The Effects of Viral Structural Proteins on Acidic Phospholipids in Host Membranes
Abstract
:1. Introduction
2. Classification, Structure, and Cellular Distribution of Phospholipids
3. The Functions and Lateral Distributions of Plasma Membrane Acidic Phospholipids
4. Influenza A Virus (IAV) Assembly and Acidic Phospholipids
4.1. Influenza A Virus Assembly
4.2. The Interplay Between Acidic Phospholipids and Influenza A Virus Structural Proteins
4.2.1. Hemagglutinin (HA) and the Acidic Phospholipids
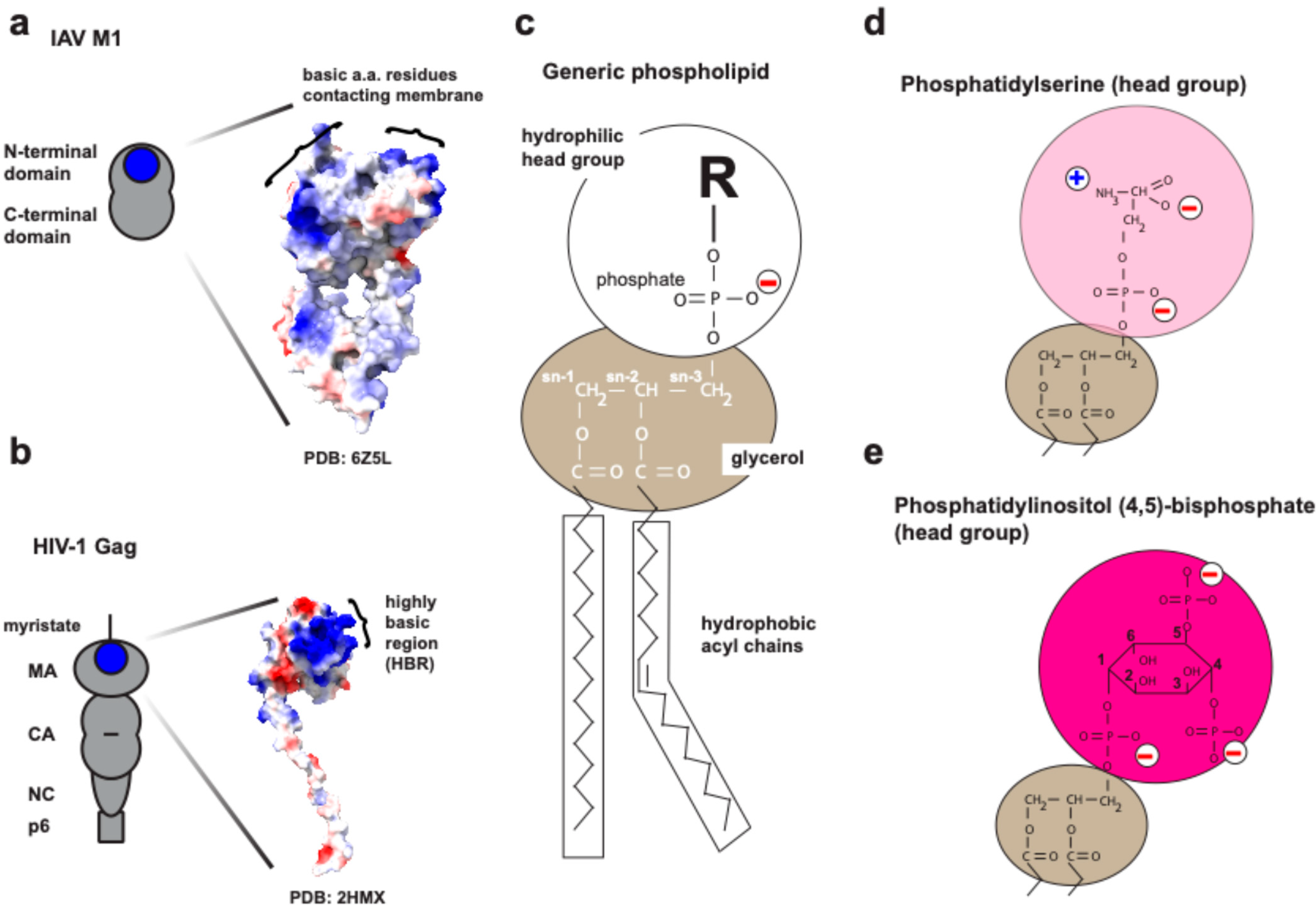
4.2.2. Matrix Protein-1 (M1) and the Acidic Phospholipids
5. Human Immunodeficiency Virus Type 1 (HIV-1) Assembly and Acidic Phospholipids
5.1. HIV-1 Assembly
5.2. The Interplay Between Cellular Phospholipids and HIV-1 Structural Protein Gag
5.2.1. Gag and PI(4,5)P2
5.2.2. HIV-1 and PS
6. Future Directions
- (1)
- Are viral structural proteins recruited to pre-existing acidic phospholipid-rich areas, or do they cause acidic phospholipids clustering or both?
- (2)
- Do acidic phospholipids play a role in the incorporation of viral transmembrane proteins and the packaging of viral genomes into nascent virus particles?
- (3)
- Do acidic phospholipids regulate the recruitment of host cellular proteins to the assembly sites, and if so, what roles do these host proteins play in the assembly process or virion infectivity?
- (4)
- What is the role, if any, of the incorporated acidic phospholipids for IAV and HIV in viral spread?
- (5)
- How do viral proteins regulate acidic phospholipid distribution locally and globally?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Jones, M.K.; Young, P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 1996, 220, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Barnard, T.R.; Abram, Q.H.; Lin, Q.F.; Wang, A.B.; Sagan, S.M. Molecular Determinants of Flavivirus Virion Assembly. Trends Biochem. Sci. 2021, 46, 378–390. [Google Scholar] [CrossRef]
- Barbosa, N.S.; Mendonca, L.R.; Dias, M.V.S.; Pontelli, M.C.; da Silva, E.Z.M.; Criado, M.F.; da Silva-Januario, M.E.; Schindler, M.; Jamur, M.C.; Oliver, C.; et al. ESCRT machinery components are required for Orthobunyavirus particle production in Golgi compartments. PLoS Pathog. 2018, 14, e1007047. [Google Scholar] [CrossRef] [PubMed]
- Barker, J.; daSilva, L.L.P.; Crump, C.M. Mechanisms of bunyavirus morphogenesis and egress. J. Gen. Virol. 2023, 104, 001845. [Google Scholar] [CrossRef] [PubMed]
- Draganova, E.B.; Valentin, J.; Heldwein, E.E. The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies. Viruses 2021, 13, 1913. [Google Scholar] [CrossRef]
- Selzer, L.; Zlotnick, A. Assembly and Release of Hepatitis B Virus. Cold Spring Harb. Perspect. Med. 2015, 5, a021394. [Google Scholar] [CrossRef]
- Roingeard, P.; Eymieux, S.; Burlaud-Gaillard, J.; Hourioux, C.; Patient, R.; Blanchard, E. The double-membrane vesicle (DMV): A virus-induced organelle dedicated to the replication of SARS-CoV-2 and other positive-sense single-stranded RNA viruses. Cell. Mol. Life Sci. 2022, 79, 425. [Google Scholar] [CrossRef]
- Roberts, S.R.; Compans, R.W.; Wertz, G.W. Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J. Virol. 1995, 69, 2667–2673. [Google Scholar] [CrossRef]
- Cardoso, R.S.; Tavares, L.A.; Jesus, B.L.S.; Criado, M.F.; de Carvalho, A.N.; Souza, J.P.; Bedi, S.; de Souza, M.M.; Silva, M.L.; Lanfredi, G.P.; et al. Host Retromer Protein Sorting Nexin 2 Interacts with Human Respiratory Syncytial Virus Structural Proteins and is Required for Efficient Viral Production. mBio 2020, 11, e01869-20. [Google Scholar] [CrossRef]
- Sugrue, R.J.; Tan, B.H. Defining the Assembleome of the Respiratory Syncytial Virus. In Virus Infected Cells; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2023; Volume 106, pp. 227–249. [Google Scholar]
- Rossman, J.S.; Lamb, R.A. Influenza virus assembly and budding. Virology 2011, 411, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Bedi, S.; Noda, T.; Kawaoka, Y.; Ono, A. A Defect in Influenza A Virus Particle Assembly Specific to Primary Human Macrophages. mBio 2018, 9, e01916-18. [Google Scholar] [CrossRef]
- Nayak, D.P.; Balogun, R.A.; Yamada, H.; Zhou, Z.H.; Barman, S. Influenza virus morphogenesis and budding. Virus Res. 2009, 143, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Geisbert, T.W.; Jahrling, P.B. Differentiation of filoviruses by electron microscopy. Virus Res. 1995, 39, 129–150. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.; Ebihara, H.; Muramoto, Y.; Fujii, K.; Takada, A.; Sagara, H.; Kim, J.H.; Kida, H.; Feldmann, H.; Kawaoka, Y. Assembly and budding of Ebolavirus. PLoS Pathog. 2006, 2, e99. [Google Scholar] [CrossRef]
- Dolnik, O.; Becker, S. Assembly and transport of filovirus nucleocapsids. PLoS Pathog. 2022, 18, e1010616. [Google Scholar] [CrossRef]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef]
- Jouvenet, N.; Neil, S.J.; Bess, C.; Johnson, M.C.; Virgen, C.A.; Simon, S.M.; Bieniasz, P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006, 4, e435. [Google Scholar] [CrossRef]
- Finzi, A.; Orthwein, A.; Mercier, J.; Cohen, E.A. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 2007, 81, 7476–7490. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- IUPAC-IUB Commission on Biochemical Nomenclature. Nomenclature of phosphorus-containing compounds of biochemical importance (Recommendations 1976). Proc. Natl. Acad. Sci. USA 1977, 74, 2222–2230. [CrossRef] [PubMed]
- Vance, J.E. Historical perspective: Phosphatidylserine and phosphatidylethanolamine from the 1800s to the present. J. Lipid Res. 2018, 59, 923–944. [Google Scholar] [CrossRef] [PubMed]
- Posor, Y.; Jang, W.; Haucke, V. Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell Biol. 2022, 23, 797–816. [Google Scholar] [CrossRef] [PubMed]
- Kay, J.G.; Fairn, G.D. Distribution, dynamics and functional roles of phosphatidylserine within the cell. Cell Commun. Signal. 2019, 17, 126. [Google Scholar] [CrossRef]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Lolicato, F.; Nickel, W.; Haucke, V.; Ebner, M. Phosphoinositide switches in cell physiology—From molecular mechanisms to disease. J. Biol. Chem. 2024, 300, 105757. [Google Scholar] [CrossRef]
- Nicolson, G.L. The Fluid-Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta 2014, 1838, 1451–1466. [Google Scholar] [CrossRef]
- Sakuragi, T.; Nagata, S. Regulation of phospholipid distribution in the lipid bilayer by flippases and scramblases. Nat. Rev. Mol. Cell Biol. 2023, 24, 576–596. [Google Scholar] [CrossRef]
- Kervin, T.A.; Overduin, M. Membranes are functionalized by a proteolipid code. BMC Biol. 2024, 22, 46. [Google Scholar] [CrossRef]
