Current State of Therapeutics for HTLV-1
Abstract
1. Introduction
1.1. Epidemiology
1.2. HTLV-1 Genome
| Gene | Protein | Function | Localisation |
|---|---|---|---|
| Gag | MA | Matrix [30] | Virion [31] |
| CA | Capsid [30] | Virion [31] | |
| NC | Nucleocapsid [30] | Virion [31] | |
| Pro | PR | Protease; cleavage of Gag and Pol [32] | Virion [32] |
| Pol | RT | Reverse Transcriptase; reverse transcription of HTLV-1 RNA into DNA [30] | Virion [33] |
| RH | RT-RNase H; mediates RNA cleavage during replication and repair [30] | Virion [33] | |
| IN | Integrase; DNA provirus integration [30] | Virion [33] | |
| Env | SU | Surface subunits bind to host cell surface receptors to facilitate fusion of viral and host cell membranes [30] | Plasma membrane [34] |
| TM | Transmembrane proteins [30] | Plasma membrane [34] | |
| Rex | Rex (p27) | RNA-binding post-transcriptional regulator; promotes export of spliced viral RNA from the nucleus to the cytoplasm [35] | Primarily in the nucleoli/nucleus exported to the cytoplasm [35] |
| Tax | Tax (p40) | Activates viral transcription by interacting with enhancer elements in the 5′LTR and activating CREB/ATF, NF-κB and AP1 pathways [36] | Primarily in the nucleoli/nucleus exported to the cytoplasm [37] |
| HBZ | HBZ | Negative regulator of TAX-mediated transcription. Expression of HBZ mRNA stimulates lymphocyte proliferation [38] | Nucleus/cytoplasm [39,40] |
| p12 | p12 | Multiple functions to promote escape of immune surveillance and T cell proliferation [41] | Endoplasmic Reticulum [42] |
| p13 | p13 | A truncated form of p30 which alters the membrane potential and reactive oxygen species (ROS) production [43] | Mitochondria [42] |
| p30 | p30 | Inhibits viral expression and promotes repression by regulating tax/rex mRNA [44] | Nucleus [45] |
| Name | Sequence |
|---|---|
| NF-κB/NFAT [46] | AA…GGGGCTCCT…CA |
| NF-YB/CBEPβ [47] | GG…CCAAT…GT |
| Sp1/Sp3 [48] | AA…CCACCC…AT |
| TRE-1 repeat I (vCRE-1) [49] | AGGC…TGACGTCT…CCCC |
| TRE-1 repeat II (vCRE-2) [49] | AGGC…TGACGTGT…CCCC |
| TRE-1 repeat III (vCRE-3) [49] | AGGC…TGACGACA…CCCC |
| TF-IIA/TF-II D [50] | TC…TATAA…AA |
| Elk-1/SRF [51] | CCGGGAA…CCGGGAA…CCATGTTTGT |
| AP-1 [52] | CA…TGAG…CC |
1.3. HTLV-1 Transmission and Life Cycle
1.3.1. Transmission
1.3.2. Life Cycle
1.4. HTLV-1 Persistence
1.5. HTLV-1-Associated Diseases
2. In Vivo Assessment of Therapeutics
2.1. Cell Models
2.2. Animal Models
2.2.1. Mice Models
Xenograft Mouse Models for ATL
Transgenic Mouse Models
Humanised Mouse Models
2.2.2. Non-Human Primate Models
2.2.3. Other Animal Models
3. Current Treatments
3.1. Antiretroviral Therapies and Limitations for HTLV-1
3.1.1. HTLV-1 Entry Inhibitors
3.1.2. Nucleoside Reverse Transcriptase Inhibitors (NRTIs) and Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs)
3.1.3. Integrase (IN) Inhibitors
3.1.4. Protease Inhibitors (Ritonavir)
3.2. Treatment of HTLV-1 Associated Diseases
3.2.1. ATL
3.2.2. HAM
3.2.3. Other Inflammatory Diseases
3.3. HTLV-1 Pre-Exposure Prophylaxis (PrEP)
3.4. Biomarkers for Disease
4. Future of HTLV-1 Treatments
4.1. Gene Editing
4.2. Gene Silencing by RNA Interference (RNAi)
4.2.1. RNAi Pathways
4.2.2. Controlling Chronic Viral Infections
4.2.3. Challenges in RNAi for HTLV-1
4.3. The BLV Vaccine: A Model for HTLV-1 Immunisation and Vaccination
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Minna, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef] [PubMed]
- Hinuma, Y.; Nagata, K.; Hanaoka, M.; Nakai, M.; Matsumoto, T.; Kinoshita, K.I.; Shirakawa, S.; Miyoshi, I. Adult T-cell leukemia: Antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. USA 1981, 78, 6476–6480. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Temin, H.M.; Mizutani, S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef]
- Verdonck, K.; González, E.; Van Dooren, S.; Vandamme, A.-M.; Vanham, G.; Gotuzzo, E. Human T-lymphotropic virus 1: Recent knowledge about an ancient infection. Lancet Infect. Dis. 2007, 7, 266–281. [Google Scholar] [CrossRef]
- Zihlmann, K.F.; de Alvarenga, A.T.; Casseb, J. Living invisible: HTLV-1-infected persons and the lack of care in public health. PLoS Negl. Trop. Dis. 2012, 6, e1705. [Google Scholar] [CrossRef]
- Fowler, F.; Einsiedel, L. A Qualitative Study Exploring Perceptions to the Human T Cell Leukaemia Virus Type 1 in Central Australia: Barriers to Preventing Transmission in a Remote Aboriginal Population. Front. Med. 2022, 9, 845594. [Google Scholar] [CrossRef]
- Carneiro-Proietti, A.B.; Catalan-Soares, B.C.; Castro-Costa, C.M.; Murphy, E.L.; Sabino, E.C.; Hisada, M.; Galvao-Castro, B.; Alcantara, L.C.; Remondegui, C.; Verdonck, K.; et al. HTLV in the Americas: Challenges and perspectives. Rev. Panam. Salud Publica 2006, 19, 44–53. [Google Scholar] [CrossRef]
- Taylor, G.P.; Bodeus, M.; Courtois, F.; Pauli, G.; Del Mistro, A.; Machuca, A.; Padua, E.; Andersson, S.; Goubau, P.; Chieco-Bianchi, L.; et al. The seroepidemiology of human T-lymphotropic viruses: Types I and II in Europe: A prospective study of pregnant women. J. Acquir. Immune Defic. Syndr. 2005, 38, 104–109. [Google Scholar] [CrossRef]
- Yoshida, M.; Seiki, M.; Yamaguchi, K.; Takatsuki, K. Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc. Natl. Acad. Sci. USA 1984, 81, 2534–2537. [Google Scholar] [CrossRef]
- Williams, C.K.O. (Ed.) Global HTLV-1/2 Burden and Associated Diseases. In Cancer and AIDS: Part II: Cancer Pathogenesis and Epidemiology; Springer International Publishing: Cham, Switzerland, 2019; pp. 21–57. [Google Scholar]
- Shimoyama, M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984–1987). Br. J. Haematol. 1991, 79, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Schierhout, G.; McGregor, S.; Gessain, A.; Einsiedel, L.; Martinello, M.; Kaldor, J. Association between HTLV-1 infection and adverse health outcomes: A systematic review and meta-analysis of epidemiological studies. Lancet Infect. Dis. 2020, 20, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Bastos Ferreira, A.P.; do Nascimento, A.; Sampaio Rocha-Filho, P.A. Cerebral and spinal cord changes observed through magnetic resonance imaging in patients with HTLV-1-associated myelopathy/tropical spastic paraparesis: A systematic review. J. Neurovirol. 2022, 28, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Carneiro-Proietti, A.B.; Ribas, J.G.; Catalan-Soares, B.C.; Martins, M.L.; Brito-Melo, G.E.; Martins-Filho, O.A.; Pinheiro, S.R.; Araujo Ade, Q.; Galvao-Castro, B.; de Oliveira, M.S.; et al. Infection and disease caused by the human T cell lymphotropic viruses type I and II in Brazil. Rev. Soc. Bras. Med. Trop. 2002, 35, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Taylor, G.P.; Rosadas, C. Human T-Cell Lymphotropic Virus Type 1 and Strongyloides stercoralis Co-infection: A Systematic Review and Meta-Analysis. Front. Med. 2022, 9, 832430. [Google Scholar] [CrossRef]
- Einsiedel, L.; Chiong, F.; Jersmann, H.; Taylor, G.P. Human T-cell leukaemia virus type 1 associated pulmonary disease: Clinical and pathological features of an under-recognised complication of HTLV-1 infection. Retrovirology 2021, 18, 1. [Google Scholar] [CrossRef]
- Gessain, A.; Mahieux, R. Epidemiology, origin and genetic diversity of HTLV-1 retrovirus and STLV-1 simian affiliated retrovirus. Bull. Soc. Pathol. Exot. 2000, 93, 163–171. [Google Scholar]
- Afonso, P.V.; Cassar, O.; Gessain, A. Molecular epidemiology, genetic variability and evolution of HTLV-1 with special emphasis on African genotypes. Retrovirology 2019, 16, 39. [Google Scholar] [CrossRef]
- Kaidarova, Z.; Murphy, E.L. HTLV-I and –II seroprevalence among United States blood donors, 2000–2009. Retrovirology 2011, 8, A74. [Google Scholar] [CrossRef][Green Version]
- Vandamme, A.-M.; Salemi, M.; Desmyter, J. The simian origins of the pathogenic human T-cell lymphotropic virus type I. Trends Microbiol. 1998, 6, 477–483. [Google Scholar] [CrossRef]
- Gessain, A. Human retrovirus HTLV-1: Descriptive and molecular epidemiology, origin, evolution, diagnosis and associated diseases. Bull. Soc. Pathol. Exot. 2011, 104, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Kusuhara, K.; Tokugawa, K.; Miyazaki, C.; Yoshida, C.; Tokumura, K.; Sonoda, S.; Takahashi, K. Cohort effect on HTLV-I seroprevalence in southern Japan. Lancet 1989, 2, 979. [Google Scholar] [CrossRef] [PubMed]
- Blank, A.; Yamaguchi, K.; Blank, M.; Zaninovic, V.; Sonoda, S.; Takatsuki, K. Six Colombian patients with adult T-cell leukemia/lymphoma. Leuk. Lymphoma 1993, 9, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.L.; Figueroa, J.P.; Gibbs, W.N.; Holding-Cobham, M.; Cranston, B.; Malley, K.; Bodner, A.J.; Alexander, S.S.; Blattner, W.A. Human T-lymphotropic virus type I (HTLV-I) seroprevalence in Jamaica. I. Demographic determinants. Am. J. Epidemiol. 1991, 133, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Cassar, O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front. Microbiol. 2012, 3, 388. [Google Scholar] [CrossRef] [PubMed]
- Van Dooren, S. Central Africa: Cradle of divergent PTLV types. AIDS Rev. 2005, 7, 126–127. [Google Scholar]
- Manns, A.; Blattner, W.A. The epidemiology of the human T-cell lymphotrophic virus type I and type II: Etiologic role in human disease. Transfusion 1991, 31, 67–75. [Google Scholar] [CrossRef]
- Felber, B.K.; Paskalis, H.; Kleinman-Ewing, C.; Wong-Staal, F.; Pavlakis, G.N. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science 1985, 229, 675–679. [Google Scholar] [CrossRef]
- Seiki, M.; Hattori, S.; Hirayama, Y.; Yoshida, M. Human adult T-cell leukemia virus: Complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 1983, 80, 3618–3622. [Google Scholar] [CrossRef]
- Delchambre, M.; Gheysen, D.; Thines, D.; Thiriart, C.; Jacobs, E.; Verdin, E.; Horth, M.; Burny, A.; Bex, F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989, 8, 2653–2660. [Google Scholar] [CrossRef]
- Kohl, N.E.; Emini, E.A.; Schleif, W.A.; Davis, L.J.; Heimbach, J.C.; Dixon, R.A.; Scolnick, E.M.; Sigal, I.S. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 1988, 85, 4686–4690. [Google Scholar] [CrossRef] [PubMed]
- Trentin, B.; Rebeyrotte, N.; Mamoun, R.Z. Human T-cell leukemia virus type 1 reverse transcriptase (RT) originates from the pro and pol open reading frames and requires the presence of RT-RNase H (RH) and RT-RH-integrase proteins for its activity. J. Virol. 1998, 72, 6504–6510. [Google Scholar] [CrossRef] [PubMed]
- Willey, R.L.; Bonifacino, J.S.; Potts, B.J.; Martin, M.A.; Klausner, R.D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 1988, 85, 9580–9584. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, M.; Inoue, J.; Yoshida, M.; Seiki, M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988, 7, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Boxus, M.; Twizere, J.C.; Legros, S.; Dewulf, J.F.; Kettmann, R.; Willems, L. The HTLV-1 Tax interactome. Retrovirology 2008, 5, 76. [Google Scholar] [CrossRef]
- Smith, M.R.; Greene, W.C. Characterization of a novel nuclear localization signal in the HTLV-I tax transactivator protein. Virology 1992, 187, 316–320. [Google Scholar] [CrossRef]
- Satou, Y.; Yasunaga, J.; Yoshida, M.; Matsuoka, M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. USA 2006, 103, 720–725. [Google Scholar] [CrossRef]
- Baratella, M.; Forlani, G.; Raval, G.U.; Tedeschi, A.; Gout, O.; Gessain, A.; Tosi, G.; Accolla, R.S. Cytoplasmic Localization of HTLV-1 HBZ Protein: A Biomarker of HTLV-1-Associated Myelopathy/Tropical Spastic Paraparesis (HAM/TSP). PLoS Negl. Trop. Dis. 2017, 11, e0005285. [Google Scholar] [CrossRef]
- Gaudray, G.; Gachon, F.; Basbous, J.; Biard-Piechaczyk, M.; Devaux, C.; Mesnard, J.M. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 2002, 76, 12813–12822. [Google Scholar] [CrossRef]
- Fukumoto, R.; Andresen, V.; Bialuk, I.; Cecchinato, V.; Walser, J.C.; Valeri, V.W.; Nauroth, J.M.; Gessain, A.; Nicot, C.; Franchini, G. In vivo genetic mutations define predominant functions of the human T-cell leukemia/lymphoma virus p12I protein. Blood 2009, 113, 3726–3734. [Google Scholar] [CrossRef]
