Abstract
Despite the efforts of practical medicine and virology, influenza viruses remain the most important pathogens affecting human and animal health. Swine are exposed to infection with all types of influenza A, B, C, and D viruses. Influenza viruses have low pathogenicity for swine, but in the case of co-infection with other pathogens, the outcome can be much more serious, even fatal. Having a high zoonotic potential, swine play an important role in the ecology and spread of influenza to humans. In this study, we review the state of the scientific literature on the zoonotic spread of swine influenza A viruses among humans, their circulation in swine populations worldwide, reverse zoonosis from humans to swine, and their role in interspecies transmission. The analysis covers a long period to trace the ecology and evolutionary history of influenza A viruses in swine. The following databases were used to search the literature: Scopus, Web of Science, Google Scholar, and PubMed. In this review, 314 papers are considered: n = 107 from Asia, n = 93 from the U.S., n = 86 from Europe, n = 20 from Africa, and n = 8 from Australia. According to the date of publication, they are conditionally divided into three groups: contemporary, released from 2011 to the present (n = 121); 2000–2010 (n = 108); and 1919–1999 (n = 85).
1. Introduction
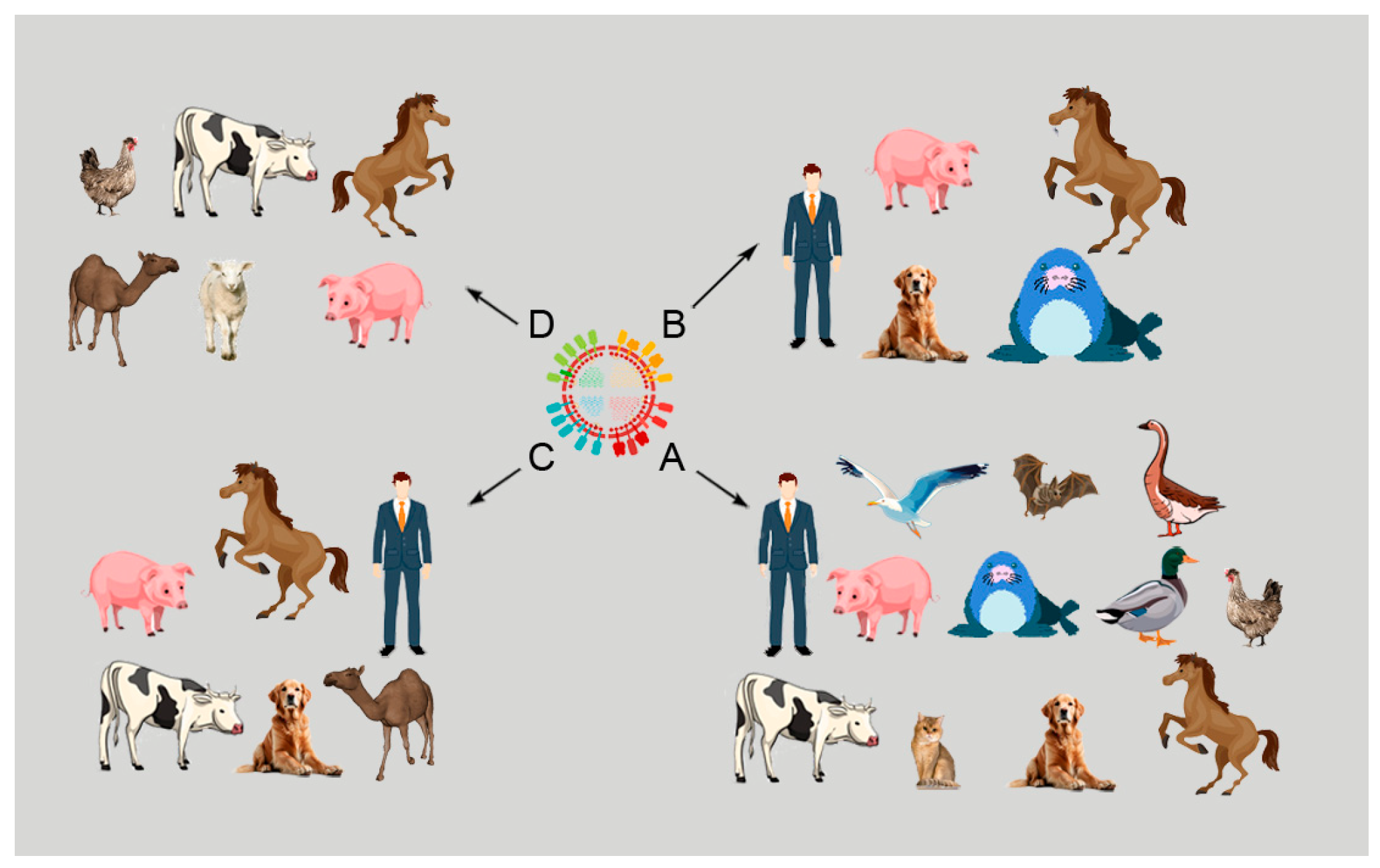