- Thakur, R.; Naik, A.; Panda, A.; Raghu, P. Regulation of Membrane Turnover by Phosphatidic Acid: Cellular Functions and Disease Implications. Front. Cell Dev. Biol. 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S.; Murray, D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 2005, 438, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.J.H.; Loewen, C.J.R. Putting the pH into phosphatidic acid signaling. BMC Biol. 2011, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Hirama, T.; Lu, S.M.; Kay, J.G.; Maekawa, M.; Kozlov, M.M.; Grinstein, S.; Fairn, G.D. Membrane curvature induced by proximity of anionic phospholipids can initiate endocytosis. Nat. Commun. 2017, 8, 1393. [Google Scholar] [CrossRef]
- Bills, B.L.; Knowles, M.K. Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes. Biomolecules 2022, 12, 1707. [Google Scholar] [CrossRef]
- Gericke, A. Is Calcium Fine-Tuning Phosphoinositide-Mediated Signaling Events Through Clustering? Biophys. J. 2018, 114, 2483–2484. [Google Scholar] [CrossRef]
- Schink, K.O.; Tan, K.W.; Stenmark, H. Phosphoinositides in Control of Membrane Dynamics. Annu. Rev. Cell Dev. Biol. 2016, 32, 143–171. [Google Scholar] [CrossRef]
- Beziau, A.; Brand, D.; Piver, E. The Role of Phosphatidylinositol Phosphate Kinases during Viral Infection. Viruses 2020, 12, 1124. [Google Scholar] [CrossRef]
- Adu-Gyamfi, E.; Johnson, K.A.; Fraser, M.E.; Scott, J.L.; Soni, S.P.; Jones, K.R.; Digman, M.A.; Gratton, E.; Tessier, C.R.; Stahelin, R.V. Host Cell Plasma Membrane Phosphatidylserine Regulates the Assembly and Budding of Ebola Virus. J. Virol. 2015, 89, 9440–9453. [Google Scholar] [CrossRef]
- Gc, J.B.; Gerstman, B.S.; Stahelin, R.V.; Chapagain, P.P. The Ebola virus protein VP40 hexamer enhances the clustering of PI(4,5)P(2) lipids in the plasma membrane. Phys. Chem. Chem. Phys. 2016, 18, 28409–28417. [Google Scholar] [CrossRef]
- Johnson, K.A.; Taghon, G.J.; Scott, J.L.; Stahelin, R.V. The Ebola Virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) for extensive oligomerization at the plasma membrane and viral egress. Sci. Rep. 2016, 6, 19125. [Google Scholar] [CrossRef] [PubMed]
- Acciani, M.D.; Lay Mendoza, M.F.; Havranek, K.E.; Duncan, A.M.; Iyer, H.; Linn, O.L.; Brindley, M.A. Ebola Virus Requires Phosphatidylserine Scrambling Activity for Efficient Budding and Optimal Infectivity. J. Virol. 2021, 95, e0116521. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.D.; Husby, M.L.; Gerstman, B.S.; Stahelin, R.V.; Chapagain, P.P. Role of phosphatidic acid lipids on plasma membrane association of the Ebola virus matrix protein VP40. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2024, 1869, 159464. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Budicini, M.R.; Bhattarai, N.; Sharma, T.; Urata, S.; Gerstman, B.S.; Chapagain, P.P.; Li, S.; Stahelin, R.V. PI(4,5)P(2) binding sites in the Ebola virus matrix protein VP40 modulate assembly and budding. J. Lipid Res. 2024, 65, 100512. [Google Scholar] [CrossRef] [PubMed]
- Norris, M.J.; Husby, M.L.; Kiosses, W.B.; Yin, J.; Saxena, R.; Rennick, L.J.; Heiner, A.; Harkins, S.S.; Pokhrel, R.; Schendel, S.L.; et al. Measles and Nipah virus assembly: Specific lipid binding drives matrix polymerization. Sci. Adv. 2022, 8, eabn1440. [Google Scholar] [CrossRef]
- Eisenberg, S.; Haimov, E.; Walpole, G.F.W.; Plumb, J.; Kozlov, M.M.; Grinstein, S. Mapping the electrostatic profiles of cellular membranes. Mol. Biol. Cell 2021, 32, 301–310. [Google Scholar] [CrossRef]
- Yeung, T.; Gilbert, G.E.; Shi, J.; Silvius, J.; Kapus, A.; Grinstein, S. Membrane phosphatidylserine regulates surface charge and protein localization. Science 2008, 319, 210–213. [Google Scholar] [CrossRef]
- McLaughlin, S. The electrostatic properties of membranes. Annu. Rev. Biophys. Biophys. Chem. 1989, 18, 113–136. [Google Scholar] [CrossRef]
- Raghupathy, R.; Anilkumar, A.A.; Polley, A.; Singh, P.P.; Yadav, M.; Johnson, C.; Suryawanshi, S.; Saikam, V.; Sawant, S.D.; Panda, A.; et al. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 2015, 161, 581–594. [Google Scholar] [CrossRef]
- Leventis, P.A.; Grinstein, S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 2010, 39, 407–427. [Google Scholar] [CrossRef]
- Varga, K.; Jiang, Z.J.; Gong, L.W. Phosphatidylserine is critical for vesicle fission during clathrin-mediated endocytosis. J. Neurochem. 2020, 152, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Hallett, M.B. Localisation of Intracellular Signals and Responses during Phagocytosis. Int. J. Mol. Sci. 2023, 24, 2825. [Google Scholar] [CrossRef]
- Bohdanowicz, M.; Grinstein, S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol. Rev. 2013, 93, 69–106. [Google Scholar] [CrossRef]
- Uchida, Y.; Hasegawa, J.; Chinnapen, D.; Inoue, T.; Okazaki, S.; Kato, R.; Wakatsuki, S.; Misaki, R.; Koike, M.; Uchiyama, Y.; et al. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. USA 2011, 108, 15846–15851. [Google Scholar] [CrossRef]
- Fairn, G.D.; Hermansson, M.; Somerharju, P.; Grinstein, S. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat. Cell Biol. 2011, 13, 1424–1430. [Google Scholar] [CrossRef]
- Ammar, M.R.; Kassas, N.; Chasserot-Golaz, S.; Bader, M.F.; Vitale, N. Lipids in Regulated Exocytosis: What are They Doing? Front. Endocrinol. 2013, 4, 125. [Google Scholar] [CrossRef]
- McLaughlin, S.; Wang, J.; Gambhir, A.; Murray, D. PIP(2) and proteins: Interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 151–175. [Google Scholar] [CrossRef]
- Katan, M.; Cockcroft, S. Phosphatidylinositol(4,5)bisphosphate: Diverse functions at the plasma membrane. Essays Biochem. 2020, 64, 513–531. [Google Scholar]
- Jost, M.; Simpson, F.; Kavran, J.M.; Lemmon, M.A.; Schmid, S.L. Phosphatidylinositol-4,5-bisphosphate is required for endocytic coated vesicle formation. Curr. Biol. 1998, 8, 1399–1402. [Google Scholar] [CrossRef]
- Crul, T.; Maleth, J. Endoplasmic Reticulum-Plasma Membrane Contact Sites as an Organizing Principle for Compartmentalized Calcium and cAMP Signaling. Int. J. Mol. Sci. 2021, 22, 4703. [Google Scholar] [CrossRef]
- Lahiri, S.; Toulmay, A.; Prinz, W.A. Membrane contact sites, gateways for lipid homeostasis. Curr. Opin. Cell Biol. 2015, 33, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Thallmair, V.; Schultz, L.; Evers, S.; Jolie, T.; Goecke, C.; Leitner, M.G.; Thallmair, S.; Oliver, D. Localization of the tubby domain, a PI(4,5)P2 biosensor, to E-Syt3-rich endoplasmic reticulum-plasma membrane junctions. J. Cell Sci. 2023, 136, jcs260848. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S.; Raghu, P. Phospholipid transport protein function at organelle contact sites. Curr. Opin. Cell Biol. 2018, 53, 52–60. [Google Scholar] [CrossRef]