- Koralnik, I.J.; Fullen, J.; Franchini, G. The p12I, p13II, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type I open reading frames I and II are localized in three different cellular compartments. J. Virol. 1993, 67, 2360–2366. [Google Scholar] [CrossRef] [PubMed]
- Silic-Benussi, M.; Cannizzaro, E.; Venerando, A.; Cavallari, I.; Petronilli, V.; La Rocca, N.; Marin, O.; Chieco-Bianchi, L.; Di Lisa, F.; D’Agostino, D.M.; et al. Modulation of mitochondrial K+ permeability and reactive oxygen species production by the p13 protein of human T-cell leukemia virus type 1. Biochim. Biophys. Acta 2009, 1787, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Baydoun, H.H.; Bellon, M.; Nicot, C. HTLV-1 Yin and Yang: Rex and p30 master regulators of viral mRNA trafficking. AIDS Rev. 2008, 10, 195–204. [Google Scholar] [PubMed]
- Ghorbel, S.; Sinha-Datta, U.; Dundr, M.; Brown, M.; Franchini, G.; Nicot, C. Human T-cell leukemia virus type I p30 nuclear/nucleolar retention is mediated through interactions with RNA and a constituent of the 60 S ribosomal subunit. J. Biol. Chem. 2006, 281, 37150–37158. [Google Scholar] [CrossRef]
- Azimi, N.; Brown, K.; Bamford, R.N.; Tagaya, Y.; Siebenlist, U.; Waldmann, T.A. Human T cell lymphotropic virus type I Tax protein trans-activates interleukin 15 gene transcription through an NF-kappaB site. Proc. Natl. Acad. Sci. USA 1998, 95, 2452–2457. [Google Scholar] [CrossRef]
- Pise-Masison, C.A.; Dittmer, J.; Clemens, K.E.; Brady, J.N. Physical and functional interaction between the human T-cell lymphotropic virus type 1 Tax1 protein and the CCAAT binding protein NF-Y. Mol. Cell. Biol. 1997, 17, 1236–1243. [Google Scholar] [CrossRef][Green Version]
- Livengood, J.A.; Nyborg, J.K. The high-affinity Sp1 binding site in the HTLV-1 promoter contributes to Tax-independent basal expression. Nucleic Acids Res. 2004, 32, 2829–2837. [Google Scholar] [CrossRef]
- Yao, J.; Grant, C.; Harhaj, E.; Nonnemacher, M.; Alefantis, T.; Martin, J.; Jain, P.; Wigdahl, B. Regulation of human T-cell leukemia virus type 1 gene expression by Sp1 and Sp3 interaction with TRE-1 repeat III. DNA Cell Biol. 2006, 25, 262–276. [Google Scholar] [CrossRef]
- Caron, C.; Rousset, R.; Béraud, C.; Moncollin, V.; Egly, J.M.; Jalinot, P. Functional and biochemical interaction of the HTLV-I Tax1 transactivator with TBP. EMBO J. 1993, 12, 4269–4278. [Google Scholar] [CrossRef]
- Winter, H.Y.; Dayaram, T.; Marriott, S.J. Activation of the human T-cell leukemia virus type 1 long terminal repeat by the ternary complex factor Elk-1. J. Virol. 2007, 81, 13075–13081. [Google Scholar] [CrossRef][Green Version]
- Matsumoto, J.; Ohshima, T.; Isono, O.; Shimotohno, K. HTLV-1 HBZ suppresses AP-1 activity by impairing both the DNA-binding ability and the stability of c-Jun protein. Oncogene 2005, 24, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Kiyokawa, T.; Seiki, M.; Iwashita, S.; Imagawa, K.; Shimizu, F.; Yoshida, M. p27x-III and p21x-III, proteins encoded by the pX sequence of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 1985, 82, 8359–8363. [Google Scholar] [CrossRef] [PubMed]
- Berneman, Z.N.; Gartenhaus, R.B.; Reitz, M.S., Jr.; Blattner, W.A.; Manns, A.; Hanchard, B.; Ikehara, O.; Gallo, R.C.; Klotman, M.E. Expression of alternatively spliced human T-lymphotropic virus type I pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc. Natl. Acad. Sci. USA 1992, 89, 3005–3009. [Google Scholar] [CrossRef] [PubMed]
- Mozhgani, S.H.; Jahantigh, H.R.; Rafatpanah, H.; Valizadeh, N.; Mohammadi, A.; Basharkhah, S.; Rezaee, S.A. Interferon Lambda Family along with HTLV-1 Proviral Load, Tax, and HBZ Implicated in the Pathogenesis of Myelopathy/Tropical Spastic Paraparesis. Neurodegener. Dis. 2018, 18, 150–155. [Google Scholar] [CrossRef]
- Hirons, A.; Khoury, G.; Purcell, D.F.J. Human T-cell lymphotropic virus type-1: A lifelong persistent infection, yet never truly silent. Lancet Infect. Dis. 2021, 21, e2–e10. [Google Scholar] [CrossRef]
- Arnold, J.; Yamamoto, B.; Li, M.; Phipps, A.J.; Younis, I.; Lairmore, M.D.; Green, P.L. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 2006, 107, 3976–3982. [Google Scholar] [CrossRef]
- Arnold, J.; Zimmerman, B.; Li, M.; Lairmore, M.D.; Green, P.L. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood 2008, 112, 3788–3797. [Google Scholar] [CrossRef]
- Panfil, A.R.; Dissinger, N.J.; Howard, C.M.; Murphy, B.M.; Landes, K.; Fernandez, S.A.; Green, P.L. Functional Comparison of HBZ and the Related APH-2 Protein Provides Insight into Human T-Cell Leukemia Virus Type 1 Pathogenesis. J. Virol. 2016, 90, 3760–3772. [Google Scholar] [CrossRef]
- Hivin, P.; Frederic, M.; Arpin-Andre, C.; Basbous, J.; Gay, B.; Thebault, S.; Mesnard, J.M. Nuclear localization of HTLV-I bZIP factor (HBZ) is mediated by three distinct motifs. J. Cell Sci. 2005, 118, 1355–1362. [Google Scholar] [CrossRef]
- Yamamoto, N.; Okada, M.; Koyanagi, Y.; Kannagi, M.; Hinuma, Y. Transformation of human leukocytes by cocultivation with an adult T cell leukemia virus producer cell line. Science 1982, 217, 737–739. [Google Scholar] [CrossRef]
- Willems, L.; Hasegawa, H.; Accolla, R.; Bangham, C.; Bazarbachi, A.; Bertazzoni, U.; Carneiro-Proietti, A.B.; Cheng, H.; Chieco-Bianchi, L.; Ciminale, V.; et al. Reducing the global burden of HTLV-1 infection: An agenda for research and action. Antivir. Res. 2017, 137, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Takezaki, T.; Oki, T.; Kawakami, K.; Yashiki, S.; Fujiyoshi, T.; Usuku, K.; Mueller, N.; Osame, M.; Miyata, K.; et al. Inhibitory effect of maternal antibody on mother-to-child transmission of human T-lymphotropic virus type I. The Mother-to-Child Transmission Study Group. Int. J. Cancer 1991, 49, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Li, H.C.; Biggar, R.J.; Miley, W.J.; Maloney, E.M.; Cranston, B.; Hanchard, B.; Hisada, M. Provirus load in breast milk and risk of mother-to-child transmission of human T lymphotropic virus type I. J. Infect. Dis. 2004, 190, 1275–1278. [Google Scholar] [CrossRef] [PubMed]
- Futsch, N.; Mahieux, R.; Dutartre, H. HTLV-1, the Other Pathogenic Yet Neglected Human Retrovirus: From Transmission to Therapeutic Treatment. Viruses 2017, 10, 1. [Google Scholar] [CrossRef]
- Tarasevich, A.; Filatov, A.; Pichugin, A.; Mazurov, D. Monoclonal antibody profiling of cell surface proteins associated with the viral biofilms on HTLV-1 transformed cells. Acta Virol. 2015, 59, 247–256. [Google Scholar] [CrossRef]
- Igakura, T.; Stinchcombe, J.C.; Goon, P.K.; Taylor, G.P.; Weber, J.N.; Griffiths, G.M.; Tanaka, Y.; Osame, M.; Bangham, C.R. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science 2003, 299, 1713–1716. [Google Scholar] [CrossRef]
- Nejmeddine, M.; Negi, V.S.; Mukherjee, S.; Tanaka, Y.; Orth, K.; Taylor, G.P.; Bangham, C.R. HTLV-1-Tax and ICAM-1 act on T-cell signal pathways to polarize the microtubule-organizing center at the virological synapse. Blood 2009, 114, 1016–1025. [Google Scholar] [CrossRef]
- Fazio, A.L.; Kendle, W.; Hoang, K.; Korleski, E.; Lemasson, I.; Polakowski, N. Human T-Cell Leukemia Virus Type 1 (HTLV-1) bZIP Factor Upregulates the Expression of ICAM-1 To Facilitate HTLV-1 Infection. J. Virol. 2019, 93, e00608-19. [Google Scholar] [CrossRef]
- Suzuki, K.; Hattori, S.; Marks, K.; Ahlenstiel, C.; Maeda, Y.; Ishida, T.; Millington, M.; Boyd, M.; Symonds, G.; Cooper, D.A.; et al. Promoter Targeting shRNA Suppresses HIV-1 Infection In vivo Through Transcriptional Gene Silencing. Mol. Ther. Nucleic Acids 2013, 2, e137. [Google Scholar] [CrossRef]
- Pinto, D.O.; DeMarino, C.; Pleet, M.L.; Cowen, M.; Branscome, H.; Al Sharif, S.; Jones, J.; Dutartre, H.; Lepene, B.; Liotta, L.A.; et al. HTLV-1 Extracellular Vesicles Promote Cell-to-Cell Contact. Front. Microbiol. 2019, 10, 2147. [Google Scholar] [CrossRef]
- Coskun, A.K.; Sutton, R.E. Expression of glucose transporter 1 confers susceptibility to human T-cell leukemia virus envelope-mediated fusion. J. Virol. 2005, 79, 4150–4158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Soltani, A.; Hashemy, S.I.; Zahedi Avval, F.; Soleimani, A.; Rafatpanah, H.; Rezaee, S.A.; Griffith, R.; Mashkani, B. Molecular targeting for treatment of human T-lymphotropic virus type 1 infection. Biomed. Pharmacother. 2019, 109, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Ghez, D.; Lepelletier, Y.; Jones, K.S.; Pique, C.; Hermine, O. Current concepts regarding the HTLV-1 receptor complex. Retrovirology 2010, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Grunwald, D.; Sardo, L.; Galli, A.; Plisov, S.; Nikolaitchik, O.A.; Chen, D.; Lockett, S.; Larson, D.R.; Pathak, V.K.; et al. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc. Natl. Acad. Sci. USA 2014, 111, E5205–E5213. [Google Scholar] [CrossRef] [PubMed]
- Demontis, M.A.; Sadiq, M.T.; Golz, S.; Taylor, G.P. HTLV-1 viral RNA is detected rarely in plasma of HTLV-1 infected subjects. J. Med. Virol. 2015, 87, 2130–2134. [Google Scholar] [CrossRef]
- Yurick, D.; Khoury, G.; Clemens, B.; Loh, L.; Pham, H.; Kedzierska, K.; Einsiedel, L.; Purcell, D. Multiplex Droplet Digital PCR Assay for Quantification of Human T-Cell Leukemia Virus Type 1 Subtype c DNA Proviral Load and T Cells from Blood and Respiratory Exudates Sampled in a Remote Setting. J. Clin. Microbiol. 2019, 57, e01063-18. [Google Scholar] [CrossRef]
- Furuta, R.; Yasunaga, J.I.; Miura, M.; Sugata, K.; Saito, A.; Akari, H.; Ueno, T.; Takenouchi, N.; Fujisawa, J.I.; Koh, K.R.; et al. Human T-cell leukemia virus type 1 infects multiple lineage hematopoietic cells in vivo. PLoS Pathog. 2017, 13, e1006722. [Google Scholar] [CrossRef]
- Richardson, J.H.; Edwards, A.J.; Cruickshank, J.K.; Rudge, P.; Dalgleish, A.G. In vivo cellular tropism of human T-cell leukemia virus type 1. J. Virol. 1990, 64, 5682–5687. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Nosaka, K.; Yasunaga, J.; Maeda, M.; Mueller, N.; Okayama, A.; Matsuoka, M. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 2005, 2, 64. [Google Scholar] [CrossRef]
- Koiwa, T.; Hamano-Usami, A.; Ishida, T.; Okayama, A.; Yamaguchi, K.; Kamihira, S.; Watanabe, T. 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 2002, 76, 9389–9397. [Google Scholar] [CrossRef]
- Tamiya, S.; Matsuoka, M.; Etoh, K.; Watanabe, T.; Kamihira, S.; Yamaguchi, K.; Takatsuki, K. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood 1996, 88, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Yasunaga, J.; Taniguchi, Y.; Tamiya, S.; Nakahata, T.; Matsuoka, M. Preferential selection of human T-cell leukemia virus type 1 provirus lacking the 5′ long terminal repeat during oncogenesis. J. Virol. 2007, 81, 5714–5723. [Google Scholar] [CrossRef] [PubMed]
- Usui, T.; Yanagihara, K.; Tsukasaki, K.; Murata, K.; Hasegawa, H.; Yamada, Y.; Kamihira, S. Characteristic expression of HTLV-1 basic zipper factor (HBZ) transcripts in HTLV-1 provirus-positive cells. Retrovirology 2008, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Matsuzaki, T.; Satou, Y.; Yasunaga, J.; Saito, K.; Arimura, K.; Matsuoka, M.; Ohara, Y. In vivo expression of the HBZ gene of HTLV-1 correlates with proviral load, inflammatory markers and disease severity in HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). Retrovirology 2009, 6, 19. [Google Scholar] [CrossRef]
- Andrade, R.G.; Goncalves Pde, C.; Ribeiro, M.A.; Romanelli, L.C.; Ribas, J.G.; Torres, E.B.; Carneiro-Proietti, A.B.; Barbosa-Stancioli, E.F.; Martins, M.L. Strong correlation between tax and HBZ mRNA expression in HAM/TSP patients: Distinct markers for the neurologic disease. J. Clin. Virol. 2013, 56, 135–140. [Google Scholar] [CrossRef]
- Matsuoka, M.; Jeang, K.T. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 2007, 7, 270–280. [Google Scholar] [CrossRef]
- Finzi, D.; Blankson, J.; Siliciano, J.D.; Margolick, J.B.; Chadwick, K.; Pierson, T.; Smith, K.; Lisziewicz, J.; Lori, F.; Flexner, C.; et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999, 5, 512–517. [Google Scholar] [CrossRef]
- Farber, D.L.; Yudanin, N.A.; Restifo, N.P. Human memory T cells: Generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 2014, 14, 24–35. [Google Scholar] [CrossRef]
- Mbonye, U.; Karn, J. The Molecular Basis for Human Immunodeficiency Virus Latency. Annu. Rev. Virol. 2017, 4, 261–285. [Google Scholar] [CrossRef]