Influenza viruses (IVs) belong to the Orthomyxoviridae family and have a segmented genome with single-stranded, negative-sense RNA. Based on genetic and antigenic differences, IVs are divided into four genera, A, B, C, and D, which infect different species of mammals and birds with the four types of IV that occur in swine [1,2,3,4]. Among the four types of IVs, influenza A viruses (IAVs) are highly significant pathogens responsible for serious epidemics in humans and domestic animals. Figure 1 presents different species of animals infected with the four types of IVs.
Figure 1.
Influenza A, B, C, and D in different species of mammals.
The IAV genome consists of eight RNA segments: PB2, PB1, PA, hemagglutinin (HA), NP, neuraminidase (NA), M, and NS [1]. Two surface antigens, HA and NA, are mainly subject to antigenic variability, while internal proteins (NP and M) are relatively conservative. HA and NA play an essential role in the initial stages of cell infection [5]. HA binds to host cell receptors that contain terminal sialic acid residues (-2,6-SA or -2,3-SA). The HA gene is in charge of attaching viral pieces to the host cell receptor. NA removes the cell surface receptor (sialic acid); this is crucial for releasing viral particles from the cell surface and virus spillover [5].
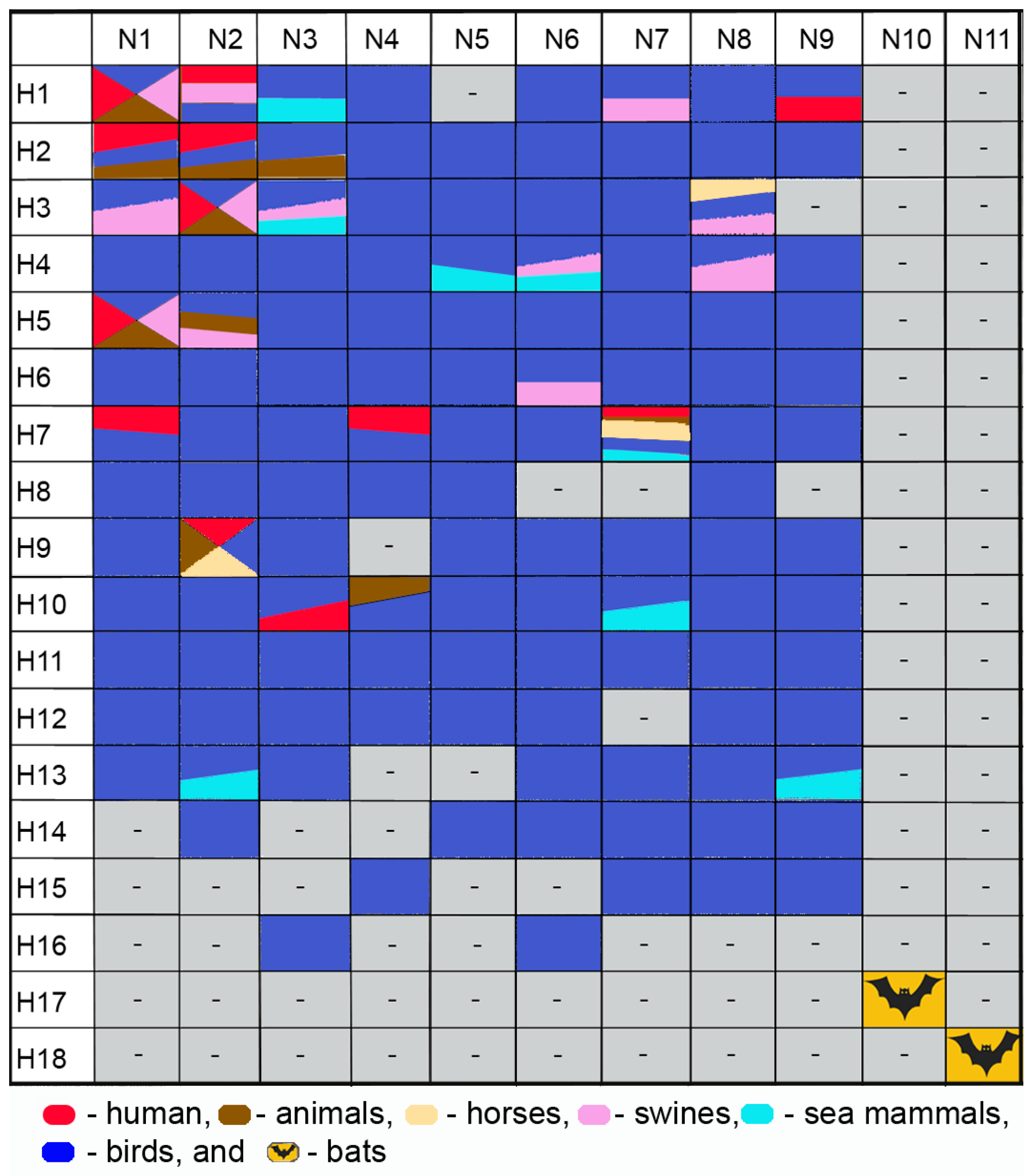