- Seki, K.; Sheu, F.S.; Huang, K.P. Binding of myristoylated alanine-rich protein kinase C substrate to phosphoinositides attenuates the phosphorylation by protein kinase C. Arch. Biochem. Biophys. 1996, 326, 193–201. [Google Scholar] [CrossRef]
- Denisov, G.; Wanaski, S.; Luan, P.; Glaser, M.; McLaughlin, S. Binding of basic peptides to membranes produces lateral domains enriched in the acidic lipids phosphatidylserine and phosphatidylinositol 4,5-bisphosphate: An electrostatic model and experimental results. Biophys. J. 1998, 74 Pt 1, 731–744. [Google Scholar] [CrossRef]
- Laux, T.; Fukami, K.; Thelen, M.; Golub, T.; Frey, D.; Caroni, P. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol. 2000, 149, 1455–1472. [Google Scholar] [CrossRef]
- Rauch, M.E.; Ferguson, C.G.; Prestwich, G.D.; Cafiso, D.S. Myristoylated alanine-rich C kinase substrate (MARCKS) sequesters spin-labeled phosphatidylinositol 4,5-bisphosphate in lipid bilayers. J. Biol. Chem. 2002, 277, 14068–14076. [Google Scholar] [CrossRef]
- Pemberton, J.G.; Balla, T. Polyphosphoinositide-Binding Domains: Insights from Peripheral Membrane and Lipid-Transfer Proteins. Adv. Exp. Med. Biol. 2019, 1111, 77–137. [Google Scholar]
- Wills, R.C.; Hammond, G.R.V. PI(4,5)P2: Signaling the plasma membrane. Biochem. J. 2022, 479, 2311–2325. [Google Scholar] [CrossRef]
- Harlan, J.E.; Hajduk, P.J.; Yoon, H.S.; Fesik, S.W. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 1994, 371, 168–170. [Google Scholar] [CrossRef]
- Lomasney, J.W.; Cheng, H.F.; Wang, L.P.; Kuan, Y.; Liu, S.; Fesik, S.W.; King, K. Phosphatidylinositol 4,5-bisphosphate binding to the pleckstrin homology domain of phospholipase C-delta1 enhances enzyme activity. J. Biol. Chem. 1996, 271, 25316–25326. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 2007, 74, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Powis, G.; Meuillet, E.J.; Indarte, M.; Booher, G.; Kirkpatrick, L. Pleckstrin Homology [PH] domain, structure, mechanism, and contribution to human disease. Biomed. Pharmacother. 2023, 165, 115024. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Reyes-Ordonez, A.; Compagnone, M.A.; Moreno, J.F.; Leslie, B.J.; Ha, T.; Chen, J. Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 2021, 12, 4339. [Google Scholar] [CrossRef]
- Mattila, P.K.; Pykalainen, A.; Saarikangas, J.; Paavilainen, V.O.; Vihinen, H.; Jokitalo, E.; Lappalainen, P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 2007, 176, 953–964. [Google Scholar] [CrossRef]
- Ahmed, S.; Bu, W.; Lee, R.T.; Maurer-Stroh, S.; Goh, W.I. F-BAR domain proteins: Families and function. Commun. Integr. Biol. 2010, 3, 116–121. [Google Scholar] [CrossRef]
- Itoh, T.; Koshiba, S.; Kigawa, T.; Kikuchi, A.; Yokoyama, S.; Takenawa, T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 2001, 291, 1047–1051. [Google Scholar] [CrossRef]
- Ford, M.G.; Pearse, B.M.; Higgins, M.K.; Vallis, Y.; Owen, D.J.; Gibson, A.; Hopkins, C.R.; Evans, P.R.; McMahon, H.T. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 2001, 291, 1051–1055. [Google Scholar] [CrossRef]
- Roux, A.; Koster, G.; Lenz, M.; Sorre, B.; Manneville, J.B.; Nassoy, P.; Bassereau, P. Membrane curvature controls dynamin polymerization. Proc. Natl. Acad. Sci. USA 2010, 107, 4141–4146. [Google Scholar] [CrossRef]
- Kadlecova, Z.; Spielman, S.J.; Loerke, D.; Mohanakrishnan, A.; Reed, D.K.; Schmid, S.L. Regulation of clathrin-mediated endocytosis by hierarchical allosteric activation of AP2. J. Cell Biol. 2017, 216, 167–179. [Google Scholar] [CrossRef]
- Krauss, M.; Kukhtina, V.; Pechstein, A.; Haucke, V. Stimulation of phosphatidylinositol kinase type I-mediated phosphatidylinositol (4,5)-bisphosphate synthesis by AP-2mu-cargo complexes. Proc. Natl. Acad. Sci. USA 2006, 103, 11934–11939. [Google Scholar] [CrossRef] [PubMed]
- Mu, L.; Tu, Z.; Miao, L.; Ruan, H.; Kang, N.; Hei, Y.; Chen, J.; Wei, W.; Gong, F.; Wang, B.; et al. A phosphatidylinositol 4,5-bisphosphate redistribution-based sensing mechanism initiates a phagocytosis programing. Nat. Commun. 2018, 9, 4259. [Google Scholar] [CrossRef] [PubMed]
- Flannagan, R.S.; Jaumouille, V.; Grinstein, S. The cell biology of phagocytosis. Annu. Rev. Pathol. 2012, 7, 61–98. [Google Scholar] [CrossRef] [PubMed]
- Golebiewska, U.; Kay, J.G.; Masters, T.; Grinstein, S.; Im, W.; Pastor, R.W.; Scarlata, S.; McLaughlin, S. Evidence for a fence that impedes the diffusion of phosphatidylinositol 4,5-bisphosphate out of the forming phagosomes of macrophages. Mol. Biol. Cell 2011, 22, 3498–3507. [Google Scholar] [CrossRef]
- Swanson, J.A. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 2008, 9, 639–649. [Google Scholar] [CrossRef]
- Yang, X.; Tu, W.; Gao, X.; Zhang, Q.; Guan, J.; Zhang, J. Functional regulation of syntaxin-1: An underlying mechanism mediating exocytosis in neuroendocrine cells. Front. Endocrinol. 2023, 14, 1096365. [Google Scholar] [CrossRef]
- Liang, T.; Xie, L.; Chao, C.; Kang, Y.; Lin, X.; Qin, T.; Xie, H.; Feng, Z.P.; Gaisano, H.Y. Phosphatidylinositol 4,5-biphosphate (PI(4,5)P2) modulates interaction of syntaxin-1A with sulfonylurea receptor 1 to regulate pancreatic beta-cell ATP-sensitive potassium channels. J. Biol. Chem. 2014, 289, 6028–6040. [Google Scholar] [CrossRef]
- Lam, A.D.; Tryoen-Toth, P.; Tsai, B.; Vitale, N.; Stuenkel, E.L. SNARE-catalyzed fusion events are regulated by Syntaxin1A-lipid interactions. Mol. Biol. Cell 2008, 19, 485–497. [Google Scholar] [CrossRef]
- Honigmann, A.; van den Bogaart, G.; Iraheta, E.; Risselada, H.J.; Milovanovic, D.; Mueller, V.; Mullar, S.; Diederichsen, U.; Fasshauer, D.; Grubmuller, H.; et al. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 2013, 20, 679–686. [Google Scholar] [CrossRef]
- van den Bogaart, G.; Meyenberg, K.; Risselada, H.J.; Amin, H.; Willig, K.I.; Hubrich, B.E.; Dier, M.; Hell, S.W.; Grubmuller, H.; Diederichsen, U.; et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature 2011, 479, 552–555. [Google Scholar] [CrossRef]
- Hammond, G.R. Does PtdIns(4,5)P2 concentrate so it can multi-task? Biochem. Soc. Trans. 2016, 44, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Kim, S.H.; Venable, R.M.; Pastor, R.W. Design principles of PI(4,5)P(2) clustering under protein-free conditions: Specific cation effects and calcium-potassium synergy. Proc. Natl. Acad. Sci. USA 2022, 119, e2202647119. [Google Scholar] [CrossRef] [PubMed]
- Golebiewska, U.; Nyako, M.; Woturski, W.; Zaitseva, I.; McLaughlin, S. Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol. Biol. Cell 2008, 19, 1663–1669. [Google Scholar] [CrossRef]