- Koga, Y.; Iwanaga, M.; Soda, M.; Inokuchi, N.; Sasaki, D.; Hasegawa, H.; Yanagihara, K.; Yamaguchi, K.; Kamihira, S.; Yamada, Y. Trends in HTLV-1 prevalence and incidence of adult T-cell leukemia/lymphoma in Nagasaki, Japan. J. Med. Virol. 2010, 82, 668–674. [Google Scholar] [CrossRef]
- Tanajura, D.; Castro, N.; Oliveira, P.; Neto, A.; Muniz, A.; Carvalho, N.B.; Orge, G.; Santos, S.; Glesby, M.J.; Carvalho, E.M. Neurological Manifestations in Human T-Cell Lymphotropic Virus Type 1 (HTLV-1)-Infected Individuals without HTLV-1-Associated Myelopathy/Tropical Spastic Paraparesis: A Longitudinal Cohort Study. Clin. Infect. Dis. 2015, 61, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, M.; Yamaguchi, K.; Takatsuki, K.; Watanabe, T.; Mori, S.; Tajima, K. HTLV-I and uveitis. Lancet 1992, 339, 1110. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Eguchi, K.; Nakamura, T.; Mizokami, A.; Shirabe, S.; Kawakami, A.; Matsuoka, N.; Migita, K.; Kawabe, Y.; Nagataki, S. High prevalence of Sjogren’s syndrome in patients with HTLV-I associated myelopathy. Ann. Rheum. Dis. 1997, 56, 167–172. [Google Scholar] [CrossRef] [PubMed]
- LaGrenade, L.; Hanchard, B.; Fletcher, V.; Cranston, B.; Blattner, W. Infective dermatitis of Jamaican children: A marker for HTLV-I infection. Lancet 1990, 336, 1345–1347. [Google Scholar] [CrossRef] [PubMed]
- Einsiedel, L.; Fernandes, L.; Spelman, T.; Steinfort, D.; Gotuzzo, E. Bronchiectasis is associated with human T-lymphotropic virus 1 infection in an Indigenous Australian population. Clin. Infect. Dis. 2012, 54, 43–50. [Google Scholar] [CrossRef]
- Guerin, B.; Arfi, S.; Numeric, P.; Jean-Baptiste, G.; Le Parc, J.M.; Smadja, D.; Grollier-Bois, L. Polyarthritis in HTLV-1-infected patients. A review of 17 cases. Rev. Rhum. Engl. Ed. 1995, 62, 21–28. [Google Scholar]
- Kawai, H.; Inui, T.; Kashiwagi, S.; Tsuchihashi, T.; Masuda, K.; Kondo, A.; Niki, S.; Iwasa, M.; Saito, S. HTLV-I infection in patients with autoimmune thyroiditis (Hashimoto’s thyroiditis). J. Med. Virol. 1992, 38, 138–141. [Google Scholar] [CrossRef]
- Einsiedel, L.J.; Pham, H.; Woodman, R.J.; Pepperill, C.; Taylor, K.A. The prevalence and clinical associations of HTLV-1 infection in a remote Indigenous community. Med. J. Aust. 2016, 205, 305–309. [Google Scholar] [CrossRef]
- Enose-Akahata, Y.; Jacobson, S. Immunovirological markers in HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). Retrovirology 2019, 16, 35. [Google Scholar] [CrossRef]
- Haziot, M.E.; Gascon, M.R.; Assone, T.; Fonseca, L.A.M.; Luiz, O.D.C.; Smid, J.; Paiva, A.M.; Marcusso, R.; de Oliveira, A.C.P.; Casseb, J. Detection of clinical and neurological signs in apparently asymptomatic HTLV-1 infected carriers: Association with high proviral load. PLoS Negl. Trop. Dis. 2019, 13, e0006967. [Google Scholar] [CrossRef]
- Maeda, M.; Tanabe-Shibuya, J.; Miyazato, P.; Masutani, H.; Yasunaga, J.I.; Usami, K.; Shimizu, A.; Matsuoka, M. IL-2/IL-2 Receptor Pathway Plays a Crucial Role in the Growth and Malignant Transformation of HTLV-1-Infected T Cells to Develop Adult T-Cell Leukemia. Front. Microbiol. 2020, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Sarin, P.S.; Robert-Gurroff, M.; Kalyanaraman, V.S.; Mann, D.; Minowada, J.; Gallo, R.C. Isolation and transmission of human retrovirus (human t-cell leukemia virus). Science 1983, 219, 856–859. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Miyoshi, I.; Hinuma, Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 1982, 79, 2031–2035. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.V.; Delviks-Frankenberry, K.A.; Scheiblin, D.A.; Happel, C.; Pathak, V.K.; Freed, E.O. Authentication Analysis of MT-4 Cells Distributed by the National Institutes of Health AIDS Reagent Program. J. Virol. 2019, 93, e01390-19. [Google Scholar] [CrossRef]
- Salahuddin, S.Z.; Markham, P.D.; Wong-Staal, F.; Franchini, G.; Kalyanaraman, V.S.; Gallo, R.C. Restricted expression of human T-cell leukemia--lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 1983, 129, 51–64. [Google Scholar] [CrossRef]
- Mitsuya, H.; Matis, L.A.; Megson, M.; Bunn, P.A.; Murray, C.; Mann, D.L.; Gallo, R.C.; Broder, S. Generation of an HLA-restricted cytotoxic T cell line reactive against cultured tumor cells from a patient infected with human T cell leukemia/lymphoma virus. J. Exp. Med. 1983, 158, 994–999. [Google Scholar] [CrossRef]
- Hamano, R.; Wu, X.; Wang, Y.; Oppenheim, J.J.; Chen, X. Characterization of MT-2 cells as a human regulatory T cell-like cell line. Cell. Mol. Immunol. 2015, 12, 780–782. [Google Scholar] [CrossRef]
- Miyoshi, I.; Kubonishi, I.; Sumida, M.; Hiraki, S.; Tsubota, T.; Kimura, I.; Miyamoto, K.; Sato, J. A novel T-cell line derived from adult T-cell leukemia. Gan 1980, 71, 155–156. [Google Scholar]
- Shioda, S.; Kasai, F.; Watanabe, K.; Kawakami, K.; Ohtani, A.; Iemura, M.; Ozawa, M.; Arakawa, A.; Hirayama, N.; Kawaguchi, E.; et al. Screening for 15 pathogenic viruses in human cell lines registered at the JCRB Cell Bank: Characterization of in vitro human cells by viral infection. R. Soc. Open Sci. 2018, 5, 172472. [Google Scholar] [CrossRef]
- Maeda, M.; Shimizu, A.; Ikuta, K.; Okamoto, H.; Kashihara, M.; Uchiyama, T.; Honjo, T.; Yodoi, J. Origin of human T-lymphotrophic virus I-positive T cell lines in adult T cell leukemia. Analysis of T cell receptor gene rearrangement. J. Exp. Med. 1985, 162, 2169–2174. [Google Scholar] [CrossRef]
- Shimizu, K.; Hirano, T.; Ishibashi, K.; Nakano, N.; Taga, T.; Sugamura, K.; Yamamura, Y.; Kishimoto, T. Immortalization of BGDF (BCGF II)- and BCDF-producing T cells by human T cell leukemia virus (HTLV) and characterization of human BGDF (BCGF II). J. Immunol. 1985, 134, 1728–1733. [Google Scholar] [CrossRef] [PubMed]
- Arima, N.; Molitor, J.A.; Smith, M.R.; Kim, J.H.; Daitoku, Y.; Greene, W.C. Human T-cell leukemia virus type I Tax induces expression of the Rel-related family of kappa B enhancer-binding proteins: Evidence for a pretranslational component of regulation. J. Virol. 1991, 65, 6892–6899. [Google Scholar] [CrossRef] [PubMed]
- Katoh, T.; Harada, T.; Morikawa, S.; Wakutani, T. IL-2- and IL-2-R- independent proliferation of T-cell lines from adult T-cell leukemia/lymphoma patients. Int. J. Cancer 1986, 38, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.K.; Masutani, H.; Yamaguchi, Y.; Kim, Y.C.; Nosaka, K.; Matsuoka, M.; Nishinaka, Y.; Maeda, M.; Yodoi, J. Loss of interleukin-2-dependency in HTLV-I-infected T cells on gene silencing of thioredoxin-binding protein-2. Oncogene 2006, 25, 2181–2191. [Google Scholar] [CrossRef][Green Version]
- Maeda, M.; Arima, N.; Daitoku, Y.; Kashihara, M.; Okamoto, H.; Uchiyama, T.; Shirono, K.; Matsuoka, M.; Hattori, T.; Takatsuki, K.; et al. Evidence for the interleukin-2 dependent expansion of leukemic cells in adult T cell leukemia. Blood 1987, 70, 1407–1411. [Google Scholar] [CrossRef][Green Version]
- Hirons, A.; Yurick, D.; Jansz, N.; Ellenberg, P.; Franchini, G.; Einsiedel, L.; Khoury, G.; Purcell, D.F.J. High level of genomic divergence in orf-I p12 and hbz genes of HTLV-1 subtype-C in Central Australia. Retrovirology 2024, 21, 14. [Google Scholar] [CrossRef]
- Talukder, M.R.; Pham, H.; Woodman, R.; Wilson, K.; Taylor, K.; Kaldor, J.; Einsiedel, L. The Association between Diabetes and Human T-Cell Leukaemia Virus Type-1 (HTLV-1) with Strongyloides stercoralis: Results of a Community-Based, Cross-Sectional Survey in Central Australia. Int. J. Environ. Res. Public Health 2022, 19, 2084. [Google Scholar] [CrossRef]
- Talukder, M.R.; Clauss, C.S.; Cherian, S.; Woodman, R.; Einsiedel, L. Risk factors for HTLV-1, acute kidney injury, and urinary tract infection among aboriginal adults with end stage kidney disease in central Australia. J. Med. Virol. 2021, 93, 6362–6370. [Google Scholar] [CrossRef]
- Talukder, M.R.; Woodman, R.; Pham, H.; Wilson, K.; Gessain, A.; Kaldor, J.; Einsiedel, L. High Human T-Cell Leukemia Virus Type 1c Proviral Loads Are Associated With Diabetes and Chronic Kidney Disease: Results of a Cross-Sectional Community Survey in Central Australia. Clin. Infect. Dis. 2023, 76, e820–e826. [Google Scholar] [CrossRef]
- Balestrieri, E.; Ascolani, A.; Igarashi, Y.; Oki, T.; Mastino, A.; Balzarini, J.; Macchi, B. Inhibition of cell-to-cell transmission of human T-cell lymphotropic virus type 1 in vitro by carbohydrate-binding agents. Antimicrob. Agents Chemother. 2008, 52, 2771–2779. [Google Scholar] [CrossRef][Green Version]
- Qari, S.H.; Magre, S.; Garcia-Lerma, J.G.; Hussain, A.I.; Takeuchi, Y.; Patience, C.; Weiss, R.A.; Heneine, W. Susceptibility of the porcine endogenous retrovirus to reverse transcriptase and protease inhibitors. J. Virol. 2001, 75, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Kushida, S.; Maeda, N.; Fang, J.; Uchida, K.; Miwa, M. Establishment of HTLV-1 carrier mice by injection with HTLV-1-producing T cells. Leukemia 1997, 11 (Suppl. 3), 260–262. [Google Scholar] [PubMed]
- Fang, J.; Kushida, S.; Feng, R.; Tanaka, M.; Kawamura, T.; Abe, H.; Maeda, N.; Onobori, M.; Hori, M.; Uchida, K.; et al. Transmission of human T-cell leukemia virus type 1 to mice. J. Virol. 1998, 72, 3952–3957. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Tanaka, M.; Abe, H.; Arashi, N.; Sun, B.; Uchida, K.; Miwa, M. Human T-cell leukemia virus type 1 can infect a wide variety of cells in mice. Jpn. J. Cancer Res. 1999, 90, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Nitta, T.; Yoshida, T.; Konishi, T.; Kawazu, Y.; Fujisawa, J.; Miwa, M. Clonal proliferation of HTLV-1-infected cells is associated with spontaneous malignant tumor formation in mice. Int. J. Oncol. 2009, 35, 701–707. [Google Scholar] [CrossRef][Green Version]
- Bosma, G.C.; Custer, R.P.; Bosma, M.J. A severe combined immunodeficiency mutation in the mouse. Nature 1983, 301, 527–530. [Google Scholar] [CrossRef]
- Mosier, D.E.; Gulizia, R.J.; Baird, S.M.; Wilson, D.B. Transfer of a functional human immune system to mice with severe combined immunodeficiency. Nature 1988, 335, 256–259. [Google Scholar] [CrossRef]
- Ishihara, S.; Tachibana, N.; Okayama, A.; Murai, K.; Tsuda, K.; Mueller, N. Successful graft of HTLV-I-transformed human T-cells (MT-2) in severe combined immunodeficiency mice treated with anti-asialo GM-1 antibody. Jpn. J. Cancer Res. 1992, 83, 320–323. [Google Scholar] [CrossRef]
- Imada, K.; Takaori-Kondo, A.; Akagi, T.; Shimotohno, K.; Sugamura, K.; Hattori, T.; Yamabe, H.; Okuma, M.; Uchiyama, T. Tumorigenicity of human T-cell leukemia virus type I-infected cell lines in severe combined immunodeficient mice and characterization of the cells proliferating in vivo. Blood 1995, 86, 2350–2357. [Google Scholar] [CrossRef]
- Kondo, A.; Imada, K.; Hattori, T.; Yamabe, H.; Tanaka, T.; Miyasaka, M.; Okuma, M.; Uchiyama, T. A model of in vivo cell proliferation of adult T-cell leukemia. Blood 1993, 82, 2501–2509. [Google Scholar] [CrossRef]
- Hutchison, T.; Malu, A.; Yapindi, L.; Bergeson, R.; Peck, K.; Romeo, M.; Harrod, C.; Pope, J.; Smitherman, L.; Gwinn, W.; et al. The TP53-Induced Glycolysis and Apoptosis Regulator mediates cooperation between HTLV-1 p30(II) and the retroviral oncoproteins Tax and HBZ and is highly expressed in an in vivo xenograft model of HTLV-1-induced lymphoma. Virology 2018, 520, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, M.; Ju, W.; Waldmann, T.A. Effective treatment of a murine model of adult T-cell leukemia using depsipeptide and its combination with unmodified daclizumab directed toward CD25. Blood 2009, 113, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.D.; Schweitzer, P.A.; Christianson, S.W.; Gott, B.; Schweitzer, I.B.; Tennent, B.; McKenna, S.; Mobraaten, L.; Rajan, T.V.; Greiner, D.L.; et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J. Immunol. 1995, 154, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Martella, C.; Tollenaere, A.I.; Waast, L.; Lacombe, B.; Groussaud, D.; Margottin-Goguet, F.; Ramirez, B.C.; Pique, C. Human T-Cell Lymphotropic Virus Type 1 Transactivator Tax Exploits the XPB Subunit of TFIIH during Viral Transcription. J. Virol. 2020, 94, e02171-19. [Google Scholar] [CrossRef] [PubMed]