Based on major antigenic differences in surface proteins (HA and NA), 18 HA subtypes (H1–H18) and 11 NA subtypes (N1–N11) have been identified. The H1–H16 and N1–N9 virus subtypes have been identified in waterfowl, which are believed to constitute the natural IAV reservoir [6,7,8], while the H17–H18 and N10–N11 subtype sequences have been found in bats [9,10,11]. Only a limited number of subtypes have been found in mammals, such as H1 and H3 viral subtypes, which we identified and circulate in both humans and swine [12,13,14,15,16,17,18,19]. Figure 2 presents subtypes of H1–H18 hemagglutinins and N1–N11 neuraminidases of influenza viruses in different species of mammals.
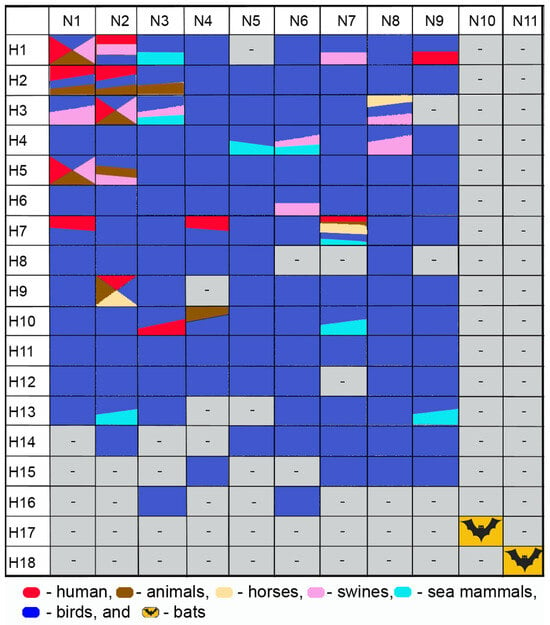
Figure 2.
Influenza subtypes of viruses H1–H18 and N1–N11 in different species of mammals.
IAVs’ epidemiology and ecology are complicated by their multi-host and segmented genome. IAVs are among the main pathogens in swine that give rise to acute respiratory infections and cause significant economic losses for swine farms. In addition to swine IAVs, human and avian IAVs can also infect swine. Swine are believed to serve as an intermediate “mixing vessel” for generating a variety of novel IVs [20]. The co-infection of swine with two or more IAV strains can induce reassortment [21,22], which in turn can promote the emergence of new IV strains [23,24,25]. There are numerous reports of zoonotic infections in humans with swine IAVs and reverse zoonotic infections in swine with human IAVs [26,27,28,29,30].