- Gambhir, A.; Hangyas-Mihalyne, G.; Zaitseva, I.; Cafiso, D.S.; Wang, J.; Murray, D.; Pentyala, S.N.; Smith, S.O.; McLaughlin, S. Electrostatic sequestration of PI(4,5)P2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys. J. 2004, 86, 2188–2207. [Google Scholar] [CrossRef] [PubMed]
- Koldso, H.; Shorthouse, D.; Helie, J.; Sansom, M.S. Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput. Biol. 2014, 10, e1003911. [Google Scholar] [CrossRef]
- Dou, D.; Revol, R.; Ostbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef]
- Carter, T.; Iqbal, M. The Influenza A Virus Replication Cycle: A Comprehensive Review. Viruses 2024, 16, 316. [Google Scholar] [CrossRef]
- Wu, N.C.; Wilson, I.A. Influenza Hemagglutinin Structures and Antibody Recognition. Cold Spring Harb. Perspect. Med. 2020, 10, a038778. [Google Scholar] [CrossRef]
- Chlanda, P.; Mekhedov, E.; Waters, H.; Sodt, A.; Schwartz, C.; Nair, V.; Blank, P.S.; Zimmerberg, J. Palmitoylation Contributes to Membrane Curvature in Influenza A Virus Assembly and Hemagglutinin-Mediated Membrane Fusion. J. Virol. 2017, 91, e00947-17. [Google Scholar] [CrossRef]
- McAuley, J.L.; Gilbertson, B.P.; Trifkovic, S.; Brown, L.E.; McKimm-Breschkin, J.L. Influenza Virus Neuraminidase Structure and Functions. Front. Microbiol. 2019, 10, 39. [Google Scholar] [CrossRef]
- Martin, K.; Helenius, A. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell 1991, 67, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gu, M.; Zheng, Q.; Gao, R.; Liu, X. Packaging signal of influenza A virus. Virol. J. 2021, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Petrich, A.; Dunsing, V.; Bobone, S.; Chiantia, S. Influenza A M2 recruits M1 to the plasma membrane: A fluorescence fluctuation microscopy study. Biophys. J. 2021, 120, 5478–5490. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef]
- Tam, V.C.; Quehenberger, O.; Oshansky, C.M.; Suen, R.; Armando, A.M.; Treuting, P.M.; Thomas, P.G.; Dennis, E.A.; Aderem, A. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell 2013, 154, 213–227. [Google Scholar] [CrossRef]
- Lin, S.; Liu, N.; Yang, Z.; Song, W.; Wang, P.; Chen, H.; Lucio, M.; Schmitt-Kopplin, P.; Chen, G.; Cai, Z. GC/MS-based metabolomics reveals fatty acid biosynthesis and cholesterol metabolism in cell lines infected with influenza A virus. Talanta 2010, 83, 262–268. [Google Scholar] [CrossRef]
- Ivanova, P.T.; Myers, D.S.; Milne, S.B.; McClaren, J.L.; Thomas, P.G.; Brown, H.A. Lipid composition of viral envelope of three strains of influenza virus—Not all viruses are created equal. ACS Infect. Dis. 2015, 1, 399–452. [Google Scholar] [CrossRef]
- Tanner, L.B.; Chng, C.; Guan, X.L.; Lei, Z.; Rozen, S.G.; Wenk, M.R. Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J. Lipid Res. 2014, 55, 1357–1365. [Google Scholar] [CrossRef]
- Tisoncik-Go, J.; Gasper, D.J.; Kyle, J.E.; Eisfeld, A.J.; Selinger, C.; Hatta, M.; Morrison, J.; Korth, M.J.; Zink, E.M.; Kim, Y.M.; et al. Integrated Omics Analysis of Pathogenic Host Responses during Pandemic H1N1 Influenza Virus Infection: The Crucial Role of Lipid Metabolism. Cell Host Microbe 2016, 19, 254–266. [Google Scholar] [CrossRef]
- Woods, P.S.; Doolittle, L.M.; Rosas, L.E.; Joseph, L.M.; Calomeni, E.P.; Davis, I.C. Lethal H1N1 influenza A virus infection alters the murine alveolar type II cell surfactant lipidome. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 311, L1160–L1169. [Google Scholar] [CrossRef]
- Schmitt, A.P.; Lamb, R.A. Influenza virus assembly and budding at the viral budozone. Adv. Virus Res. 2005, 64, 383–416. [Google Scholar] [PubMed]
- Leser, G.P.; Lamb, R.A. Lateral Organization of Influenza Virus Proteins in the Budozone Region of the Plasma Membrane. J. Virol. 2017, 91, e02104-16. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Chen, C.Y.; Yang, J.H.; Chiu, Y.F. Modulating cholesterol-rich lipid rafts to disrupt influenza A virus infection. Front. Immunol. 2022, 13, 982264. [Google Scholar] [CrossRef] [PubMed]
- Veit, M.; Engel, S.; Thaa, B.; Scolari, S.; Herrmann, A. Lipid domain association of influenza virus proteins detected by dynamic fluorescence microscopy techniques. Cell. Microbiol. 2013, 15, 179–189. [Google Scholar] [CrossRef]
- Ono, A.; Freed, E.O. Role of lipid rafts in virus replication. Adv. Virus Res. 2005, 64, 311–358. [Google Scholar]
- Zhang, J.; Pekosz, A.; Lamb, R.A. Influenza virus assembly and lipid raft microdomains: A role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 2000, 74, 4634–4644. [Google Scholar] [CrossRef]
- Skibbens, J.E.; Roth, M.G.; Matlin, K.S. Differential extractability of influenza virus hemagglutinin during intracellular transport in polarized epithelial cells and nonpolar fibroblasts. J. Cell Biol. 1989, 108, 821–832. [Google Scholar] [CrossRef]
- Barman, S.; Nayak, D.P. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 2000, 74, 6538–6545. [Google Scholar] [CrossRef]
- Kundu, A.; Avalos, R.T.; Sanderson, C.M.; Nayak, D.P. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 1996, 70, 6508–6515. [Google Scholar] [CrossRef]
- Ali, A.; Avalos, R.T.; Ponimaskin, E.; Nayak, D.P. Influenza virus assembly: Effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 2000, 74, 8709–8719. [Google Scholar] [CrossRef]
- Schroeder, C. Cholesterol-binding viral proteins in virus entry and morphogenesis. In Cholesterol Binding and Cholesterol Transport Proteins; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2010; Volume 51, pp. 77–108. [Google Scholar]
- Leser, G.P.; Lamb, R.A. Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 2005, 342, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Petrich, A.; Chiantia, S. Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane. Viruses 2023, 15, 1830. [Google Scholar] [CrossRef] [PubMed]
- Scheiffele, P.; Rietveld, A.; Wilk, T.; Simons, K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 1999, 274, 2038–2044. [Google Scholar] [CrossRef]
- Veit, M.; Thaa, B. Association of influenza virus proteins with membrane rafts. Adv. Virol. 2011, 2011, 370606. [Google Scholar] [CrossRef] [PubMed]
- Gerl, M.J.; Sampaio, J.L.; Urban, S.; Kalvodova, L.; Verbavatz, J.M.; Binnington, B.; Lindemann, D.; Lingwood, C.A.; Shevchenko, A.; Schroeder, C.; et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012, 196, 213–221. [Google Scholar] [CrossRef]
- Raut, P.; Weller, S.R.; Obeng, B.; Soos, B.L.; West, B.E.; Potts, C.M.; Sangroula, S.; Kinney, M.S.; Burnell, J.E.; King, B.L.; et al. Cetylpyridinium chloride (CPC) reduces zebrafish mortality from influenza infection: Super-resolution microscopy reveals CPC interference with multiple protein interactions with phosphatidylinositol 4,5-bisphosphate in immune function. Toxicol. Appl. Pharmacol. 2022, 440, 115913. [Google Scholar] [CrossRef]