- Nicot, C. HTLV-I Tax-Mediated Inactivation of Cell Cycle Checkpoints and DNA Repair Pathways Contribute to Cellular Transformation: “A Random Mutagenesis Model”. J. Cancer Sci. 2015, 2, 6. [Google Scholar] [CrossRef]
- Hleihel, R.; Skayneh, H.; de The, H.; Hermine, O.; Bazarbachi, A. Primary cells from patients with adult T cell leukemia/lymphoma depend on HTLV-1 Tax expression for NF-kappaB activation and survival. Blood Cancer J. 2023, 13, 67. [Google Scholar] [CrossRef]
- Nerenberg, M.; Hinrichs, S.H.; Reynolds, R.K.; Khoury, G.; Jay, G. The tat gene of human T-lymphotropic virus type 1 induces mesenchymal tumors in transgenic mice. Science 1987, 237, 1324–1329. [Google Scholar] [CrossRef]
- Hinrichs, S.H.; Nerenberg, M.; Reynolds, R.K.; Khoury, G.; Jay, G. A transgenic mouse model for human neurofibromatosis. Science 1987, 237, 1340–1343. [Google Scholar] [CrossRef]
- Nerenberg, M.I.; Wiley, C.A. Degeneration of oxidative muscle fibers in HTLV-1 tax transgenic mice. Am. J. Pathol. 1989, 135, 1025–1033. [Google Scholar]
- Coscoy, L.; Gonzalez-Dunia, D.; Tangy, F.; Syan, S.; Brahic, M.; Ozden, S. Molecular mechanism of tumorigenesis in mice transgenic for the human T cell leukemia virus Tax gene. Virology 1998, 248, 332–341. [Google Scholar] [CrossRef]
- Grossman, W.J.; Kimata, J.T.; Wong, F.H.; Zutter, M.; Ley, T.J.; Ratner, L. Development of leukemia in mice transgenic for the tax gene of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 1995, 92, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Benvenisty, N.; Ornitz, D.M.; Bennett, G.L.; Sahagan, B.G.; Kuo, A.; Cardiff, R.D.; Leder, P. Brain tumours and lymphomas in transgenic mice that carry HTLV-I LTR/c-myc and Ig/tax genes. Oncogene 1992, 7, 2399–2405. [Google Scholar] [PubMed]
- Gao, L.; Deng, H.; Zhao, H.; Hirbe, A.; Harding, J.; Ratner, L.; Weilbaecher, K. HTLV-1 Tax transgenic mice develop spontaneous osteolytic bone metastases prevented by osteoclast inhibition. Blood 2005, 106, 4294–4302. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Yasunaga, J.; Zhao, T.; Yoshida, M.; Miyazato, P.; Takai, K.; Shimizu, K.; Ohshima, K.; Green, P.L.; Ohkura, N.; et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011, 7, e1001274. [Google Scholar] [CrossRef] [PubMed]
- Villaudy, J.; Wencker, M.; Gadot, N.; Gillet, N.A.; Scoazec, J.Y.; Gazzolo, L.; Manz, M.G.; Bangham, C.R.; Dodon, M.D. HTLV-1 propels thymic human T cell development in “human immune system” Rag2−/− gamma c−/− mice. PLoS Pathog. 2011, 7, e1002231. [Google Scholar] [CrossRef]
- Tezuka, K.; Xun, R.; Tei, M.; Ueno, T.; Tanaka, M.; Takenouchi, N.; Fujisawa, J. An animal model of adult T-cell leukemia: Humanized mice with HTLV-1-specific immunity. Blood 2014, 123, 346–355. [Google Scholar] [CrossRef]
- Ginwala, R.; Caruso, B.; Khan, Z.K.; Pattekar, A.; Chew, G.M.; Corley, M.J.; Loonawat, R.; Jacobson, S.; Sreedhar, S.; Ndhlovu, L.C.; et al. HTLV-1 Infection and Neuropathogenesis in the Context of Rag1−/− gammac−/− (RAG1-Hu) and BLT Mice. J. Neuroimmune Pharmacol. 2017, 12, 504–520. [Google Scholar] [CrossRef]
- Dodon, M.D.; Villaudy, J.; Gazzolo, L.; Haines, R.; Lairmore, M. What we are learning on HTLV-1 pathogenesis from animal models. Front. Microbiol. 2012, 3, 320. [Google Scholar] [CrossRef]
- Miyoshi, I.; Yoshimoto, S.; Fujishita, M.; Kubonishi, I.; Taguchi, H.; Ohtsuki, Y. Infectious transmission of human T-cell leukemia virus to animals. Princess Takamatsu Symp. 1984, 15, 121–127. [Google Scholar]
- Nakamura, H.; Tanaka, Y.; Komuro-Tsujimoto, A.; Ishikawa, K.; Takadaya, K.; Tozawa, H.; Tsujimoto, H.; Honjo, S.; Hayami, M. Experimental inoculation of monkeys with autologous lymphoid cell lines immortalized by and producing human T-cell leukemia virus type-I. Int. J. Cancer 1986, 38, 867–875. [Google Scholar] [CrossRef]
- Beilke, M.A.; Traina-Dorge, V.; England, J.D.; Blanchard, J.L. Polymyositis, arthritis, and uveitis in a macaque experimentally infected with human T lymphotropic virus type I. Arthritis Rheum. 1996, 39, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Kazanji, M.; Tartaglia, J.; Franchini, G.; de Thoisy, B.; Talarmin, A.; Contamin, H.; Gessain, A.; de The, G. Immunogenicity and protective efficacy of recombinant human T-cell leukemia/lymphoma virus type 1 NYVAC and naked DNA vaccine candidates in squirrel monkeys (Saimiri sciureus). J. Virol. 2001, 75, 5939–5948. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, I.; Yoshimoto, S.; Kubonishi, I.; Fujishita, M.; Ohtsuki, Y.; Yamashita, M.; Yamato, K.; Hirose, S.; Taguchi, H.; Niiya, K.; et al. Infectious transmission of human T-cell leukemia virus to rabbits. Int. J. Cancer 1985, 35, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Kannian, P.; Yin, H.; Doueiri, R.; Lairmore, M.D.; Fernandez, S.; Green, P.L. Distinct transformation tropism exhibited by human T lymphotropic virus type 1 (HTLV-1) and HTLV-2 is the result of postinfection T cell clonal expansion. J. Virol. 2012, 86, 3757–3766. [Google Scholar] [CrossRef]
- Shirinian, M.; Kambris, Z.; Hamadeh, L.; Grabbe, C.; Journo, C.; Mahieux, R.; Bazarbachi, A. A Transgenic Drosophila melanogaster Model To Study Human T-Lymphotropic Virus Oncoprotein Tax-1-Driven Transformation In Vivo. J. Virol. 2015, 89, 8092–8095. [Google Scholar] [CrossRef]
- Akkouche, A.; Moodad, S.; Hleihel, R.; Skayneh, H.; Chambeyron, S.; El Hajj, H.; Bazarbachi, A. In vivo antagonistic role of the Human T-Cell Leukemia Virus Type 1 regulatory proteins Tax and HBZ. PLoS Pathog. 2021, 17, e1009219. [Google Scholar] [CrossRef]
- Montaner, J.S.; Reiss, P.; Cooper, D.; Vella, S.; Harris, M.; Conway, B.; Wainberg, M.A.; Smith, D.; Robinson, P.; Hall, D.; et al. A randomized, double-blind trial comparing combinations of nevirapine, didanosine, and zidovudine for HIV-infected patients: The INCAS Trial. Italy, The Netherlands, Canada and Australia Study. JAMA 1998, 279, 930–937. [Google Scholar] [CrossRef]
- Taylor, G.J. 150 Practice ECGs; John Wiley & Sons: Hoboken, NJ, USA, 2006; pp. 63–213. [Google Scholar]
- Trevino, A.; Parra, P.; Bar-Magen, T.; Garrido, C.; de Mendoza, C.; Soriano, V. Antiviral effect of raltegravir on HTLV-1 carriers. J. Antimicrob. Chemother. 2012, 67, 218–221. [Google Scholar] [CrossRef]
- Taylor, G.P.; Goon, P.; Furukawa, Y.; Green, H.; Barfield, A.; Mosley, A.; Nose, H.; Babiker, A.; Rudge, P.; Usuku, K.; et al. Zidovudine plus lamivudine in Human T-Lymphotropic Virus type-I-associated myelopathy: A randomised trial. Retrovirology 2006, 3, 63. [Google Scholar] [CrossRef]
- Murray, J.S.; Elashoff, M.R.; Iacono-Connors, L.C.; Cvetkovich, T.A.; Struble, K.A. The use of plasma HIV RNA as a study endpoint in efficacy trials of antiretroviral drugs. AIDS 1999, 13, 797–804. [Google Scholar] [CrossRef]
- Mocroft, A.; Phillips, A.N.; Gatell, J.; Ledergerber, B.; Fisher, M.; Clumeck, N.; Losso, M.; Lazzarin, A.; Fatkenheuer, G.; Lundgren, J.D. Normalisation of CD4 counts in patients with HIV-1 infection and maximum virological suppression who are taking combination antiretroviral therapy: An observational cohort study. Lancet 2007, 370, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Hamlyn, E.; Ewings, F.M.; Porter, K.; Cooper, D.A.; Tambussi, G.; Schechter, M.; Pedersen, C.; Okulicz, J.F.; McClure, M.; Babiker, A.; et al. Plasma HIV viral rebound following protocol-indicated cessation of ART commenced in primary and chronic HIV infection. PLoS ONE 2012, 7, e43754. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.W.; Justement, J.S.; Murray, D.; Hallahan, C.W.; Maenza, J.; Collier, A.C.; Sheth, P.M.; Kaul, R.; Ostrowski, M.; Moir, S.; et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: Implications for eradication. AIDS 2010, 24, 2803–2808. [Google Scholar] [CrossRef] [PubMed]
- Colven, R.; Harrington, R.D.; Spach, D.H.; Cohen, C.J.; Hooton, T.M. Retroviral rebound syndrome after cessation of suppressive antiretroviral therapy in three patients with chronic HIV infection. Ann. Intern. Med. 2000, 133, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Mirsaliotis, A.; Lamb, D.; Brighty, D.W. Nonhelical leash and alpha-helical structures determine the potency of a peptide antagonist of human T-cell leukemia virus entry. J. Virol. 2008, 82, 4965–4973. [Google Scholar] [CrossRef]
- Mirsaliotis, A.; Nurkiyanova, K.; Lamb, D.; Kuo, C.S.; Brighty, D.W. An antibody that blocks human T-cell leukemia virus type 1 six-helix-bundle formation in vitro identified by a novel assay for inhibitors of envelope function. J. Gen. Virol. 2007, 88, 660–669. [Google Scholar] [CrossRef]
- Argyris, E.G.; Acheampong, E.; Nunnari, G.; Mukhtar, M.; Williams, K.J.; Pomerantz, R.J. Human immunodeficiency virus type 1 enters primary human brain microvascular endothelial cells by a mechanism involving cell surface proteoglycans independent of lipid rafts. J. Virol. 2003, 77, 12140–12151. [Google Scholar] [CrossRef]
- Ida, H.; Kurata, A.; Eguchi, K.; Yamashita, I.; Nakashima, M.; Sakai, M.; Kawabe, Y.; Nakamura, T.; Nagataki, S. Mechanism of inhibitory effect of dextran sulfate and heparin on human T-cell lymphotropic virus type I (HTLV-I)-induced syncytium formation in vitro: Role of cell-to-cell contact. Antivir. Res. 1994, 23, 143–159. [Google Scholar] [CrossRef]
- Moulard, M.; Lortat-Jacob, H.; Mondor, I.; Roca, G.; Wyatt, R.; Sodroski, J.; Zhao, L.; Olson, W.; Kwong, P.D.; Sattentau, Q.J. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J. Virol. 2000, 74, 1948–1960. [Google Scholar] [CrossRef]
- Okuma, K.; Nakamura, M.; Nakano, S.; Niho, Y.; Matsuura, Y. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 1999, 254, 235–244. [Google Scholar] [CrossRef]
- Secchiero, P.; Sun, D.; De Vico, A.L.; Crowley, R.W.; Reitz, M.S., Jr.; Zauli, G.; Lusso, P.; Gallo, R.C. Role of the extracellular domain of human herpesvirus 7 glycoprotein B in virus binding to cell surface heparan sulfate proteoglycans. J. Virol. 1997, 71, 4571–4580. [Google Scholar] [CrossRef] [PubMed]
- Jinno-Oue, A.; Tanaka, A.; Shimizu, N.; Mori, T.; Sugiura, N.; Kimata, K.; Isomura, H.; Hoshino, H. Inhibitory effect of chondroitin sulfate type E on the binding step of human T-cell leukemia virus type 1. AIDS Res. Hum. Retroviruses 2013, 29, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Sluis-Cremer, N.; Arion, D.; Parniak, M.A. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell. Mol. Life Sci. 2000, 57, 1408–1422. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Chen, X.; Li, D.; Fang, Z.; De Clercq, E.; Liu, X. HIV-1 NNRTIs: Structural diversity, pharmacophore similarity, and implications for drug design. Med. Res. Rev. 2013, 33 (Suppl. 1), E1–E72. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, H.; Matsui, T.; Harada, S.; Kobayashi, N.; Matsuda, A.; Ueda, T.; Yamamoto, N. Inhibition of replication and cytopathic effect of human T cell lymphotropic virus type III/lymphadenopathy-associated virus by 3′-azido-3′-deoxythymidine in vitro. Antimicrob. Agents Chemother. 1986, 30, 933–937. [Google Scholar] [CrossRef]
- Zhang, J.; Balestrieri, E.; Grelli, S.; Matteucci, C.; Pagnini, V.; D’Agostini, C.; Mastino, A.; Macchi, B. Efficacy of 3′-azido 3′deoxythymidine (AZT) in preventing HTLV-1 transmission to human cord blood mononuclear cells. Virus Res. 2001, 78, 67–78. [Google Scholar] [CrossRef]
- Macchi, B.; Faraoni, I.; Zhang, J.; Grelli, S.; Favalli, C.; Mastino, A.; Bonmassar, E. AZT inhibits the transmission of human T cell leukaemia/lymphoma virus type I to adult peripheral blood mononuclear cells in vitro. J. Gen. Virol. 1997, 78 Pt 5, 1007–1016. [Google Scholar] [CrossRef]
- Balestrieri, E.; Sciortino, M.T.; Mastino, A.; Macchi, B. Protective effect of the acyclic nucleoside phosphonate tenofovir toward human T-cell leukemia/lymphotropic virus type 1 infection of human peripheral blood mononuclear cells in vitro. Antivir. Res. 2005, 68, 154–162. [Google Scholar] [CrossRef]
- Miyazato, P.; Yasunaga, J.; Taniguchi, Y.; Koyanagi, Y.; Mitsuya, H.; Matsuoka, M. De novo human T-cell leukemia virus type 1 infection of human lymphocytes in NOD-SCID, common gamma-chain knockout mice. J. Virol. 2006, 80, 10683–10691. [Google Scholar] [CrossRef]