A literature search on the review topic was conducted on knowledge-intensive databases, including Scopus, Web of Science, Google Scholar, and PubMed. The following words and phrases were used as criteria for the literature search: “influenza viruses”, “classification and structure of the influenza virus”, “swine flu”; “avian flu viruses”, “reassortment”; “epizootology”; “zoonotic infection”; “transmission”, “swine flu in humans”, and “swine flu in horses”.
The following sections of this review assess swine influenza A viruses, their ecology, and their geographic distribution.
2. Ecology of Influenza Virus
Due to recurrent outbreaks and fast spillover, differing both genetically and antigenically, IAVs pose a serious problem for both swine production and public health. This issue is considered not only from the point of view of estimating and controlling the presently developing strains in swine but also in terms of describing their zoonotic potential [31].
IAVs infect various animal species, and swine (Sus scrofa) are among the natural hosts for these viruses. Swine influenza (SI) is an extremely dangerous respiratory disease in swine. The major causative viruses include the H1N1, H1N2, and H3N2 subtypes [32], which are antigenically related to human IV.
The development of IAV proceeds very rapidly due to two key mechanisms: antigenic drift and antigenic shift. Antigenic drift occurs due to the gradual accumulation of mutations in the surface proteins HA and NA, which leads to antigenic changes in IAV. Antigenic drift is responsible for human seasonal influenza viruses. Antigenic shift, otherwise known as reassortment, happens due to the segmented IV genome. When two or more different IAVs infect a cell or a host, the viral gene segments can be intermixed and reassorted, leading to the appearance of different reassortant viruses. Examples of antigenic shift include the emergence of three of the four pandemic IVs that have occurred in human history, which caused the 1957 H2N2 Asian flu pandemic, the 1968 Hong Kong H3N2 flu pandemic, and the 2009 H1N1 flu pandemic [33,34].
It was discovered that swine tissues express both α-2,6-SA (human IAV receptor preference) and α-2,3-SA (avian IAV receptor preference) [20]. Genetic reassortment between human and/or avian and/or swine IAVs can take place in swine. A prime example of this reassortment is the 2009 influenza pandemic caused by the H1N1 swine influenza reassortant virus containing the NA and M genes from the Eurasian swine virus and six remaining genes from the North American triple-reassortant virus [35,36]. The possibility of creating novel IAVs has led to swine becoming known as “mixing vessels” [24,34,37]. However, whether swine are necessary intermediate hosts for bird-to-human infections is a debatable matter because people and certain land birds have also been found to express both types of sialic acids [38,39,40]. At the same time, swine play a major role in IAV ecology, and this is especially true of human–swine interactions. Infection in swine can nevertheless cause viruses to acquire the adaptation needed either to maintain infection inside swine, thereby ensuring an additional reservoir for human infection, or to provide the mutations necessary for transmission to humans, which will lead to the establishment of that virus in the human population [27,41,42,43].