- Hess, S.T.; Gould, T.J.; Gudheti, M.V.; Maas, S.A.; Mills, K.D.; Zimmerberg, J. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA 2007, 104, 17370–17375. [Google Scholar] [CrossRef]
- Hess, S.T.; Kumar, M.; Verma, A.; Farrington, J.; Kenworthy, A.; Zimmerberg, J. Quantitative electron microscopy and fluorescence spectroscopy of the membrane distribution of influenza hemagglutinin. J. Cell Biol. 2005, 169, 965–976. [Google Scholar] [CrossRef]
- Scolari, S.; Engel, S.; Krebs, N.; Plazzo, A.P.; De Almeida, R.F.; Prieto, M.; Veit, M.; Herrmann, A. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 2009, 284, 15708–15716. [Google Scholar] [CrossRef]
- Wilson, R.L.; Frisz, J.F.; Klitzing, H.A.; Zimmerberg, J.; Weber, P.K.; Kraft, M.L. Hemagglutinin clusters in the plasma membrane are not enriched with cholesterol and sphingolipids. Biophys. J. 2015, 108, 1652–1659. [Google Scholar] [CrossRef]
- Chlanda, P.; Zimmerberg, J. Protein-lipid interactions critical to replication of the influenza A virus. FEBS Lett. 2016, 590, 1940–1954. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Casey, L.; Pike, L.J. Compartmentalization of phosphatidylinositol 4,5-bisphosphate in low-density membrane domains in the absence of caveolin. Biochem. Biophys. Res. Commun. 1998, 245, 684–690. [Google Scholar] [CrossRef] [PubMed]
- Hope, H.R.; Pike, L.J. Phosphoinositides and phosphoinositide-utilizing enzymes in detergent-insoluble lipid domains. Mol. Biol. Cell 1996, 7, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J. Lipid rafts: Bringing order to chaos. J. Lipid Res. 2003, 44, 655–667. [Google Scholar] [CrossRef]
- Raut, P.; Obeng, B.; Waters, H.; Zimmerberg, J.; Gosse, J.A.; Hess, S.T. Phosphatidylinositol 4,5-Bisphosphate Mediates the Co-Distribution of Influenza A Hemagglutinin and Matrix Protein M1 at the Plasma Membrane. Viruses 2022, 14, 2509. [Google Scholar] [CrossRef]
- Curthoys, N.M.; Mlodzianoski, M.J.; Parent, M.; Butler, M.B.; Raut, P.; Wallace, J.; Lilieholm, J.; Mehmood, K.; Maginnis, M.S.; Waters, H.; et al. Influenza Hemagglutinin Modulates Phosphatidylinositol 4,5-Bisphosphate Membrane Clustering. Biophys. J. 2019, 116, 893–909. [Google Scholar] [CrossRef]
- Tatulian, S.A.; Tamm, L.K. Secondary structure, orientation, oligomerization, and lipid interactions of the transmembrane domain of influenza hemagglutinin. Biochemistry 2000, 39, 496–507. [Google Scholar] [CrossRef]
- Mineev, K.S.; Lyukmanova, E.N.; Krabben, L.; Serebryakova, M.V.; Shulepko, M.A.; Arseniev, A.S.; Kordyukova, L.V.; Veit, M. Structural investigation of influenza virus hemagglutinin membrane-anchoring peptide. Protein Eng. Des. Sel. 2013, 26, 547–552. [Google Scholar] [CrossRef]
- Ngo, V.N.; Winski, D.P.; Aho, B.; Kamath, P.L.; King, B.L.; Waters, H.; Zimmerberg, J.; Sodt, A.; Hess, S.T. Conserved sequence features in intracellular domains of viral spike proteins. Virology 2024, 599, 110198. [Google Scholar] [CrossRef]
- Gudheti, M.V.; Curthoys, N.M.; Gould, T.J.; Kim, D.; Gunewardene, M.S.; Gabor, K.A.; Gosse, J.A.; Kim, C.H.; Zimmerberg, J.; Hess, S.T. Actin mediates the nanoscale membrane organization of the clustered membrane protein influenza hemagglutinin. Biophys. J. 2013, 104, 2182–2192. [Google Scholar] [CrossRef]
- Zhang, J.; Lamb, R.A. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology 1996, 225, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Peukes, J.; Xiong, X.; Erlendsson, S.; Qu, K.; Wan, W.; Calder, L.J.; Schraidt, O.; Kummer, S.; Freund, S.M.V.; Krausslich, H.G.; et al. The native structure of the assembled matrix protein 1 of influenza A virus. Nature 2020, 587, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Hofer, C.T.; Di Lella, S.; Dahmani, I.; Jungnick, N.; Bordag, N.; Bobone, S.; Huang, Q.; Keller, S.; Herrmann, A.; Chiantia, S. Structural determinants of the interaction between influenza A virus matrix protein M1 and lipid membranes. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Kerviel, A.; Dash, S.; Moncorge, O.; Panthu, B.; Prchal, J.; Decimo, D.; Ohlmann, T.; Lina, B.; Favard, C.; Decroly, E.; et al. Involvement of an Arginine Triplet in M1 Matrix Protein Interaction with Membranes and in M1 Recruitment into Virus-Like Particles of the Influenza A(H1N1)pdm09 Virus. PLoS ONE 2016, 11, e0165421. [Google Scholar] [CrossRef]
- Ruigrok, R.W.; Barge, A.; Durrer, P.; Brunner, J.; Ma, K.; Whittaker, G.R. Membrane interaction of influenza virus M1 protein. Virology 2000, 267, 289–298. [Google Scholar] [CrossRef]
- Baudin, F.; Petit, I.; Weissenhorn, W.; Ruigrok, R.W. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 2001, 281, 102–108. [Google Scholar] [CrossRef]
- Das, S.C.; Watanabe, S.; Hatta, M.; Noda, T.; Neumann, G.; Ozawa, M.; Kawaoka, Y. The highly conserved arginine residues at positions 76 through 78 of influenza A virus matrix protein M1 play an important role in viral replication by affecting the intracellular localization of M1. J. Virol. 2012, 86, 1522–1530. [Google Scholar] [CrossRef]
- Thaa, B.; Herrmann, A.; Veit, M. The polybasic region is not essential for membrane binding of the matrix protein M1 of influenza virus. Virology 2009, 383, 150–155. [Google Scholar] [CrossRef]
- Dahmani, I.; Ludwig, K.; Chiantia, S. Influenza A matrix protein M1 induces lipid membrane deformation via protein multimerization. Biosci. Rep. 2019, 39, BSR20191024. [Google Scholar] [CrossRef]
- Shtykova, E.V.; Dadinova, L.A.; Fedorova, N.V.; Golanikov, A.E.; Bogacheva, E.N.; Ksenofontov, A.L.; Baratova, L.A.; Shilova, L.A.; Tashkin, V.Y.; Galimzyanov, T.R.; et al. Influenza virus Matrix Protein M1 preserves its conformation with pH, changing multimerization state at the priming stage due to electrostatics. Sci. Rep. 2017, 7, 16793. [Google Scholar] [CrossRef]
- Hilsch, M.; Goldenbogen, B.; Sieben, C.; Hofer, C.T.; Rabe, J.P.; Klipp, E.; Herrmann, A.; Chiantia, S. Influenza A matrix protein M1 multimerizes upon binding to lipid membranes. Biophys. J. 2014, 107, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, L.V.; Konarev, P.V.; Fedorova, N.V.; Shtykova, E.V.; Ksenofontov, A.L.; Loshkarev, N.A.; Dadinova, L.A.; Timofeeva, T.A.; Abramchuk, S.S.; Moiseenko, A.V.; et al. The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer. Membranes 2021, 11, 772. [Google Scholar] [CrossRef] [PubMed]
- Bobone, S.; Hilsch, M.; Storm, J.; Dunsing, V.; Herrmann, A.; Chiantia, S. Phosphatidylserine Lateral Organization Influences the Interaction of Influenza Virus Matrix Protein 1 with Lipid Membranes. J. Virol. 2017, 91, e00267-17. [Google Scholar] [CrossRef] [PubMed]