- Balestrieri, E.; Forte, G.; Matteucci, C.; Mastino, A.; Macchi, B. Effect of lamivudine on transmission of human T-cell lymphotropic virus type 1 to adult peripheral blood mononuclear cells in vitro. Antimicrob. Agents Chemother. 2002, 46, 3080–3083. [Google Scholar] [CrossRef]
- Datta, A.; Bellon, M.; Sinha-Datta, U.; Bazarbachi, A.; Lepelletier, Y.; Canioni, D.; Waldmann, T.A.; Hermine, O.; Nicot, C. Persistent inhibition of telomerase reprograms adult T-cell leukemia to p53-dependent senescence. Blood 2006, 108, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Hukezalie, K.R.; Thumati, N.R.; Cote, H.C.; Wong, J.M. In vitro and ex vivo inhibition of human telomerase by anti-HIV nucleoside reverse transcriptase inhibitors (NRTIs) but not by non-NRTIs. PLoS ONE 2012, 7, e47505. [Google Scholar] [CrossRef] [PubMed]
- Leeansyah, E.; Cameron, P.U.; Solomon, A.; Tennakoon, S.; Velayudham, P.; Gouillou, M.; Spelman, T.; Hearps, A.; Fairley, C.; de Smit, V.; et al. Inhibition of telomerase activity by human immunodeficiency virus (HIV) nucleos(t)ide reverse transcriptase inhibitors: A potential factor contributing to HIV-associated accelerated aging. J. Infect. Dis. 2013, 207, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Barski, M.S.; Minnell, J.J.; Maertens, G.N. Inhibition of HTLV-1 Infection by HIV-1 First- and Second-Generation Integrase Strand Transfer Inhibitors. Front. Microbiol. 2019, 10, 1877. [Google Scholar] [CrossRef] [PubMed]
- Rabaaoui, S.; Zouhiri, F.; Lancon, A.; Leh, H.; d’Angelo, J.; Wattel, E. Inhibitors of strand transfer that prevent integration and inhibit human T-cell leukemia virus type 1 early replication. Antimicrob. Agents Chemother. 2008, 52, 3532–3541. [Google Scholar] [CrossRef] [PubMed]
- Seegulam, M.E.; Ratner, L. Integrase inhibitors effective against human T-cell leukemia virus type 1. Antimicrob. Agents Chemother. 2011, 55, 2011–2017. [Google Scholar] [CrossRef]
- Kuhnert, M.; Steuber, H.; Diederich, W.E. Structural basis for HTLV-1 protease inhibition by the HIV-1 protease inhibitor indinavir. J. Med. Chem. 2014, 57, 6266–6272. [Google Scholar] [CrossRef]
- Dewan, M.Z.; Uchihara, J.N.; Terashima, K.; Honda, M.; Sata, T.; Ito, M.; Fujii, N.; Uozumi, K.; Tsukasaki, K.; Tomonaga, M.; et al. Efficient intervention of growth and infiltration of primary adult T-cell leukemia cells by an HIV protease inhibitor, ritonavir. Blood 2006, 107, 716–724. [Google Scholar] [CrossRef]
- Kondo, T.; Kono, H.; Miyamoto, N.; Yoshida, R.; Toki, H.; Matsumoto, I.; Hara, M.; Inoue, H.; Inatsuki, A.; Funatsu, T.; et al. Age- and sex-specific cumulative rate and risk of ATLL for HTLV-I carriers. Int. J. Cancer 1989, 43, 1061–1064. [Google Scholar] [CrossRef]
- Takasaki, Y.; Iwanaga, M.; Imaizumi, Y.; Tawara, M.; Joh, T.; Kohno, T.; Yamada, Y.; Kamihira, S.; Ikeda, S.; Miyazaki, Y.; et al. Long-term study of indolent adult T-cell leukemia-lymphoma. Blood 2010, 115, 4337–4343. [Google Scholar] [CrossRef]
- Utsunomiya, A.; Choi, I.; Chihara, D.; Seto, M. Recent advances in the treatment of adult T-cell leukemia-lymphomas. Cancer Sci. 2015, 106, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.B.; Fuji, S.; Hermine, O.; Bazarbachi, A.; Ramos, J.C.; Ratner, L.; Horwitz, S.; Fields, P.; Tanase, A.; Bumbea, H.; et al. Revised Adult T-Cell Leukemia-Lymphoma International Consensus Meeting Report. J. Clin. Oncol. 2019, 37, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Fuji, S.; Fujiwara, H.; Nakano, N.; Wake, A.; Inoue, Y.; Fukuda, T.; Hidaka, M.; Moriuchi, Y.; Miyamoto, T.; Uike, N.; et al. Early application of related SCT might improve clinical outcome in adult T-cell leukemia/lymphoma. Bone Marrow Transplant. 2016, 51, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Bazarbachi, A.; Cwynarski, K.; Boumendil, A.; Finel, H.; Fields, P.; Raj, K.; Nagler, A.; Mohty, M.; Sureda, A.; Dreger, P.; et al. Outcome of patients with HTLV-1-associated adult T-cell leukemia/lymphoma after SCT: A retrospective study by the EBMT LWP. Bone Marrow Transplant. 2014, 49, 1266–1268. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, J.M.; Whiteside, G.; McKeage, K.; Croxtall, J.C. Mogamulizumab: First global approval. Drugs 2012, 72, 1293–1298. [Google Scholar] [CrossRef]
- Yoshie, O.; Fujisawa, R.; Nakayama, T.; Harasawa, H.; Tago, H.; Izawa, D.; Hieshima, K.; Tatsumi, Y.; Matsushima, K.; Hasegawa, H.; et al. Frequent expression of CCR4 in adult T-cell leukemia and human T-cell leukemia virus type 1-transformed T cells. Blood 2002, 99, 1505–1511. [Google Scholar] [CrossRef]
- Ishida, T.; Utsunomiya, A.; Iida, S.; Inagaki, H.; Takatsuka, Y.; Kusumoto, S.; Takeuchi, G.; Shimizu, S.; Ito, M.; Komatsu, H.; et al. Clinical significance of CCR4 expression in adult T-cell leukemia/lymphoma: Its close association with skin involvement and unfavorable outcome. Clin. Cancer Res. 2003, 9, 3625–3634. [Google Scholar]
- Yamano, Y.; Araya, N.; Sato, T.; Utsunomiya, A.; Azakami, K.; Hasegawa, D.; Izumi, T.; Fujita, H.; Aratani, S.; Yagishita, N.; et al. Abnormally high levels of virus-infected IFN-gamma+ CCR4+ CD4+ CD25+ T cells in a retrovirus-associated neuroinflammatory disorder. PLoS ONE 2009, 4, e6517. [Google Scholar] [CrossRef]
- Ishida, T.; Joh, T.; Uike, N.; Yamamoto, K.; Utsunomiya, A.; Yoshida, S.; Saburi, Y.; Miyamoto, T.; Takemoto, S.; Suzushima, H.; et al. Defucosylated anti-CCR4 monoclonal antibody (KW-0761) for relapsed adult T-cell leukemia-lymphoma: A multicenter phase II study. J. Clin. Oncol. 2012, 30, 837–842. [Google Scholar] [CrossRef]
- Phillips, A.A.; Fields, P.A.; Hermine, O.; Ramos, J.C.; Beltran, B.E.; Pereira, J.; Wandroo, F.; Feldman, T.; Taylor, G.P.; Sawas, A.; et al. Mogamulizumab versus investigator’s choice of chemotherapy regimen in relapsed/refractory adult T-cell leukemia/lymphoma. Haematologica 2019, 104, 993–1003. [Google Scholar] [CrossRef]
- Sato, T.; Coler-Reilly, A.L.G.; Yagishita, N.; Araya, N.; Inoue, E.; Furuta, R.; Watanabe, T.; Uchimaru, K.; Matsuoka, M.; Matsumoto, N.; et al. Mogamulizumab (Anti-CCR4) in HTLV-1-Associated Myelopathy. N. Engl. J. Med. 2018, 378, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Jo, T.; Takemoto, S.; Suzushima, H.; Uozumi, K.; Yamamoto, K.; Uike, N.; Saburi, Y.; Nosaka, K.; Utsunomiya, A.; et al. Dose-intensified chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T-cell leukaemia-lymphoma: A randomized phase II study. Br. J. Haematol. 2015, 169, 672–682. [Google Scholar] [CrossRef]
- Fuji, S.; Inoue, Y.; Utsunomiya, A.; Moriuchi, Y.; Uchimaru, K.; Choi, I.; Otsuka, E.; Henzan, H.; Kato, K.; Tomoyose, T.; et al. Pretransplantation Anti-CCR4 Antibody Mogamulizumab Against Adult T-Cell Leukemia/Lymphoma Is Associated With Significantly Increased Risks of Severe and Corticosteroid-Refractory Graft-Versus-Host Disease, Nonrelapse Mortality, and Overall Mortality. J. Clin. Oncol. 2016, 34, 3426–3433. [Google Scholar] [CrossRef] [PubMed]
- Vidal, C.; Matsushita, S.; Colamonici, O.R.; Trepel, J.B.; Mitsuya, H.; Neckers, L.M. Human T lymphotropic virus I infection deregulates surface expression of the transferrin receptor. J. Immunol. 1988, 141, 984–988. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.C.; Lepelletier, Y.; Arnulf, B.; England, P.; Baude, C.; Beaumont, C.; Bazarbachi, A.; Benhamou, M.; Monteiro, R.C.; Hermine, O. A neutralizing monoclonal antibody (mAb A24) directed against the transferrin receptor induces apoptosis of tumor T lymphocytes from ATL patients. Blood 2004, 103, 1838–1845. [Google Scholar] [CrossRef] [PubMed]
- Raedler, L.A. Revlimid (Lenalidomide) Now FDA Approved as First-Line Therapy for Patients with Multiple Myeloma. Am. Health Drug Benefits 2016, 9, 140–143. [Google Scholar] [PubMed]
- Ishida, T.; Fujiwara, H.; Nosaka, K.; Taira, N.; Abe, Y.; Imaizumi, Y.; Moriuchi, Y.; Jo, T.; Ishizawa, K.; Tobinai, K.; et al. Multicenter Phase II Study of Lenalidomide in Relapsed or Recurrent Adult T-Cell Leukemia/Lymphoma: ATLL-002. J. Clin. Oncol. 2016, 34, 4086–4093. [Google Scholar] [CrossRef]
- Oka, S.; Ono, K.; Nohgawa, M. Effective maintenance treatment with lenalidomide for a patient with aggressive adult T cell leukemia after chemotherapy. Leuk. Res. Rep. 2019, 11, 21–23. [Google Scholar] [CrossRef]
- Lee, S.H.; Li, Y.; Kim, H.; Eum, S.; Park, K.; Lee, C.H. The role of EZH1 and EZH2 in development and cancer. BMB Rep. 2022, 55, 595–601. [Google Scholar] [CrossRef]
- Yamagishi, M.; Kuze, Y.; Kobayashi, S.; Nakashima, M.; Morishima, S.; Kawamata, T.; Makiyama, J.; Suzuki, K.; Seki, M.; Abe, K.; et al. Mechanisms of action and resistance in histone methylation-targeted therapy. Nature 2024, 627, 221–228. [Google Scholar] [CrossRef]
- Izutsu, K.; Makita, S.; Nosaka, K.; Yoshimitsu, M.; Utsunomiya, A.; Kusumoto, S.; Morishima, S.; Tsukasaki, K.; Kawamata, T.; Ono, T.; et al. An open-label, single-arm phase 2 trial of valemetostat for relapsed or refractory adult T-cell leukemia/lymphoma. Blood 2023, 141, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Koseki, A.; Araya, N.; Yamagishi, M.; Yamauchi, J.; Yagishita, N.; Takao, N.; Takahashi, K.; Kunitomo, Y.; Honma, D.; Araki, K.; et al. EZH1/2 dual inhibitors suppress HTLV-1-infected cell proliferation and hyperimmune response in HTLV-1-associated myelopathy. Front. Microbiol. 2023, 14, 1175762. [Google Scholar] [CrossRef] [PubMed]
- Enose-Akahata, Y.; Vellucci, A.; Jacobson, S. Role of HTLV-1 Tax and HBZ in the Pathogenesis of HAM/TSP. Front. Microbiol. 2017, 8, 2563. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Yamano, Y.; Brennan, M.B.; Mora, C.A.; Jacobson, S. Increased HTLV-I proviral load and preferential expansion of HTLV-I Tax-specific CD8+ T cells in cerebrospinal fluid from patients with HAM/TSP. Ann. Neurol. 2001, 50, 807–812. [Google Scholar] [CrossRef] [PubMed]
- El Hajj, H.; Khalil, B.; Ghandour, B.; Nasr, R.; Shahine, S.; Ghantous, A.; Abdel-Samad, R.; Sinjab, A.; Hasegawa, H.; Jabbour, M.; et al. Preclinical efficacy of the synthetic retinoid ST1926 for treating adult T-cell leukemia/lymphoma. Blood 2014, 124, 2072–2080. [Google Scholar] [CrossRef]
- Ozaki, A.; Arima, N.; Matsushita, K.; Uozumi, K.; Akimoto, M.; Hamada, H.; Kawada, H.; Horai, S.; Tanaka, Y.; Tei, C. Cyclosporin A inhibits HTLV-I tax expression and shows anti-tumor effects in combination with VP-16. J. Med. Virol. 2007, 79, 1906–1913. [Google Scholar] [CrossRef]
- Xiang, D.; Yuan, Y.; Chen, L.; Liu, X.; Belani, C.; Cheng, H. Niclosamide, an anti-helminthic molecule, downregulates the retroviral oncoprotein Tax and pro-survival Bcl-2 proteins in HTLV-1-transformed T lymphocytes. Biochem. Biophys. Res. Commun. 2015, 464, 221–228. [Google Scholar] [CrossRef][Green Version]
- Jacobson, S.; Shida, H.; McFarlin, D.E.; Fauci, A.S.; Koenig, S. Circulating CD8+ cytotoxic T lymphocytes specific for HTLV-I pX in patients with HTLV-I associated neurological disease. Nature 1990, 348, 245–248. [Google Scholar] [CrossRef]
- Kannagi, M.; Harada, S.; Maruyama, I.; Inoko, H.; Igarashi, H.; Kuwashima, G.; Sato, S.; Morita, M.; Kidokoro, M.; Sugimoto, M.; et al. Predominant recognition of human T cell leukemia virus type I (HTLV-I) pX gene products by human CD8+ cytotoxic T cells directed against HTLV-I-infected cells. Int. Immunol. 1991, 3, 761–767. [Google Scholar] [CrossRef]
- Mahgoub, M.; Yasunaga, J.I.; Iwami, S.; Nakaoka, S.; Koizumi, Y.; Shimura, K.; Matsuoka, M. Sporadic on/off switching of HTLV-1 Tax expression is crucial to maintain the whole population of virus-induced leukemic cells. Proc. Natl. Acad. Sci. USA 2018, 115, E1269–E1278. [Google Scholar] [CrossRef]
- Kurihara, K.; Harashima, N.; Hanabuchi, S.; Masuda, M.; Utsunomiya, A.; Tanosaki, R.; Tomonaga, M.; Ohashi, T.; Hasegawa, A.; Masuda, T.; et al. Potential immunogenicity of adult T cell leukemia cells in vivo. Int. J. Cancer 2005, 114, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Bellon, M.; Nicot, C. Feedback Loop Regulation between Pim Kinases and Tax Keeps Human T-Cell Leukemia Virus Type 1 Viral Replication in Check. J. Virol. 2022, 96, e01960-21. [Google Scholar] [CrossRef] [PubMed]