3. Geographical Distribution of Swine Influenza Viruses
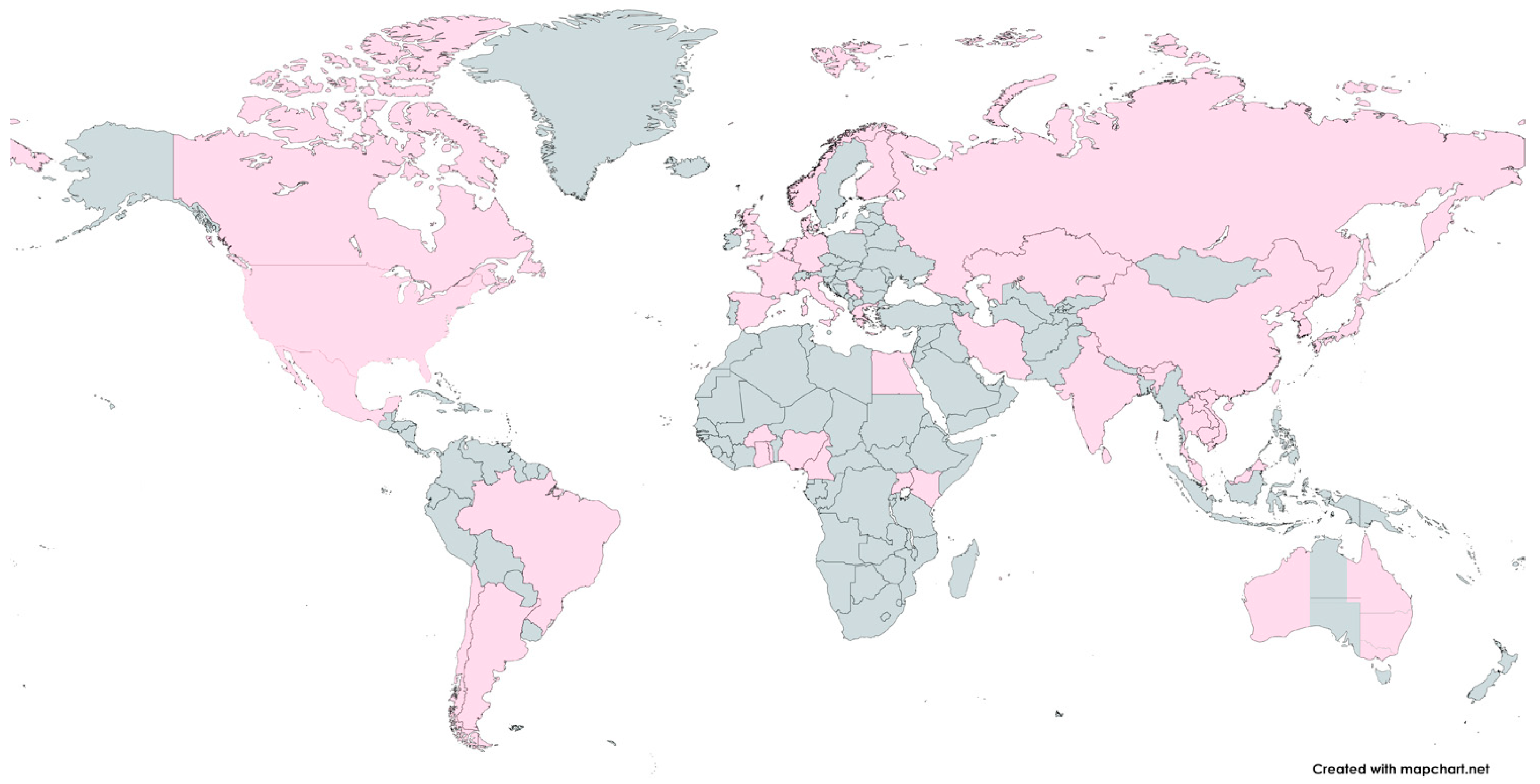
There is a huge variety of H1N1/N2 and H3N2 IAV subtypes in swine worldwide, and it has been established that different lineages are circulating in various regions of the Earth. A common characteristic of swine IAV ecology is the periodic spread of seasonal human viruses. The introduction of seasonal H1N1 and H3N2 IAVs from humans to swine at different times and in various geographical areas has led to the establishment of novel IAV lineages in swine hosts and contributed to the genetic and antigenic diversity of influenza observed in swine [27,44,45,46]. Figure 3 presents the geographical distribution of swine influenza viruses worldwide.
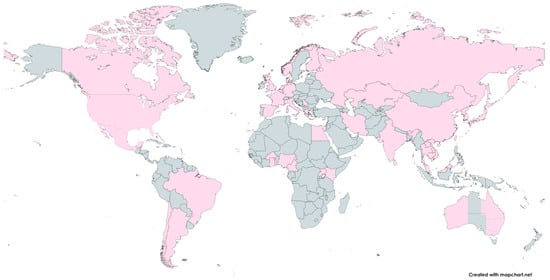
Figure 3.
The geographical distribution of swine influenza viruses. The locations where influenza viruses were detected in pigs are marked in pink. The world map was created online at https://mapchart.net (accessed on 11 October 2024).
Swine surveillance levels have increased worldwide following the emergence of the pandemic H1N1 virus (H1N1pdm09), as well as still-detectable IAV and/or novel gene lineages not previously recognized in swine, in regions that are not IAV-endemic [22,25,47,48].