- Loshkareva, A.S.; Popova, M.M.; Shilova, L.A.; Fedorova, N.V.; Timofeeva, T.A.; Galimzyanov, T.R.; Kuzmin, P.I.; Knyazev, D.G.; Batishchev, O.V. Influenza A Virus M1 Protein Non-Specifically Deforms Charged Lipid Membranes and Specifically Interacts with the Raft Boundary. Membranes 2023, 13, 76. [Google Scholar] [CrossRef]
- Sha, B.; Luo, M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat. Struct. Biol. 1997, 4, 239–244. [Google Scholar] [CrossRef]
- Kakisaka, M.; Yamada, K.; Yamaji-Hasegawa, A.; Kobayashi, T.; Aida, Y. Intrinsically disordered region of influenza A NP regulates viral genome packaging via interactions with viral RNA and host PI(4,5)P2. Virology 2016, 496, 116–126. [Google Scholar] [CrossRef]
- Gottlinger, H.G.; Sodroski, J.G.; Haseltine, W.A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 1989, 86, 5781–5785. [Google Scholar] [CrossRef]
- Bryant, M.; Ratner, L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 1990, 87, 523–527. [Google Scholar] [CrossRef]
- Zhou, W.; Parent, L.J.; Wills, J.W.; Resh, M.D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 1994, 68, 2556–2569. [Google Scholar] [CrossRef]
- Yuan, X.; Yu, X.; Lee, T.H.; Essex, M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 1993, 67, 6387–6394. [Google Scholar] [CrossRef]
- Hermida-Matsumoto, L.; Resh, M.D. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55(gag) and p17MA. J. Virol. 1999, 73, 1902–1908. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Gottlinger, H.G. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of gag membrane targeting. J. Virol. 1999, 73, 2604–2612. [Google Scholar] [CrossRef] [PubMed]
- Saad, J.S.; Loeliger, E.; Luncsford, P.; Liriano, M.; Tai, J.; Kim, A.; Miller, J.; Joshi, A.; Freed, E.O.; Summers, M.F. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 2007, 366, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Spearman, P.; Horton, R.; Ratner, L.; Kuli-Zade, I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 1997, 71, 6582–6592. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Freed, E.O. Binding of human immunodeficiency virus type 1 Gag to membrane: Role of the matrix amino terminus. J. Virol. 1999, 73, 4136–4144. [Google Scholar] [CrossRef]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 2004, 101, 517–522. [Google Scholar] [CrossRef]
- Freed, E.O.; Orenstein, J.M.; Buckler-White, A.J.; Martin, M.A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 1994, 68, 5311–5320. [Google Scholar] [CrossRef]
- Ono, A.; Orenstein, J.M.; Freed, E.O. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 2000, 74, 2855–2866. [Google Scholar] [CrossRef]
- Kleinpeter, A.B.; Freed, E.O. HIV-1 Maturation: Lessons Learned from Inhibitors. Viruses 2020, 12, 940. [Google Scholar] [CrossRef]
- Thornhill, D.; Murakami, T.; Ono, A. Rendezvous at Plasma Membrane: Cellular Lipids and tRNA Set up Sites of HIV-1 Particle Assembly and Incorporation of Host Transmembrane Proteins. Viruses 2020, 12, 842. [Google Scholar] [CrossRef]
- Ono, A.; Ablan, S.D.; Lockett, S.J.; Nagashima, K.; Freed, E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA 2004, 101, 14889–14894. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.; Uchil, P.D.; Jin, J.; Shui, G.; Ott, D.E.; Mothes, W.; Wenk, M.R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008, 82, 11228–11238. [Google Scholar] [CrossRef] [PubMed]
- Monde, K.; Chukkapalli, V.; Ono, A. Assembly and replication of HIV-1 in T cells with low levels of phosphatidylinositol-(4,5)-bisphosphate. J. Virol. 2011, 85, 3584–3595. [Google Scholar] [CrossRef]
- Chan, J.; Dick, R.A.; Vogt, V.M. Rous sarcoma virus gag has no specific requirement for phosphatidylinositol-(4,5)-bisphosphate for plasma membrane association in vivo or for liposome interaction in vitro. J. Virol. 2011, 85, 10851–10860. [Google Scholar] [CrossRef]
- Mucksch, F.; Laketa, V.; Muller, B.; Schultz, C.; Krausslich, H.G. Synchronized HIV assembly by tunable PIP(2) changes reveals PIP(2) requirement for stable Gag anchoring. eLife 2017, 6, e25287. [Google Scholar] [CrossRef]
- Chukkapalli, V.; Hogue, I.B.; Boyko, V.; Hu, W.S.; Ono, A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J. Virol. 2008, 82, 2405–2417. [Google Scholar] [CrossRef]
- Gerber, P.P.; Cabrini, M.; Jancic, C.; Paoletti, L.; Banchio, C.; von Bilderling, C.; Sigaut, L.; Pietrasanta, L.I.; Duette, G.; Freed, E.O.; et al. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 2015, 209, 435–452. [Google Scholar] [CrossRef]
- Gonzales, B.; de Rocquigny, H.; Beziau, A.; Durand, S.; Burlaud-Gaillard, J.; Lefebvre, A.; Krull, S.; Emond, P.; Brand, D.; Piver, E. Type I Phosphatidylinositol-4-Phosphate 5-Kinases alpha and gamma Play a Key Role in Targeting HIV-1 Pr55(Gag) to the Plasma Membrane. J. Virol. 2020, 94, e00189-20. [Google Scholar] [CrossRef]
- Thornhill, D.; Olety, B.; Ono, A. Relationships between MA-RNA Binding in Cells and Suppression of HIV-1 Gag Mislocalization to Intracellular Membranes. J. Virol. 2019, 93, e00756-19. [Google Scholar] [CrossRef]
- Chukkapalli, V.; Oh, S.J.; Ono, A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA 2010, 107, 1600–1605. [Google Scholar] [CrossRef]
- Mercredi, P.Y.; Bucca, N.; Loeliger, B.; Gaines, C.R.; Mehta, M.; Bhargava, P.; Tedbury, P.R.; Charlier, L.; Floquet, N.; Muriaux, D.; et al. Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein. J. Mol. Biol. 2016, 428, 1637–1655. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Samal, A.B.; Vlach, J.; Mas, V.; Prevelige, P.E.; Saad, J.S. Structural and biophysical characterizations of HIV-1 matrix trimer binding to lipid nanodiscs shed light on virus assembly. J. Biol. Chem. 2019, 294, 18600–18612. [Google Scholar] [CrossRef] [PubMed]
- Junkova, P.; Pleskot, R.; Prchal, J.; Sys, J.; Ruml, T. Differences and commonalities in plasma membrane recruitment of the two morphogenetically distinct retroviruses HIV-1 and MMTV. J. Biol. Chem. 2020, 295, 8819–8833. [Google Scholar] [CrossRef]
- Barros, M.; Heinrich, F.; Datta, S.A.K.; Rein, A.; Karageorgos, I.; Nanda, H.; Lösche, M. Membrane Binding of HIV-1 Matrix Protein: Dependence on Bilayer Composition and Protein Lipidation. J. Virol. 2016, 90, 4544–4555. [Google Scholar] [CrossRef]
- Keller, H.; Krausslich, H.G.; Schwille, P. Multimerizable HIV Gag derivative binds to the liquid-disordered phase in model membranes. Cell. Microbiol. 2013, 15, 237–247. [Google Scholar] [CrossRef]
- Tran, R.J.; Lalonde, M.S.; Sly, K.L.; Conboy, J.C. Mechanistic Investigation of HIV-1 Gag Association with Lipid Membranes. J. Phys. Chem. B 2019, 123, 4673–4687. [Google Scholar] [CrossRef]