- Kfoury, Y.; Nasr, R.; Hermine, O.; de The, H.; Bazarbachi, A. Proapoptotic regimes for HTLV-I-transformed cells: Targeting Tax and the NF-kappaB pathway. Cell Death Differ. 2005, 12 (Suppl. 1), 871–877. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, C.; Ikezoe, T.; Yang, J.; Komatsu, N.; Bandobashi, K.; Taniguchi, A.; Kuwayama, Y.; Togitani, K.; Koeffler, H.P.; Taguchi, H. Histone deacetylase inhibitors induce growth arrest and apoptosis of HTLV-1-infected T-cells via blockade of signaling by nuclear factor kappaB. Leuk. Res. 2008, 32, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, B.; Sargeant, A.; Landes, K.; Fernandez, S.A.; Chen, C.S.; Lairmore, M.D. Efficacy of novel histone deacetylase inhibitor, AR42, in a mouse model of, human T-lymphotropic virus type 1 adult T cell lymphoma. Leuk. Res. 2011, 35, 1491–1497. [Google Scholar] [CrossRef]
- Watanabe, M.; Ohsugi, T.; Shoda, M.; Ishida, T.; Aizawa, S.; Maruyama-Nagai, M.; Utsunomiya, A.; Koga, S.; Yamada, Y.; Kamihira, S.; et al. Dual targeting of transformed and untransformed HTLV-1-infected T cells by DHMEQ, a potent and selective inhibitor of NF-kappaB, as a strategy for chemoprevention and therapy of adult T-cell leukemia. Blood 2005, 106, 2462–2471. [Google Scholar] [CrossRef]
- Schwalb, A.; Perez-Muto, V.; Cachay, R.; Tipismana, M.; Alvarez, C.; Mejia, F.; Gonzalez-Lagos, E.; Gotuzzo, E. Early-Onset HTLV-1-Associated Myelopathy/Tropical Spastic Paraparesis. Pathogens 2020, 9, 450. [Google Scholar] [CrossRef]
- Martin, F.; Fedina, A.; Youshya, S.; Taylor, G.P. A 15-year prospective longitudinal study of disease progression in patients with HTLV-1 associated myelopathy in the UK. J. Neurol. Neurosurg. Psychiatry 2010, 81, 1336–1340. [Google Scholar] [CrossRef]
- Araujo, A.; Bangham, C.R.M.; Casseb, J.; Gotuzzo, E.; Jacobson, S.; Martin, F.; Penalva de Oliveira, A.; Puccioni-Sohler, M.; Taylor, G.P.; Yamano, Y. Management of HAM/TSP: Systematic Review and Consensus-based Recommendations 2019. Neurol. Clin. Pract. 2021, 11, 49–56. [Google Scholar] [CrossRef]
- Tsutsumi, S.; Sato, T.; Yagishita, N.; Yamauchi, J.; Araya, N.; Hasegawa, D.; Nagasaka, M.; Coler-Reilly, A.L.G.; Inoue, E.; Takata, A.; et al. Real-world clinical course of HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) in Japan. Orphanet J. Rare Dis. 2019, 14, 227. [Google Scholar] [CrossRef]
- Macchi, B.; Balestrieri, E.; Ascolani, A.; Hilburn, S.; Martin, F.; Mastino, A.; Taylor, G.P. Susceptibility of primary HTLV-1 isolates from patients with HTLV-1-associated myelopathy to reverse transcriptase inhibitors. Viruses 2011, 3, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Coler-Reilly, A.L.G.; Sato, T.; Matsuzaki, T.; Nakagawa, M.; Niino, M.; Nagai, M.; Nakamura, T.; Takenouchi, N.; Araya, N.; Yagishita, N.; et al. Effectiveness of Daily Prednisolone to Slow Progression of Human T-Lymphotropic Virus Type 1-Associated Myelopathy/Tropical Spastic Paraparesis: A Multicenter Retrospective Cohort Study. Neurotherapeutics 2017, 14, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Croda, M.G.; de Oliveira, A.C.; Vergara, M.P.; Bonasser, F.; Smid, J.; Duarte, A.J.; Casseb, J. Corticosteroid therapy in TSP/HAM patients: The results from a 10 years open cohort. J. Neurol. Sci. 2008, 269, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Nakahara, K.; Maruyama, Y.; Kawabata, M.; Higuchi, I.; Kubota, H.; Izumo, S.; Arimura, K.; Osame, M. Therapeutic trials in 200 patients with HTLV-I-associated myelopathy/tropical spastic paraparesis. J. Neurovirol. 1996, 2, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Araujo, A.Q.; Afonso, C.R.; Leite, A.C.; Dultra, S.V. Intravenous methylprednisolone in HTLV-I associated myelopathy/tropical spastic paraparesis (HAM/TSP). Arq. Neuropsiquiatr. 1993, 51, 325–328. [Google Scholar] [CrossRef]
- Kira, J.; Fujihara, K.; Itoyama, Y.; Goto, I.; Hasuo, K. Leukoencephalopathy in HTLV-I-associated myelopathy/tropical spastic paraparesis: MRI analysis and a two year follow-up study after corticosteroid therapy. J. Neurol. Sci. 1991, 106, 41–49. [Google Scholar] [CrossRef]
- Nagai, M.; Tsujii, T.; Iwaki, H.; Nishikawa, N.; Nomoto, M. Cerebrospinal fluid neopterin, but not osteopontin, is a valuable biomarker for the treatment response in patients with HTLV-I-associated myelopathy. Intern. Med. 2013, 52, 2203–2208. [Google Scholar] [CrossRef]
- Saito, M.; Nakagawa, M.; Kaseda, S.; Matsuzaki, T.; Jonosono, M.; Eiraku, N.; Kubota, R.; Takenouchi, N.; Nagai, M.; Furukawa, Y.; et al. Decreased human T lymphotropic virus type I (HTLV-I) provirus load and alteration in T cell phenotype after interferon-alpha therapy for HTLV-I-associated myelopathy/tropical spastic paraparesis. J. Infect. Dis. 2004, 189, 29–40. [Google Scholar] [CrossRef]
- Tamaki, K.; Sato, T.; Tsugawa, J.; Fujioka, S.; Yagishita, N.; Araya, N.; Yamauchi, J.; Coler-Reilly, A.L.G.; Nagasaka, M.; Hasegawa, Y.; et al. Cerebrospinal Fluid CXCL10 as a Candidate Surrogate Marker for HTLV-1-Associated Myelopathy/Tropical Spastic Paraparesis. Front. Microbiol. 2019, 10, 2110. [Google Scholar] [CrossRef]
- Yamauchi, J.; Tanabe, K.; Sato, T.; Nakagawa, M.; Matsuura, E.; Tsuboi, Y.; Tamaki, K.; Sakima, H.; Ishihara, S.; Ohta, Y.; et al. Efficacy of Corticosteroid Therapy for HTLV-1-Associated Myelopathy: A Randomized Controlled Trial (HAMLET-P). Viruses 2022, 14, 136. [Google Scholar] [CrossRef]
- Ihrke, P.J.; Norton, A.L.; Ling, G.V.; Stannard, A.A. Urinary tract infection associated with long-term corticosteroid administration in dogs with chronic skin diseases. J. Am. Vet. Med. Assoc. 1985, 186, 43–46. [Google Scholar] [PubMed]
- Izumo, S.; Goto, I.; Itoyama, Y.; Okajima, T.; Watanabe, S.; Kuroda, Y.; Araki, S.; Mori, M.; Nagataki, S.; Matsukura, S.; et al. Interferon-alpha is effective in HTLV-I-associated myelopathy: A multicenter, randomized, double-blind, controlled trial. Neurology 1996, 46, 1016–1021. [Google Scholar] [CrossRef] [PubMed]
- Arimura, K.; Nakagawa, M.; Izumo, S.; Usuku, K.; Itoyama, Y.; Kira, J.; Osame, M. Safety and efficacy of interferon-alpha in 167 patients with human T-cell lymphotropic virus type 1-associated myelopathy. J. Neurovirol. 2007, 13, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Oh, U.; Yamano, Y.; Mora, C.A.; Ohayon, J.; Bagnato, F.; Butman, J.A.; Dambrosia, J.; Leist, T.P.; McFarland, H.; Jacobson, S. Interferon-beta1a therapy in human T-lymphotropic virus type I-associated neurologic disease. Ann. Neurol. 2005, 57, 526–534. [Google Scholar] [CrossRef]
- Martin, F.; Castro, H.; Gabriel, C.; Adonis, A.; Fedina, A.; Harrison, L.; Brodnicki, L.; Demontis, M.A.; Babiker, A.G.; Weber, J.N.; et al. Ciclosporin A proof of concept study in patients with active, progressive HTLV-1 associated myelopathy/tropical spastic paraparesis. PLoS Negl. Trop. Dis. 2012, 6, e1675. [Google Scholar] [CrossRef]
- Sanchez-Montalva, A.; Salvador, F.; Caballero, E.; Molina, I. Cyclosporine for the treatment of HLTV-1-induced HAM/TSP: An experience from a case report. Medicine 2015, 94, e382. [Google Scholar] [CrossRef]
- Lehky, T.J.; Levin, M.C.; Kubota, R.; Bamford, R.N.; Flerlage, A.N.; Soldan, S.S.; Leist, T.P.; Xavier, A.; White, J.D.; Brown, M.; et al. Reduction in HTLV-I proviral load and spontaneous lymphoproliferation in HTLV-I-associated myelopathy/tropical spastic paraparesis patients treated with humanized anti-Tac. Ann. Neurol. 1998, 44, 942–947. [Google Scholar] [CrossRef]
- Sato, T.; Nagai, M.; Watanabe, O.; Misu, T.; Takenouchi, N.; Ohkubo, R.; Ishihara, S.; Tsuboi, Y.; Katsuno, M.; Nakagawa, M.; et al. Multicenter, randomized, double-blind, placebo-controlled phase 3 study of mogamulizumab with open-label extension study in a minimum number of patients with human T-cell leukemia virus type-1-associated myelopathy. J. Neurol. 2024, 271, 3471–3485. [Google Scholar] [CrossRef]
- Harrington, W.J., Jr.; Sheremata, W.A.; Snodgrass, S.R.; Emerson, S.; Phillips, S.; Berger, J.R. Tropical spastic paraparesis/HTLV-1-associated myelopathy (TSP/HAM): Treatment with an anabolic steroid danazol. AIDS Res. Hum. Retroviruses 1991, 7, 1031–1034. [Google Scholar] [CrossRef]
- Melo, A.; Moura, L.; Meireles, A.; Costa, G. Danazol. A new perspective in the treatment of HTLV-1 associated myelopathy (preliminary report). Arq. Neuropsiquiatr. 1992, 50, 402–403. [Google Scholar] [CrossRef][Green Version]
- Olindo, S.; Belrose, G.; Gillet, N.; Rodriguez, S.; Boxus, M.; Verlaeten, O.; Asquith, B.; Bangham, C.; Signate, A.; Smadja, D.; et al. Safety of long-term treatment of HAM/TSP patients with valproic acid. Blood 2011, 118, 6306–6309. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, A.; Imai, H.; Inayoshi, S.; Tsuda, T. Intermittent high-dose vitamin C therapy in patients with HTLV-I associated myelopathy. J. Neurol. Neurosurg. Psychiatry 1993, 56, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Moens, B.; Decanine, D.; Menezes, S.M.; Khouri, R.; Silva-Santos, G.; Lopez, G.; Alvarez, C.; Talledo, M.; Gotuzzo, E.; de Almeida Kruschewsky, R.; et al. Ascorbic acid has superior ex vivo antiproliferative, cell death-inducing and immunomodulatory effects over IFN-alpha in HTLV-1-associated myelopathy. PLoS Negl. Trop. Dis. 2012, 6, e1729. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, H.; Nakamura, T.; Tsujihata, M.; Kinoshita, I.; Satoh, A.; Tomita, I.; Shirabe, S.; Shibayama, K.; Nagataki, S. Plasmapheresis in treatment of human T-lymphotropic virus type-I associated myelopathy. Lancet 1988, 2, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Enose-Akahata, Y.; Ngouth, N.; Ohayon, J.; Mandel, M.; Chavin, J.; Turner, T.J.; Jacobson, S. Effect of Teriflunomide on Cells from Patients With Human T-cell Lymphotropic Virus Type 1-Associated Neurologic Disease. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e986. [Google Scholar] [CrossRef]
- Sonoda, J.; Koriyama, C.; Yamamoto, S.; Kozako, T.; Li, H.C.; Lema, C.; Yashiki, S.; Fujiyoshi, T.; Yoshinaga, M.; Nagata, Y.; et al. HTLV-1 provirus load in peripheral blood lymphocytes of HTLV-1 carriers is diminished by green tea drinking. Cancer Sci. 2004, 95, 596–601. [Google Scholar] [CrossRef]
- Matsuzaki, T.; Saito, M.; Usuku, K.; Nose, H.; Izumo, S.; Arimura, K.; Osame, M. A prospective uncontrolled trial of fermented milk drink containing viable Lactobacillus casei strain Shirota in the treatment of HTLV-1 associated myelopathy/tropical spastic paraparesis. J. Neurol. Sci. 2005, 237, 75–81. [Google Scholar] [CrossRef]
- Matsuura, E.; Yoshimura, A.; Nozuma, S.; Higuchi, I.; Kubota, R.; Takashima, H. Clinical presentation of axial myopathy in two siblings with HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). BMC Neurol. 2015, 15, 18. [Google Scholar] [CrossRef]
- Gaspar Sobrinho, F.P.; Souza-Machado, A.; Cruz, A.A.; Lessa, H.A.; Ramos, E.A. Chronic rhinitis in HTLV-1 carriers: A histopathologic study. Braz. J. Otorhinolaryngol. 2012, 78, 35–40. [Google Scholar] [CrossRef]
- Saito, M.; Kato, K.; Kondo, A.; Miyake, K. Neurogenic bladder in HAM (HTLV-I associated myelopathy). Hinyokika Kiyo 1991, 37, 1005–1008. [Google Scholar]
- Merle, H.; Cabre, P.; Olindo, S.; Merle, S.; Smadja, D. Ocular lesions in 200 patients infected by the human T-cell lymphotropic virus type 1 in martinique (French West Indies). Am. J. Ophthalmol. 2002, 134, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Taylor, G.P.; Jacobson, S. Inflammatory manifestations of HTLV-1 and their therapeutic options. Expert. Rev. Clin. Immunol. 2014, 10, 1531–1546. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Saijo, Y.; Sakai, T.; Abe, T.; Ohnuma, K.; Tezuka, F.; Terunuma, H.; Ogata, K.; Nukiwa, T. Human T-cell lymphotropic virus type I (HTLV-I) carrier with clinical manifestations characteristic of diffuse panbronchiolitis. Intern. Med. 1996, 35, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Littleton, E.T.; Man, W.D.; Holton, J.L.; Landon, D.N.; Hanna, M.G.; Polkey, M.I.; Taylor, G.P. Human T cell leukaemia virus type I associated neuromuscular disease causing respiratory failure. J. Neurol. Neurosurg. Psychiatry 2002, 72, 650–652. [Google Scholar] [CrossRef] [PubMed]
- Dalakas, M.C. Understanding the immunopathogenesis of inclusion-body myositis: Present and future prospects. Rev. Neurol. 2002, 158, 948–958. [Google Scholar] [PubMed]
