%, 1–2; 30%, 3 | 4–39%, 1–2; 28%, 2 | 11–24%, 1–2; 29%, 2 |
| HPC | 41–52%, 1–3; 33%, 4 | 6–40%, 1–2; 39%, 3 | 8–19%, 1–2; 18%, 2 |
| Sensitivity (ng/mL) and Hook effect | 54; 25~3200 ng/mL PC No hook effect | 45 | 15; 100~10,000 ng/mL PC No hook effect |
| drug tolerance | - | - | Tolerant to 109 vg/mL |
| Specificity | |||
| No cross-reactivity to 20 μg/mL anti-AAV8 MoAb | - | No cross-reactivity to 20 μg/mL anti-AAV8 MoAb | |
| Selectivity | - | ||
| hemolysis interference | 100% pass at 0, 200 ng/mL PC | - | 100% pass at 0, 200 ng/mL PC |
| lipemia interference | 100% pass at 0, 200 ng/mL PC | - | 100% pass at 0, 200 ng/mL PC |
| System suitability (%GCV, fold change in titer; GMT range) | |||
| LPC | 37%, 3; 34–135 | 41%, 3; 35–139 | 44%, 2; 74–295 |
| HPC | 33%, 3; 383–1532 | 39%, 3; 283–1132 | 16%, 2; 621–2486 |
| Robustness and Stability | |||
| Pre-incubation time: 1 h ± 10 min; HEK-293T-C340: P17–50; incubation time: 48 ± 4 h LPC is stable at 4, 25, −30, −80 °C for short- and long-term storage and six times freeze/thawing | - | Pre-incubation time: 1–2 h; HEK-293T-C340: P21–24; incubation time: 44–64 h LPC is stable at 4, 25, −30, −80 °C for short- and long-term storage and 6 times freeze/thawing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Zhao, Q.; Zhang, C.; Fu, D.; Zhu, X.; Zhou, J.; Ma, W.; Dong, Z.; Zhai, X.; Jiang, L.; et al. Methodological Validation and Inter-Laboratory Comparison of Microneutralization Assay for Detecting Anti-AAV9 Neutralizing Antibody in Human. Viruses 2024, 16, 1512. https://doi.org/10.3390/v16101512
Yu S, Zhao Q, Zhang C, Fu D, Zhu X, Zhou J, Ma W, Dong Z, Zhai X, Jiang L, et al. Methodological Validation and Inter-Laboratory Comparison of Microneutralization Assay for Detecting Anti-AAV9 Neutralizing Antibody in Human. Viruses. 2024; 16(10):1512. https://doi.org/10.3390/v16101512
Chicago/Turabian StyleYu, Shuangqing, Qian Zhao, Cengceng Zhang, Diyi Fu, Xueyang Zhu, Jianfang Zhou, Wenhao Ma, Zheyue Dong, Xiaoliang Zhai, Lijie Jiang, and et al. 2024. "Methodological Validation and Inter-Laboratory Comparison of Microneutralization Assay for Detecting Anti-AAV9 Neutralizing Antibody in Human" Viruses 16, no. 10: 1512. https://doi.org/10.3390/v16101512
APA StyleYu, S., Zhao, Q., Zhang, C., Fu, D., Zhu, X., Zhou, J., Ma, W., Dong, Z., Zhai, X., Jiang, L., Han, X., Zhang, S., Wu, X., & Dong, X. (2024). Methodological Validation and Inter-Laboratory Comparison of Microneutralization Assay for Detecting Anti-AAV9 Neutralizing Antibody in Human. Viruses, 16(10), 1512. https://doi.org/10.3390/v16101512