- Carlson, L.A.; Hurley, J.H. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc. Natl. Acad. Sci. USA 2012, 109, 16928–16933. [Google Scholar] [CrossRef]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef]
- Qu, K.; Ke, Z.; Zila, V.; Anders-Osswein, M.; Glass, B.; Mucksch, F.; Muller, R.; Schultz, C.; Muller, B.; Krausslich, H.G.; et al. Maturation of the matrix and viral membrane of HIV-1. Science 2021, 373, 700–704. [Google Scholar] [CrossRef]
- Olety, B.; Veatch, S.L.; Ono, A. Phosphatidylinositol-(4,5)-Bisphosphate Acyl Chains Differentiate Membrane Binding of HIV-1 Gag from That of the Phospholipase Cdelta1 Pleckstrin Homology Domain. J. Virol. 2015, 89, 7861–7873. [Google Scholar] [CrossRef]
- Lochrie, M.A.; Waugh, S.; Pratt, D.G., Jr.; Clever, J.; Parslow, T.G.; Polisky, B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997, 25, 2902–2910. [Google Scholar] [CrossRef] [PubMed]
- Purohit, P.; Dupont, S.; Stevenson, M.; Green, M.R. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 2001, 7, 576–584. [Google Scholar] [CrossRef]
- Cimarelli, A.; Luban, J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1999, 73, 5388–5401. [Google Scholar] [CrossRef]
- Ramalingam, D.; Duclair, S.; Datta, S.A.; Ellington, A.; Rein, A.; Prasad, V.R. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011, 85, 305–314. [Google Scholar] [CrossRef]
- Jones, C.P.; Datta, S.A.; Rein, A.; Rouzina, I.; Musier-Forsyth, K. Matrix domain modulates HIV-1 Gag’s nucleic acid chaperone activity via inositol phosphate binding. J. Virol. 2011, 85, 1594–1603. [Google Scholar] [CrossRef]
- Hearps, A.C.; Wagstaff, K.M.; Piller, S.C.; Jans, D.A. The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry 2008, 47, 2199–2210. [Google Scholar] [CrossRef]
- Alfadhli, A.; Still, A.; Barklis, E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 2009, 83, 12196–12203. [Google Scholar] [CrossRef]
- Socas, L.B.P.; Ambroggio, E.E. HIV-1 Gag specificity for PIP(2) is regulated by macromolecular electric properties of both protein and membrane local environments. Biochim. Biophys. Acta Biomembr. 2023, 1865, 184157. [Google Scholar] [CrossRef]
- Sumner, C.; Kotani, O.; Liu, S.; Musier-Forsyth, K.; Sato, H.; Ono, A. Molecular Determinants in tRNA D-arm Required for Inhibition of HIV-1 Gag Membrane Binding. J. Mol. Biol. 2022, 434, 167390. [Google Scholar] [CrossRef]
- Todd, G.C.; Duchon, A.; Inlora, J.; Olson, E.D.; Musier-Forsyth, K.; Ono, A. Inhibition of HIV-1 Gag-membrane interactions by specific RNAs. RNA 2017, 23, 395–405. [Google Scholar] [CrossRef]
- Inlora, J.; Collins, D.R.; Trubin, M.E.; Chung, J.Y.; Ono, A. Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA. mBio 2014, 5, e02202. [Google Scholar] [CrossRef]
- Chukkapalli, V.; Inlora, J.; Todd, G.C.; Ono, A. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J. Virol. 2013, 87, 7155–7159. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Inlora, J.; Chukkapalli, V.; Derse, D.; Ono, A. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J. Virol. 2011, 85, 3802–3810. [Google Scholar] [CrossRef]
- Dick, R.A.; Kamynina, E.; Vogt, V.M. Effect of multimerization on membrane association of Rous sarcoma virus and HIV-1 matrix domain proteins. J. Virol. 2013, 87, 13598–13608. [Google Scholar] [CrossRef] [PubMed]
- Alfadhli, A.; McNett, H.; Tsagli, S.; Bachinger, H.P.; Peyton, D.H.; Barklis, E. HIV-1 matrix protein binding to RNA. J. Mol. Biol. 2011, 410, 653–666. [Google Scholar] [CrossRef]
- Gaines, C.R.; Tkacik, E.; Rivera-Oven, A.; Somani, P.; Achimovich, A.; Alabi, T.; Zhu, A.; Getachew, N.; Yang, A.L.; McDonough, M.; et al. HIV-1 Matrix Protein Interactions with tRNA: Implications for Membrane Targeting. J. Mol. Biol. 2018, 430, 2113–2127. [Google Scholar] [CrossRef]
- Bou-Nader, C.; Muecksch, F.; Brown, J.B.; Gordon, J.M.; York, A.; Peng, C.; Ghirlando, R.; Summers, M.F.; Bieniasz, P.D.; Zhang, J. HIV-1 matrix-tRNA complex structure reveals basis for host control of Gag localization. Cell Host Microbe 2021, 29, 1421–1436.e7. [Google Scholar] [CrossRef]
- Charlier, L.; Louet, M.; Chaloin, L.; Fuchs, P.; Martinez, J.; Muriaux, D.; Favard, C.; Floquet, N. Coarse-grained simulations of the HIV-1 matrix protein anchoring: Revisiting its assembly on membrane domains. Biophys. J. 2014, 106, 577–585. [Google Scholar] [CrossRef]
- Banerjee, P.; Qu, K.; Briggs, J.A.G.; Voth, G.A. Molecular dynamics simulations of HIV-1 matrix-membrane interactions at different stages of viral maturation. Biophys. J. 2024, 123, 389–406. [Google Scholar] [CrossRef]
- Favard, C.; Chojnacki, J.; Merida, P.; Yandrapalli, N.; Mak, J.; Eggeling, C.; Muriaux, D. HIV-1 Gag specifically restricts PI(4,5)P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly. Sci. Adv. 2019, 5, eaaw8651. [Google Scholar] [CrossRef] [PubMed]
- Mucksch, F.; Citir, M.; Luchtenborg, C.; Glass, B.; Traynor-Kaplan, A.; Schultz, C.; Brugger, B.; Krausslich, H.G. Quantification of phosphoinositides reveals strong enrichment of PIP(2) in HIV-1 compared to producer cell membranes. Sci. Rep. 2019, 9, 17661. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Feigenson, G.W.; Vogt, V.M.; Dick, R.A. Mechanisms of PI(4,5)P2 Enrichment in HIV-1 Viral Membranes. J. Mol. Biol. 2020, 432, 5343–5364. [Google Scholar] [CrossRef]
- Yandrapalli, N.; Lubart, Q.; Tanwar, H.S.; Picart, C.; Mak, J.; Muriaux, D.; Favard, C. Self assembly of HIV-1 Gag protein on lipid membranes generates PI(4,5)P(2)/Cholesterol nanoclusters. Sci. Rep. 2016, 6, 39332. [Google Scholar] [CrossRef]
- Grover, J.R.; Veatch, S.L.; Ono, A. Basic motifs target PSGL-1, CD43, and CD44 to plasma membrane sites where HIV-1 assembles. J. Virol. 2015, 89, 454–467. [Google Scholar] [CrossRef]
- Murakami, T.; Carmona, N.; Ono, A. Virion-incorporated PSGL-1 and CD43 inhibit both cell-free infection and transinfection of HIV-1 by preventing virus-cell binding. Proc. Natl. Acad. Sci. USA 2020, 117, 8055–8063. [Google Scholar] [CrossRef]
- Murakami, T.; Kim, J.; Li, Y.; Green, G.E.; Shikanov, A.; Ono, A. Secondary lymphoid organ fibroblastic reticular cells mediate trans-infection of HIV-1 via CD44-hyaluronan interactions. Nat. Commun. 2018, 9, 2436. [Google Scholar] [CrossRef]
- Murakami, T.; Ono, A. Roles of Virion-Incorporated CD162 (PSGL-1), CD43, and CD44 in HIV-1 Infection of T Cells. Viruses 2021, 13, 1935. [Google Scholar] [CrossRef]
- Fu, Y.; He, S.; Waheed, A.A.; Dabbagh, D.; Zhou, Z.; Trinite, B.; Wang, Z.; Yu, J.; Wang, D.; Li, F.; et al. PSGL-1 restricts HIV-1 infectivity by blocking virus particle attachment to target cells. Proc. Natl. Acad. Sci. USA 2020, 117, 9537–9545. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Y.; Zhang, S.; Diao, M.; Huang, S.; Li, S.; Tan, X. PSGL-1 inhibits HIV-1 infection by restricting actin dynamics and sequestering HIV envelope proteins. Cell Discov. 2020, 6, 53. [Google Scholar] [CrossRef]