- Frenzel, L.; Moura, B.; Marcais, A.; Chapdelaine, H.; Hermine, O. HTLV-1-associated arthropathy treated with anti-TNF-alpha agent. Jt. Bone Spine 2014, 81, 360–361. [Google Scholar] [CrossRef]
- Division of HIV Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention (NCHSTP), Centers for Disease Control and Prevention. About PrEP. Available online: https://www.cdc.gov/hiv/basics/prep/about-prep.html (accessed on 29 August 2024).
- Bradshaw, D.; Taylor, G.P. HTLV-1 Transmission and HIV Pre-exposure Prophylaxis: A Scoping Review. Front. Med. 2022, 9, 881547. [Google Scholar] [CrossRef]
- Kira, J.; Koyanagi, Y.; Yamada, T.; Itoyama, Y.; Goto, I.; Yamamoto, N.; Sasaki, H.; Sakaki, Y. Increased HTLV-I proviral DNA in HTLV-I-associated myelopathy: A quantitative polymerase chain reaction study. Ann. Neurol. 1991, 29, 194–201. [Google Scholar] [CrossRef]
- Gessain, A.; Saal, F.; Gout, O.; Daniel, M.T.; Flandrin, G.; de The, G.; Peries, J.; Sigaux, F. High human T-cell lymphotropic virus type I proviral DNA load with polyclonal integration in peripheral blood mononuclear cells of French West Indian, Guianese, and African patients with tropical spastic paraparesis. Blood 1990, 75, 428–433. [Google Scholar] [CrossRef][Green Version]
- Nagai, M.; Usuku, K.; Matsumoto, W.; Kodama, D.; Takenouchi, N.; Moritoyo, T.; Hashiguchi, S.; Ichinose, M.; Bangham, C.R.; Izumo, S.; et al. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: High proviral load strongly predisposes to HAM/TSP. J. Neurovirol. 1998, 4, 586–593. [Google Scholar] [CrossRef]
- Yoshida, M.; Osame, M.; Kawai, H.; Toita, M.; Kuwasaki, N.; Nishida, Y.; Hiraki, Y.; Takahashi, K.; Nomura, K.; Sonoda, S.; et al. Increased replication of HTLV-I in HTLV-I-associated myelopathy. Ann. Neurol. 1989, 26, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Einsiedel, L.; Pham, H.; Wilson, K.; Walley, R.; Turpin, J.; Bangham, C.; Gessain, A.; Woodman, R.J. Human T-Lymphotropic Virus type 1c subtype proviral loads, chronic lung disease and survival in a prospective cohort of Indigenous Australians. PLoS Negl. Trop. Dis. 2018, 12, e0006281. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, K.J.; Siddiqui, A.A.; Bunce, M.; Lloyd, A.L.; Vine, A.M.; Witkover, A.D.; Izumo, S.; Usuku, K.; Welsh, K.I.; Osame, M.; et al. The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J. Immunol. 2000, 165, 7278–7284. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, K.J.; Usuku, K.; Hall, S.E.; Matsumoto, W.; Taylor, G.P.; Procter, J.; Bunce, M.; Ogg, G.S.; Welsh, K.I.; Weber, J.N.; et al. HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc. Natl. Acad. Sci. USA 1999, 96, 3848–3853. [Google Scholar] [CrossRef] [PubMed]
- Barmak, K.; Harhaj, E.; Grant, C.; Alefantis, T.; Wigdahl, B. Human T cell leukemia virus type I-induced disease: Pathways to cancer and neurodegeneration. Virology 2003, 308, 1–12. [Google Scholar] [CrossRef]
- Yamano, Y.; Nagai, M.; Brennan, M.; Mora, C.A.; Soldan, S.S.; Tomaru, U.; Takenouchi, N.; Izumo, S.; Osame, M.; Jacobson, S. Correlation of human T-cell lymphotropic virus type 1 (HTLV-1) mRNA with proviral DNA load, virus-specific CD8+ T cells, and disease severity in HTLV-1-associated myelopathy (HAM/TSP). Blood 2002, 99, 88–94. [Google Scholar] [CrossRef]
- Sonoda, S.; Fujiyoshi, T.; Yashiki, S. Immunogenetics of HTLV-I/II and associated diseases. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol 1996, 13 (Suppl. 1), S119–S123. [Google Scholar] [CrossRef]
- Furukawa, Y.; Kubota, R.; Tara, M.; Izumo, S.; Osame, M. Existence of escape mutant in HTLV-I tax during the development of adult T-cell leukemia. Blood 2001, 97, 987–993. [Google Scholar] [CrossRef]
- Goedert, J.J.; Li, H.C.; Gao, X.J.; Chatterjee, N.; Sonoda, S.; Biggar, R.J.; Cranston, B.; Kim, N.; Carrington, M.; Morgan, O.; et al. Risk of human T-lymphotropic virus type I-associated diseases in Jamaica with common HLA types. Int. J. Cancer 2007, 121, 1092–1097. [Google Scholar] [CrossRef]
- Queiroz, M.A.F.; Azevedo, V.N.; Amoras, E.; Moura, T.C.F.; Guimaraes Ishak, M.O.; Ishak, R.; Vallinoto, A.C.R.; Martins Feitosa, R.N. IFNG +874A/T Polymorphism among Asymptomatic HTLV-1-Infected Individuals Is Potentially Related to a Worse Prognosis. Front. Microbiol. 2018, 9, 795. [Google Scholar] [CrossRef]
- Rocha-Junior, M.C.; Haddad, R.; Ciliao Alves, D.C.; de Deus Wagatsuma, V.M.; Mendes-Junior, C.T.; Deghaide, N.H.; Takayanagui, O.M.; Covas, D.T.; Donadi, E.A.; Kashima, S. Interleukin-18 and interferon-gamma polymorphisms are implicated on proviral load and susceptibility to human T-lymphotropic virus type 1 infection. Tissue Antigens 2012, 80, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Tsukasaki, K.; Miller, C.W.; Kubota, T.; Takeuchi, S.; Fujimoto, T.; Ikeda, S.; Tomonaga, M.; Koeffler, H.P. Tumor necrosis factor alpha polymorphism associated with increased susceptibility to development of adult T-cell leukemia/lymphoma in human T-lymphotropic virus type 1 carriers. Cancer Res. 2001, 61, 3770–3774. [Google Scholar] [PubMed]
- Sabouri, A.H.; Saito, M.; Lloyd, A.L.; Vine, A.M.; Witkover, A.W.; Furukawa, Y.; Izumo, S.; Arimura, K.; Marshall, S.E.; Usuku, K.; et al. Polymorphism in the interleukin-10 promoter affects both provirus load and the risk of human T lymphotropic virus type I-associated myelopathy/tropical spastic paraparesis. J. Infect. Dis. 2004, 190, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Shirdel, A.; Azarpazhooh, M.R.; Sahebari, M.; Ghanbari, M.; Mirfeizi, S.Z.; Hutchinson, I.; Ziaee, A.; Rafatpanah, H. Association of IL-10 Gene Polymorphisms and Human T Lymphotropic Virus Type I-Associated Myelopathy/tropical Spastic Paraparesis in North-East of Iran (Mashhad). Iran. J. Basic Med. Sci. 2013, 16, 258–263. [Google Scholar] [PubMed]
- Carroll, D. Progress and prospects: Zinc-finger nucleases as gene therapy agents. Gene Ther. 2008, 15, 1463–1468. [Google Scholar] [CrossRef]
- Maier, D.A.; Brennan, A.L.; Jiang, S.; Binder-Scholl, G.K.; Lee, G.; Plesa, G.; Zheng, Z.; Cotte, J.; Carpenito, C.; Wood, T.; et al. Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum. Gene Ther. 2013, 24, 245–258. [Google Scholar] [CrossRef]
- Tebas, P.; Jadlowsky, J.K.; Shaw, P.A.; Tian, L.; Esparza, E.; Brennan, A.; Kim, S.; Naing, S.Y.; Richardson, M.W.; Vogel, A.N.; et al. CCR5-edited CD4+ T cells augment HIV-specific immunity to enable post rebound control of HIV replication. J. Clin. Investig. 2021, 131, e181576. [Google Scholar] [CrossRef]
- Yi, G.; Choi, J.G.; Bharaj, P.; Abraham, S.; Dang, Y.; Kafri, T.; Alozie, O.; Manjunath, M.N.; Shankar, P. CCR5 Gene Editing of Resting CD4+ T Cells by Transient ZFN Expression from HIV Envelope Pseudotyped Nonintegrating Lentivirus Confers HIV-1 Resistance in Humanized Mice. Mol. Ther. Nucleic Acids 2014, 3, e198. [Google Scholar] [CrossRef]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of CCR5 in autologous CD4+ T cells of persons infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef]
- Tanaka, A.; Takeda, S.; Kariya, R.; Matsuda, K.; Urano, E.; Okada, S.; Komano, J. A novel therapeutic molecule against HTLV-1 infection targeting provirus. Leukemia 2013, 27, 1621–1627. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Lai, M.; Maori, E.; Quaranta, P.; Matteoli, G.; Maggi, F.; Sgarbanti, M.; Crucitta, S.; Pacini, S.; Turriziani, O.; Antonelli, G.; et al. CRISPR/Cas9 Ablation of Integrated HIV-1 Accumulates Proviral DNA Circles with Reformed Long Terminal Repeats. J. Virol. 2021, 95, e01358-21. [Google Scholar] [CrossRef] [PubMed]
- Jurczyszak, D.; Manganaro, L.; Buta, S.; Gruber, C.; Martin-Fernandez, M.; Taft, J.; Patel, R.S.; Cipolla, M.; Alshammary, H.; Mulder, L.C.F.; et al. ISG15 deficiency restricts HIV-1 infection. PLoS Pathog. 2022, 18, e1010405. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Kaminski, R.; Bella, R.; Su, H.; Mathews, S.; Ahooyi, T.M.; Chen, C.; Mancuso, P.; Sariyer, R.; Ferrante, P.; et al. Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat. Commun. 2019, 10, 2753. [Google Scholar] [CrossRef] [PubMed]
- Panfil, A.R.; Green, P.L.; Yoder, K.E. CRISPR Genome Editing Applied to the Pathogenic Retrovirus HTLV-1. Front. Cell. Infect. Microbiol. 2020, 10, 580371. [Google Scholar] [CrossRef]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Kelleher, A.D.; Cortez-Jugo, C.; Cavalieri, F.; Qu, Y.; Glanville, A.R.; Caruso, F.; Symonds, G.; Ahlenstiel, C.L. RNAi therapeutics: An antiviral strategy for human infections. Curr. Opin. Pharmacol. 2020, 54, 121–129. [Google Scholar] [CrossRef]
- Morris, K.V.; Chan, S.W.; Jacobsen, S.E.; Looney, D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 2004, 305, 1289–1292. [Google Scholar] [CrossRef]
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006, 441, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Kayikcioglu, O.R.; Cheng, L.; Kozak, I.; Bergeron-Lynn, G.; Schulteis, C.T.; Rhoades, K.L.; Freeman, W.R. Toxicity of subretinal ribozyme to the proliferating cell nuclear antigen and 5-fluorouracil in rat eyes. Curr. Eye Res. 2006, 31, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Scacheri, P.C.; Rozenblatt-Rosen, O.; Caplen, N.J.; Wolfsberg, T.G.; Umayam, L.; Lee, J.C.; Hughes, C.M.; Shanmugam, K.S.; Bhattacharjee, A.; Meyerson, M.; et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1892–1897. [Google Scholar] [CrossRef] [PubMed]
- Ait-Ammar, A.; Kula, A.; Darcis, G.; Verdikt, R.; De Wit, S.; Gautier, V.; Mallon, P.W.G.; Marcello, A.; Rohr, O.; Van Lint, C. Current Status of Latency Reversing Agents Facing the Heterogeneity of HIV-1 Cellular and Tissue Reservoirs. Front. Microbiol. 2019, 10, 3060. [Google Scholar] [CrossRef] [PubMed]
- Spina, C.A.; Anderson, J.; Archin, N.M.; Bosque, A.; Chan, J.; Famiglietti, M.; Greene, W.C.; Kashuba, A.; Lewin, S.R.; Margolis, D.M.; et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013, 9, e1003834. [Google Scholar] [CrossRef]
- Suzuki, K.; Shijuuku, T.; Fukamachi, T.; Zaunders, J.; Guillemin, G.; Cooper, D.; Kelleher, A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J. RNAi Gene Silenc. 2005, 1, 66–78. [Google Scholar]
- Suzuki, K.; Juelich, T.; Lim, H.; Ishida, T.; Watanebe, T.; Cooper, D.A.; Rao, S.; Kelleher, A.D. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J. Biol. Chem. 2008, 283, 23353–23363. [Google Scholar] [CrossRef]
- Suzuki, K.; Ahlenstiel, C.; Marks, K.; Kelleher, A.D. Promoter Targeting RNAs: Unexpected Contributors to the Control of HIV-1 Transcription. Mol. Ther. Nucleic Acids 2015, 4, e222. [Google Scholar] [CrossRef]
- Yamagishi, M.; Ishida, T.; Miyake, A.; Cooper, D.A.; Kelleher, A.D.; Suzuki, K.; Watanabe, T. Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infect. 2009, 11, 500–508. [Google Scholar] [CrossRef]
- Suzuki, K.; Ishida, T.; Yamagishi, M.; Ahlenstiel, C.; Swaminathan, S.; Marks, K.; Murray, D.; McCartney, E.M.; Beard, M.R.; Alexander, M.; et al. Transcriptional gene silencing of HIV-1 through promoter targeted RNA is highly specific. RNA Biol. 2011, 8, 1035–1046. [Google Scholar] [CrossRef]
- Tsukamoto, T. Transcriptional gene silencing limits CXCR4-associated depletion of bone marrow CD34+ cells in HIV-1 infection. AIDS 2018, 32, 1737–1747, Erratum in AIDS 2018, 32, 2857–2858. [Google Scholar] [CrossRef] [PubMed]
- Bowden-Reid, E.; Ledger, S.; Zhang, Y.; Di Giallonardo, F.; Aggarwal, A.; Stella, A.O.; Akerman, A.; Milogiannakis, V.; Walker, G.; Rawlinson, W.; et al. Novel siRNA therapeutics demonstrate multi-variant efficacy against SARS-CoV-2. Antivir. Res. 2023, 217, 105677. [Google Scholar] [CrossRef] [PubMed]
- Ahlenstiel, C.; Mendez, C.; Lim, S.T.; Marks, K.; Turville, S.; Cooper, D.A.; Kelleher, A.D.; Suzuki, K. Novel RNA Duplex Locks HIV-1 in a Latent State via Chromatin-mediated Transcriptional Silencing. Mol. Ther. Nucleic Acids 2015, 4, e261. [Google Scholar] [CrossRef]
- Collotta, D.; Bertocchi, I.; Chiapello, E.; Collino, M. Antisense oligonucleotides: A novel Frontier in pharmacological strategy. Front. Pharmacol. 2023, 14, 1304342. [Google Scholar] [CrossRef]
- Stein, C.A.; Castanotto, D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef]
- Raal, F.J.; Santos, R.D.; Blom, D.J.; Marais, A.D.; Charng, M.J.; Cromwell, W.C.; Lachmann, R.H.; Gaudet, D.; Tan, J.L.; Chasan-Taber, S.; et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet 2010, 375, 998–1006. [Google Scholar] [CrossRef]
- Gagliardi, M.; Ashizawa, A.T. The Challenges and Strategies of Antisense Oligonucleotide Drug Delivery. Biomedicines 2021, 9, 433. [Google Scholar] [CrossRef]
- Zhang, K.; Zheludev, I.N.; Hagey, R.J.; Haslecker, R.; Hou, Y.J.; Kretsch, R.; Pintilie, G.D.; Rangan, R.; Kladwang, W.; Li, S.; et al. Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. Nat. Struct. Mol. Biol. 2021, 28, 747–754. [Google Scholar] [CrossRef]
- Hagihara, M.; Yamauchi, L.; Seo, A.; Yoneda, K.; Senda, M.; Nakatani, K. Antisense-induced guanine quadruplexes inhibit reverse transcription by HIV-1 reverse transcriptase. J. Am. Chem. Soc. 2010, 132, 11171–11178. [Google Scholar] [CrossRef]
- Takahashi, M.; Li, H.; Zhou, J.; Chomchan, P.; Aishwarya, V.; Damha, M.J.; Rossi, J.J. Dual Mechanisms of Action of Self-Delivering, Anti-HIV-1 FANA Oligonucleotides as a Potential New Approach to HIV Therapy. Mol. Ther. Nucleic Acids 2019, 17, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hong, J.; Zheng, S.; Ding, Y.; Guo, S.; Zhang, H.; Zhang, X.; Du, Q.; Liang, Z. Elimination pathways of systemically delivered siRNA. Mol. Ther. 2011, 19, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef]
- Wojnilowicz, M.; Besford, Q.A.; Wu, Y.L.; Loh, X.J.; Braunger, J.A.; Glab, A.; Cortez-Jugo, C.; Caruso, F.; Cavalieri, F. Glycogen-nucleic acid constructs for gene silencing in multicellular tumor spheroids. Biomaterials 2018, 176, 34–49. [Google Scholar] [CrossRef]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef]
- Naeye, B.; Deschout, H.; Caveliers, V.; Descamps, B.; Braeckmans, K.; Vanhove, C.; Demeester, J.; Lahoutte, T.; De Smedt, S.C.; Raemdonck, K. In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. Biomaterials 2013, 34, 2350–2358. [Google Scholar] [CrossRef]
- Zuckerman, J.E.; Choi, C.H.; Han, H.; Davis, M.E. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc. Natl. Acad. Sci. USA 2012, 109, 3137–3142. [Google Scholar] [CrossRef]
- Scott, L.J. Givosiran: First Approval. Drugs 2020, 80, 335–339. [Google Scholar] [CrossRef]
- Zuckerman, J.E.; Gritli, I.; Tolcher, A.; Heidel, J.D.; Lim, D.; Morgan, R.; Chmielowski, B.; Ribas, A.; Davis, M.E.; Yen, Y. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc. Natl. Acad. Sci. USA 2014, 111, 11449–11454. [Google Scholar] [CrossRef]
- Garrelfs, S.F.; Frishberg, Y.; Hulton, S.A.; Koren, M.J.; O’Riordan, W.D.; Cochat, P.; Deschenes, G.; Shasha-Lavsky, H.; Saland, J.M.; Van’t Hoff, W.G.; et al. Lumasiran, an RNAi Therapeutic for Primary Hyperoxaluria Type 1. N. Engl. J. Med. 2021, 384, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.K.; Troquay, R.P.T.; Visseren, F.L.J.; Leiter, L.A.; Scott Wright, R.; Vikarunnessa, S.; Talloczy, Z.; Zang, X.; Maheux, P.; Lesogor, A.; et al. Long-term efficacy and safety of inclisiran in patients with high cardiovascular risk and elevated LDL cholesterol (ORION-3): Results from the 4-year open-label extension of the ORION-1 trial. Lancet Diabetes Endocrinol. 2023, 11, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Tournev, I.L.; Taylor, M.S.; Coelho, T.; Plante-Bordeneuve, V.; Berk, J.L.; Gonzalez-Duarte, A.; Gillmore, J.D.; Low, S.C.; Sekijima, Y.; et al. Efficacy and safety of vutrisiran for patients with hereditary transthyretin-mediated amyloidosis with polyneuropathy: A randomized clinical trial. Amyloid 2023, 30, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yang, J.; Feng, J.; Liu, Z.; Dong, Y.; Luo, J.; Yu, L.; Wang, J.; Fan, H.; Ma, W.; et al. PreS/2-21-Guided siRNA Nanoparticles Target to Inhibit Hepatitis B Virus Infection and Replication. Front. Immunol. 2022, 13, 856463. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Witwer, K.W. The evolving paradigm of extracellular vesicles in intercellular signaling and delivery of therapeutic RNAs. Mol. Ther. 2022, 30, 2393–2394. [Google Scholar] [CrossRef]
- Colombo, E.; Borgiani, B.; Verderio, C.; Furlan, R. Microvesicles: Novel biomarkers for neurological disorders. Front. Physiol. 2012, 3, 63. [Google Scholar] [CrossRef]
- Aryani, A.; Denecke, B. Exosomes as a Nanodelivery System: A Key to the Future of Neuromedicine? Mol. Neurobiol. 2016, 53, 818–834. [Google Scholar] [CrossRef]
- Shrivastava, S.; Ray, R.M.; Holguin, L.; Echavarria, L.; Grepo, N.; Scott, T.A.; Burnett, J.; Morris, K.V. Exosome-mediated stable epigenetic repression of HIV-1. Nat. Commun. 2021, 12, 5541. [Google Scholar] [CrossRef]
- Scott, T.A.; Soemardy, C.; Ray, R.M.; Morris, K.V. Targeted zinc-finger repressors to the oncogenic HBZ gene inhibit adult T-cell leukemia (ATL) proliferation. NAR Cancer 2023, 5, zcac046. [Google Scholar] [CrossRef]
- Villamizar, O.; Waters, S.A.; Scott, T.; Grepo, N.; Jaffe, A.; Morris, K.V. Mesenchymal Stem Cell exosome delivered Zinc Finger Protein activation of cystic fibrosis transmembrane conductance regulator. J. Extracell. Vesicles 2021, 10, e12053. [Google Scholar] [CrossRef]
- Scott, T.A.; Supramaniam, A.; Idris, A.; Cardoso, A.A.; Shrivastava, S.; Kelly, G.; Grepo, N.A.; Soemardy, C.; Ray, R.M.; McMillan, N.A.J.; et al. Engineered extracellular vesicles directed to the spike protein inhibit SARS-CoV-2. Mol. Ther. Methods Clin. Dev. 2022, 24, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Marawan, M.A.; Alouffi, A.; El Tokhy, S.; Badawy, S.; Shirani, I.; Dawood, A.; Guo, A.; Almutairi, M.M.; Alshammari, F.A.; Selim, A. Bovine Leukaemia Virus: Current Epidemiological Circumstance and Future Prospective. Viruses 2021, 13, 2167. [Google Scholar] [CrossRef] [PubMed]
- Hajj, H.E.; Nasr, R.; Kfoury, Y.; Dassouki, Z.; Nasser, R.; Kchour, G.; Hermine, O.; de The, H.; Bazarbachi, A. Animal models on HTLV-1 and related viruses: What did we learn? Front. Microbiol. 2012, 3, 333. [Google Scholar] [CrossRef] [PubMed]
- Pluta, A.; Jaworski, J.P.; Douville, R.N. Regulation of Expression and Latency in BLV and HTLV. Viruses 2020, 12, 1079. [Google Scholar] [CrossRef] [PubMed]
- Kettmann, R.; Marbaix, G.; Cleuter, Y.; Portetelle, D.; Mammerickx, M.; Burny, A. Genomic integration of bovine leukemia provirus and lack of viral RNA expression in the target cells of cattle with different responses to BLV infection. Leuk. Res. 1980, 4, 509–519. [Google Scholar] [CrossRef]
- Merimi, M.; Klener, P.; Szynal, M.; Cleuter, Y.; Bagnis, C.; Kerkhofs, P.; Burny, A.; Martiat, P.; Van den Broeke, A. Complete suppression of viral gene expression is associated with the onset and progression of lymphoid malignancy: Observations in Bovine Leukemia Virus-infected sheep. Retrovirology 2007, 4, 51. [Google Scholar] [CrossRef]
- Merezak, C.; Pierreux, C.; Adam, E.; Lemaigre, F.; Rousseau, G.G.; Calomme, C.; Van Lint, C.; Christophe, D.; Kerkhofs, P.; Burny, A.; et al. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: Implications for viral latency. J. Virol. 2001, 75, 6977–6988. [Google Scholar] [CrossRef]
- Suarez Archilla, G.; Gutierrez, G.; Camussone, C.; Calvinho, L.; Abdala, A.; Alvarez, I.; Petersen, M.; Franco, L.; Destefano, G.; Monti, G.; et al. A safe and effective vaccine against bovine leukemia virus. Front. Immunol. 2022, 13, 980514. [Google Scholar] [CrossRef]
- Gutierrez, G.; Rodriguez, S.M.; de Brogniez, A.; Gillet, N.; Golime, R.; Burny, A.; Jaworski, J.P.; Alvarez, I.; Vagnoni, L.; Trono, K.; et al. Vaccination against delta-retroviruses: The bovine leukemia virus paradigm. Viruses 2014, 6, 2416–2427. [Google Scholar] [CrossRef]
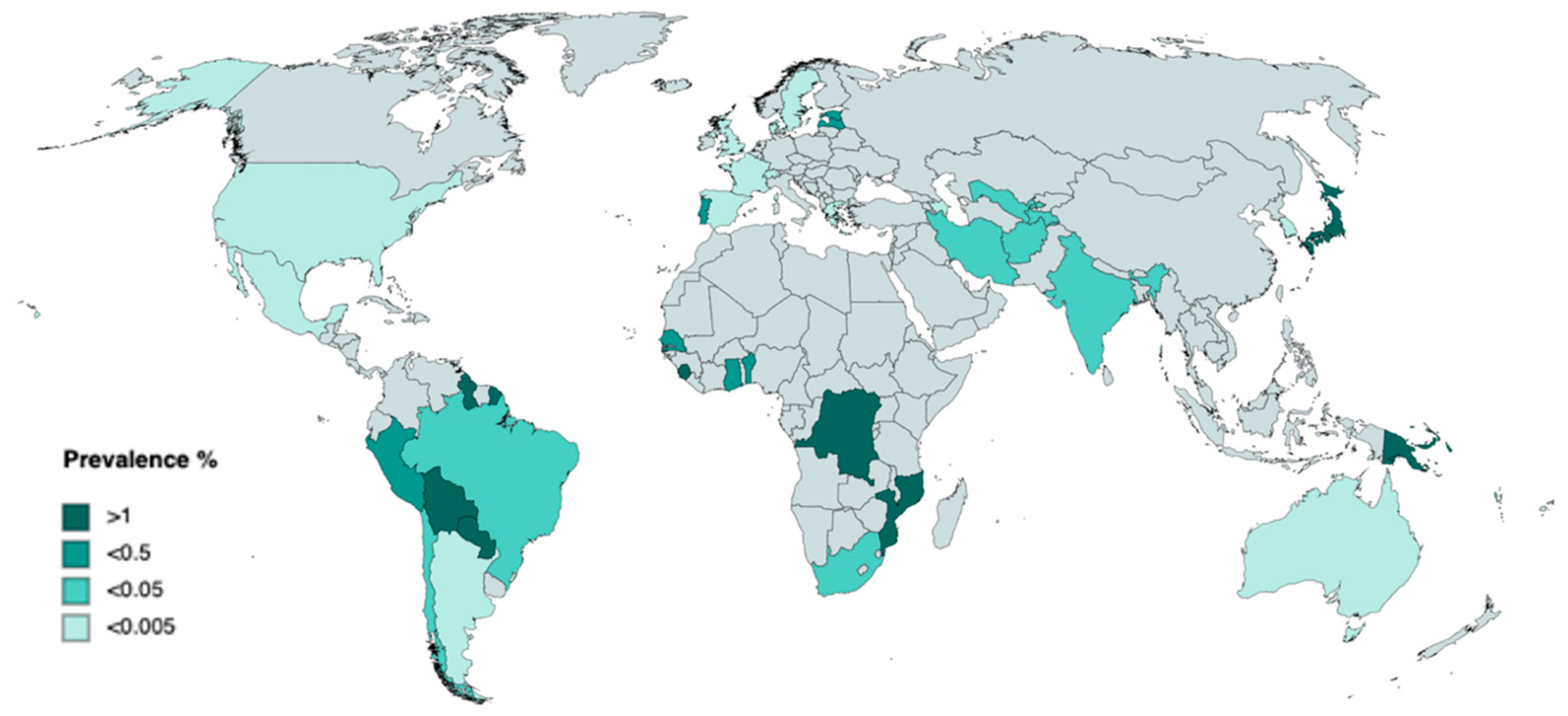

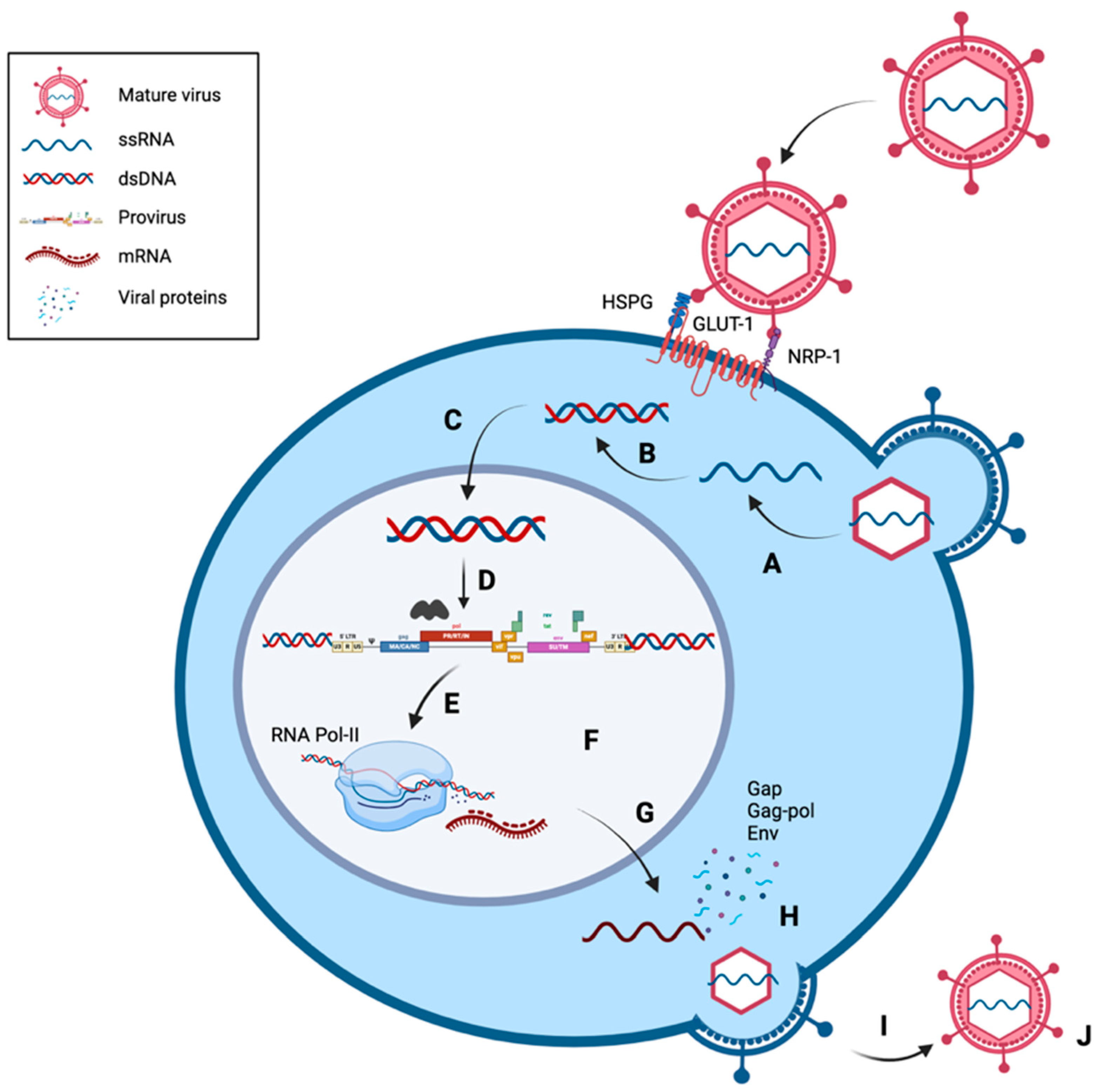
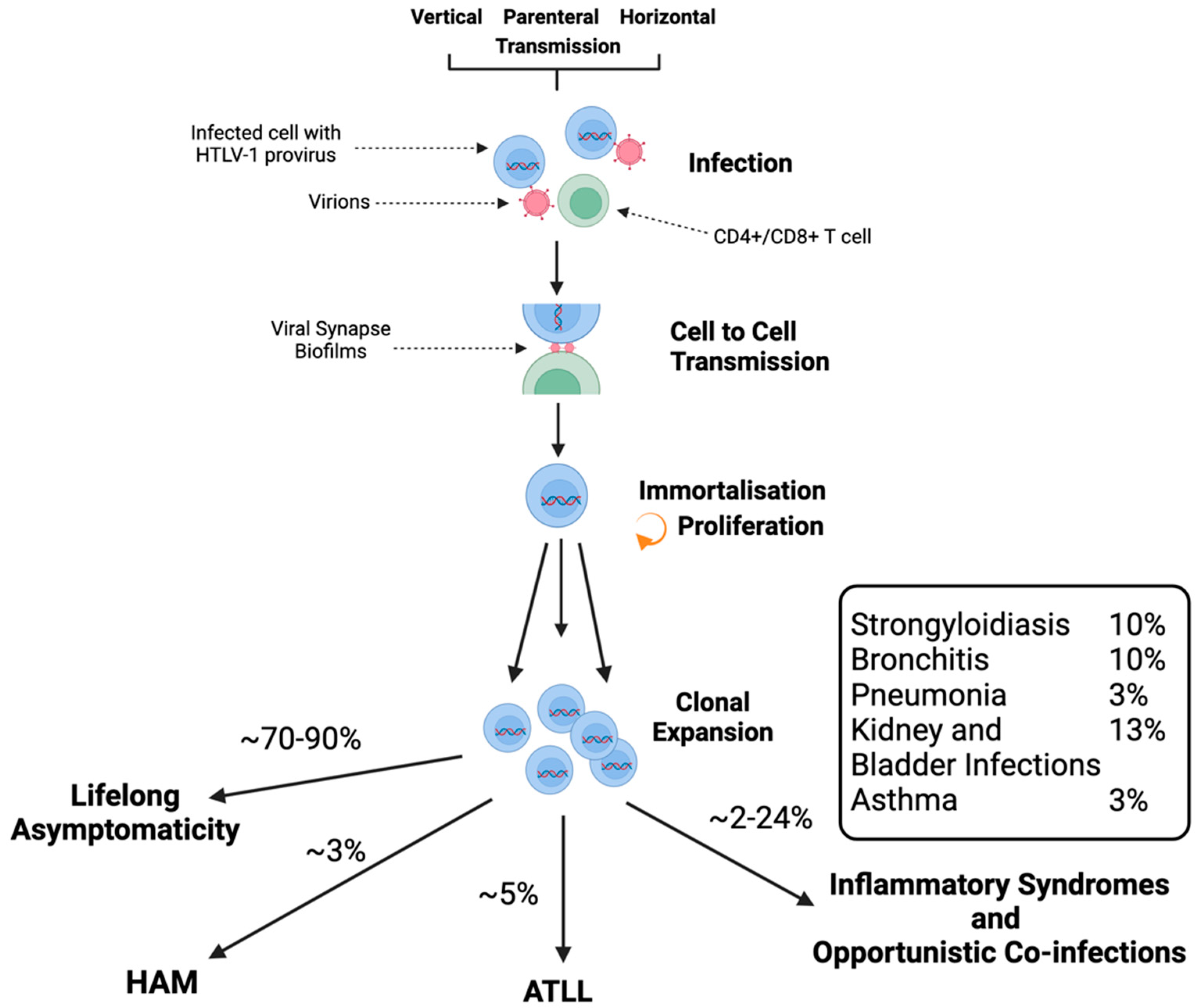

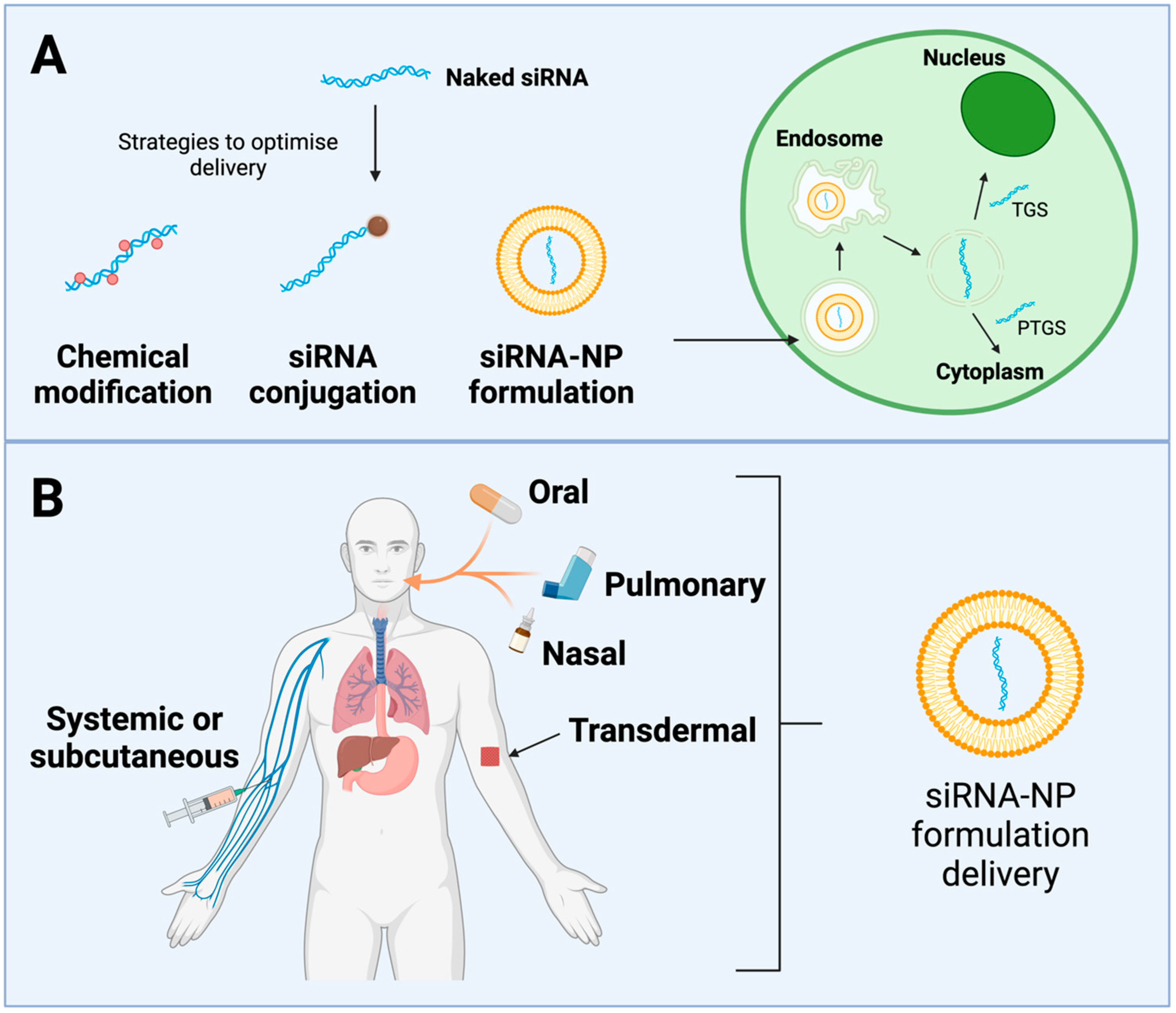
| Property | HTLV-1 | HIV-1 |
|---|---|---|
| Infectious Duration | Lifelong | Lifelong |
| Primary Immune Targets | CD4+/CD8+ T cells | CD4+ T cells |
| Transmission | Cell-to-Cell | Virus particles |
| Viral Expression | Low | High |
| Immune consequences | Overactive inflammation | Immune deficiency |
| Treatment | Few targeting symptoms | ART targeting virus |
| Tumorigenesis | Direct | Indirect |
| Cell Line | Cell Type | IL-2 Dependency | Antigen Expression | |
|---|---|---|---|---|
| HTLV-1-transformed cell lines | MJ [103] | Lymphoblast | Independent | CD2+; CD3+; CD4+ |
| MT-2 [104] | Lymphoblast | Independent | CD4+; CD25+; FoxP3+ | |
| MT-4 [105] | Lymphoblast | Independent | CD4+ | |
| HuT-102 [1] | Cutaneous T Lymphocyte | Dependent | CD4+ | |
| C81-66 [106] | Lymphoblast | Independent | CD4+ | |
| HTLV-1 chronically infected cell lines | C91/PL [103,107] | Umbilical cord-blood T cells | Independent | CD4+ |
| MS-9 [108] | Cord-blood T cells | Dependent | CD4+ | |
| ATL cell lines | MT-1 [109] | Lymphoblast | Independent | CD4+; Tax− |
| MT-3 [110] | Lymphoblast | Independent | CD4+; Tax− | |
| ATL-2 [111] | Lymphoblast | Independent | CD4+; CD3− | |
| TL-Om1 [112] | Lymphoblast | Independent | CD4+; Tax− | |
| F6T [113] | Lymphoblast | Independent | CD4+; CD25+ | |
| K3T [113] | Lymphoblast | Independent | CD4+; CD25+ | |
| S1T [113] | Lymphoblast | Independent | CD4+; CD25+ | |
| ATL-T [114] | Lymphoblast | Independent | CD4+ | |
| ATL-35T [115] | Lymphoblast | Independent | CD4+ | |
| ATL-55T [116] | Lymphoblast | Dependent | CD4+ | |
| Su9T01 [113] | Lymphoblast | Independent | CD4+ |
| Disease | Treatment |
|---|---|
| HTLV-1-associated uveitis | Topical or systemic corticosteroids and mydriatics [263] |
| HTLV-1-associated Sjögren’s Syndrome | Artificial lubricants such as saliva/tears replacement solutions coupled with systemic pharmacotherapy with pilocarpine hydrochloride [264] |
| HTLV-1-associated Hashimoto’s thyroiditis and Graves’ disease | Partial or complete thyroidectomy and hormone replacement therapy [264] |
| HTLV-1-associated infective dermatitis | Antibiotics and corticosteroids [264] |
| HTLV-1-associated pulmonary disease | Long-term corticosteroid therapy [265] |
| HTLV-1-associated inflammatory myositis | Currently no available therapeutic; myositis is resistant to corticosteroids and immunomodulatory therapies such as cyclosporin [266,267] |
| HTLV-1-associated arthritis | Immunomodulatory therapies such as TNF-α [268] |
| HTLV-1-associated conjunctivitis sicca syndrome and interstitial keratitis | Artificial lubricants such as tears, cyclosporine eye drops [264] |
| Gene | Allele/Genotype/Haplotype | Disease | Indication |
|---|---|---|---|
| HLA | DRB1*DQB1* 0101, 1502, 0803 [280] | HAM | Increased risk |
| DRB1*DQB1* 1501, 0901, 1401 [280] | ATL | Increased risk | |
| A*24 and Cw*01 [280] | ATL | Protective | |
| Cw*7, B*7 and DR1 [280] | HAM | Increased risk | |
| A*02 [281] | HAM ATL | Protective Increased risk | |
| HLA-B*5401 [276] | HAM | Increased risk | |
| A*03 and DQB1*0501 [282] | ATL | Protective | |
| B*15 and B*53 [282] | ATL | Increased risk | |
| IFN-γ | IFNG+874A/T [283] | - | High PVL |
| IL-18 | −607CC [284] | - | Protective |
| −607AC [284] | - | Increased risk | |
| TNF-α | TNFA−857C/T [285] | ATL | Increased risk |
| IL-10 | IL10−592A/C [286] | HAM | Increased risk High PVL |
| IL10−819*C/T and −592*C/A [287] | - | Increased risk |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.T.; Hirons, A.; Doerflinger, M.; Morris, K.V.; Ledger, S.; Purcell, D.F.J.; Kelleher, A.D.; Ahlenstiel, C.L. Current State of Therapeutics for HTLV-1. Viruses 2024, 16, 1616. https://doi.org/10.3390/v16101616
Wang TT, Hirons A, Doerflinger M, Morris KV, Ledger S, Purcell DFJ, Kelleher AD, Ahlenstiel CL. Current State of Therapeutics for HTLV-1. Viruses. 2024; 16(10):1616. https://doi.org/10.3390/v16101616
Chicago/Turabian StyleWang, Tiana T., Ashley Hirons, Marcel Doerflinger, Kevin V. Morris, Scott Ledger, Damian F. J. Purcell, Anthony D. Kelleher, and Chantelle L. Ahlenstiel. 2024. "Current State of Therapeutics for HTLV-1" Viruses 16, no. 10: 1616. https://doi.org/10.3390/v16101616
APA StyleWang, T. T., Hirons, A., Doerflinger, M., Morris, K. V., Ledger, S., Purcell, D. F. J., Kelleher, A. D., & Ahlenstiel, C. L. (2024). Current State of Therapeutics for HTLV-1. Viruses, 16(10), 1616. https://doi.org/10.3390/v16101616