- Liu, Y.; Fu, Y.; Wang, Q.; Li, M.; Zhou, Z.; Dabbagh, D.; Fu, C.; Zhang, H.; Li, S.; Zhang, T.; et al. Proteomic profiling of HIV-1 infection of human CD4(+) T cells identifies PSGL-1 as an HIV restriction factor. Nat. Microbiol. 2019, 4, 813–825. [Google Scholar] [CrossRef]
- Cardoso, R.d.S.; Murakami, T.; Jacobovitz, B.; Veatch, S.L.; Ono, A. PI(4,5)P2 promotes the incorporation of CD43, PSGL-1 and CD44 into nascent HIV-1 particles. bioRxiv 2024. [CrossRef]
- Dalton, A.K.; Ako-Adjei, D.; Murray, P.S.; Murray, D.; Vogt, V.M. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 2007, 81, 6434–6445. [Google Scholar] [CrossRef]
- Vlach, J.; Saad, J.S. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc. Natl. Acad. Sci. USA 2013, 110, 3525–3530. [Google Scholar] [CrossRef]
- Monje-Galvan, V.; Voth, G.A. Binding mechanism of the matrix domain of HIV-1 gag on lipid membranes. eLife 2020, 9, e58621. [Google Scholar] [CrossRef]
- Sumner, C.; Ono, A. The “basics” of HIV-1 assembly. PLoS Pathog. 2024, 20, e1011937. [Google Scholar] [CrossRef]
- Chua, B.A.; Ngo, J.A.; Situ, K.; Morizono, K. Roles of phosphatidylserine exposed on the viral envelope and cell membrane in HIV-1 replication. Cell Commun. Signal. 2019, 17, 132. [Google Scholar] [CrossRef]
- Moller-Tank, S.; Maury, W. Phosphatidylserine receptors: Enhancers of enveloped virus entry and infection. Virology 2014, 468–470, 565–580. [Google Scholar] [CrossRef]
- Callahan, M.K.; Popernack, P.M.; Tsutsui, S.; Truong, L.; Schlegel, R.A.; Henderson, A.J. Phosphatidylserine on HIV envelope is a cofactor for infection of monocytic cells. J. Immunol. 2003, 170, 4840–4845. [Google Scholar] [CrossRef]
- Fadok, V.A.; Voelker, D.R.; Campbell, P.A.; Cohen, J.J.; Bratton, D.L.; Henson, P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992, 148, 2207–2216. [Google Scholar] [CrossRef]
- Segawa, K.; Nagata, S. An Apoptotic ‘Eat Me’ Signal: Phosphatidylserine Exposure. Trends Cell Biol. 2015, 25, 639–650. [Google Scholar] [CrossRef]
- Mercer, J.; Helenius, A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 2008, 320, 531–535. [Google Scholar] [CrossRef]
- Mercer, J.; Knebel, S.; Schmidt, F.I.; Crouse, J.; Burkard, C.; Helenius, A. Vaccinia virus strains use distinct forms of macropinocytosis for host-cell entry. Proc. Natl. Acad. Sci. USA 2010, 107, 9346–9351. [Google Scholar] [CrossRef]
- Li, M.; Ablan, S.D.; Miao, C.; Zheng, Y.M.; Fuller, M.S.; Rennert, P.D.; Maury, W.; Johnson, M.C.; Freed, E.O.; Liu, S.L. TIM-family proteins inhibit HIV-1 release. Proc. Natl. Acad. Sci. USA 2014, 111, E3699–E3707. [Google Scholar] [CrossRef]
- Rosa, A.; Chande, A.; Ziglio, S.; De Sanctis, V.; Bertorelli, R.; Goh, S.L.; McCauley, S.M.; Nowosielska, A.; Antonarakis, S.E.; Luban, J.; et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 2015, 526, 212–217. [Google Scholar] [CrossRef]
- Usami, Y.; Wu, Y.; Gottlinger, H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef]
- Sid Ahmed, S.; Bajak, K.; Fackler, O.T. Beyond Impairment of Virion Infectivity: New Activities of the Anti-HIV Host Cell Factor SERINC5. Viruses 2024, 16, 284. [Google Scholar] [CrossRef]
- Leonhardt, S.A.; Purdy, M.D.; Grover, J.R.; Yang, Z.; Poulos, S.; McIntire, W.E.; Tatham, E.A.; Erramilli, S.K.; Nosol, K.; Lai, K.K.; et al. Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry. Nat. Commun. 2023, 14, 4368. [Google Scholar] [CrossRef]
- Raghunath, G.; Abbott, E.H.; Marin, M.; Wu, H.; Reyes Ballista, J.M.; Brindley, M.A.; Melikyan, G.B. Disruption of Transmembrane Phosphatidylserine Asymmetry by HIV-1 Incorporated SERINC5 Is Not Responsible for Virus Restriction. Biomolecules 2024, 14, 570. [Google Scholar] [CrossRef]
- Harrison, M.S.; Sakaguchi, T.; Schmitt, A.P. Paramyxovirus assembly and budding: Building particles that transmit infections. Int. J. Biochem. Cell Biol. 2010, 42, 1416–1429. [Google Scholar] [CrossRef]
- Swain, J.; Bierre, M.; Veyrie, L.; Richard, C.A.; Eleouet, J.F.; Muriaux, D.; Bajorek, M. Selective targeting and clustering of phosphatidylserine lipids by RSV M protein is critical for virus particle production. J. Biol. Chem. 2023, 299, 105323. [Google Scholar] [CrossRef]
- Wang, J.; Gambhir, A.; Hangyas-Mihalyne, G.; Murray, D.; Golebiewska, U.; McLaughlin, S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J. Biol. Chem. 2002, 277, 34401–34412. [Google Scholar] [CrossRef]
- Murphy, R.E.; Samal, A.B.; Vlach, J.; Saad, J.S. Solution Structure and Membrane Interaction of the Cytoplasmic Tail of HIV-1 gp41 Protein. Structure 2017, 25, 1708–1718.e05. [Google Scholar] [CrossRef]
- Harris, A.; Cardone, G.; Winkler, D.C.; Heymann, J.B.; Brecher, M.; White, J.M.; Steven, A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA 2006, 103, 19123–19127. [Google Scholar] [CrossRef]
- Calder, L.J.; Wasilewski, S.; Berriman, J.A.; Rosenthal, P.B. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. USA 2010, 107, 10685–10690. [Google Scholar] [CrossRef]
- Inamdar, K.; Tsai, F.C.; Dibsy, R.; de Poret, A.; Manzi, J.; Merida, P.; Muller, R.; Lappalainen, P.; Roingeard, P.; Mak, J.; et al. Full assembly of HIV-1 particles requires assistance of the membrane curvature factor IRSp53. eLife 2021, 10, e67321. [Google Scholar]
- Kumakura, M.; Kawaguchi, A.; Nagata, K. Actin-myosin network is required for proper assembly of influenza virus particles. Virology 2015, 476, 141–150. [Google Scholar] [CrossRef]
- Bedi, S.; Ono, A. Friend or Foe: The Role of the Cytoskeleton in Influenza A Virus Assembly. Viruses 2019, 11, 46. [Google Scholar] [CrossRef]
- Morizono, K.; Chen, I.S. Role of phosphatidylserine receptors in enveloped virus infection. J. Virol. 2014, 88, 4275–4290. [Google Scholar]
- Amara, A.; Mercer, J. Viral apoptotic mimicry. Nat. Rev. Microbiol. 2015, 13, 461–469. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Souza Cardoso, R.; Ono, A. The Effects of Viral Structural Proteins on Acidic Phospholipids in Host Membranes. Viruses 2024, 16, 1714. https://doi.org/10.3390/v16111714
de Souza Cardoso R, Ono A. The Effects of Viral Structural Proteins on Acidic Phospholipids in Host Membranes. Viruses. 2024; 16(11):1714. https://doi.org/10.3390/v16111714
Chicago/Turabian Stylede Souza Cardoso, Ricardo, and Akira Ono. 2024. "The Effects of Viral Structural Proteins on Acidic Phospholipids in Host Membranes" Viruses 16, no. 11: 1714. https://doi.org/10.3390/v16111714
APA Stylede Souza Cardoso, R., & Ono, A. (2024). The Effects of Viral Structural Proteins on Acidic Phospholipids in Host Membranes. Viruses, 16(11), 1714. https://doi.org/10.3390/v16111714