3.1. Circulation of Swine Influenza Viruses in North America
SI was first identified as a swine respiratory infection in the U.S. in 1918, coinciding with the Spanish human influenza pandemic [49,50]. IAV was first isolated from swine in 1930 by Schope, who demonstrated that the virus caused a respiratory illness similar to human influenza in swine [51]. A phylogenetic analysis of this virus, classical H1N1 (cH1N1) SIV, showed that the cH1N1 lineage was closely related to the 1918 H1N1 Spanish IVs [52] and other isolated human IAVs after the discovery of the SI virus (SIVs) in the 1930s. The cH1N1 virus circulated among swine in the U.S. for almost 80 years with minimal genetic changes [40]. However, in the late 1990s, a triple-reassortant IAV was discovered. It contained the HA, NA, and PB1 genes derived from seasonal human H3N2; PB2 and PA from avian IAV; and NP, M, and NS from IAV cH1N1 of swine [53].
Several outbreaks of respiratory diseases occurred in 1998 among swine herds in North Carolina, Texas, Minnesota, and Iowa. The disease was caused by H3N2 IAVs. Molecular genetic analyses showed that the origin of these SIVs appeared in two diverse ways. The isolate from North Carolina was a product of genetic reassortment, and it contained segments of the IAV H3N2 gene of human origin (PB1, HA, and NA) circulating in 1995 and segments of genes from the cH1N1 porcine lineage (PB2, PA, NP, M, and NS). The strains isolated in Minnesota, Iowa, and Texas constituted the triple reassortants and contained segments of the cH1N1 SIV gene (NP, M, and NS), the same segments of the human virus gene (PB1, HA, NA), and viruses of avian origin (PB2 and PA) [54]. This swine IAV genomic composition is known as the triple-reassortant internal gene (TRIG) cassette [25,55,56,57].
At the end of the 1990s in the United States, in the swine population, in contrast to double-reassortant viruses that did not take root, active circulation of triple-reassortant viruses was observed [58]. Since H3N2 IAVs were almost non-existent among the U.S. swine population, a question arose as to the in-depth study of the virus’s spread. For this purpose, 4382 swine serums collected from 23 states were serologically analyzed using the hemagglutination inhibition (HI) assay [58]. It was established that 28.3% of these animals were exposed to the cH1N1 virus, while 20.5% were exposed to the triple-reassortant-like H3N2 viruses. The HI assay results suggested that viruses antigenically related to the double-reassortment H3N2 virus have not become widespread among the U.S. swine population. In contrast, serodiagnosis of swine serum samples, as well as additional molecular sequencing analysis of six 1999 isolates that had a triple-reassortant genotype, suggested that the H3N2 viruses carrying the avian PA and PB2 genes had spread throughout most of the United States [58].
The presence of avian polymerase genes (PB2 and PA) in the triple-reassortant virus provided the majority of the diversity between the two viruses. The epidemiology of IAVs in swine from the U.S. drastically changed after 1998, when the triple-reassortant H3N2 viruses containing gene segments from the cH1N1 virus (NP, M, and NS), seasonal H3N2 human IAV subtype (PB1, HA, and NA), and avian IAV (PB2 and RA) successfully emerged in the swine population [54,58].
Since 1998, human H3N2 IAVs have been introduced into swine populations at least three times [59,60], resulting in the formation of phylogenetic clusters I, II, and III [61]. Cluster III (C-III) viruses actively spread in North America [62] and then evolved into variants of cluster IV (C-IV) over time [63], which contributed to the conservation and expansion of H3 IAV C-IV [64].
H3N2 viruses subsequently formed reassortants with swine H1N1 IAVs, acquiring the H1N1 or H1N2 subtypes [60,65]. Most fully characterized swine viruses contain the TRIG cassette regardless of their subtype. Reassortant H1 TRIG viruses, having become endemic, therefore continued to circulate in most swine-producing regions of the U.S. and Canada, including other drifting H3N2 variants [58,59,60,63,66], H1N2 [67], and reassortant IV rH1N1 [60,68,69,70,71].
Reassortments observed from 2000 to 2009 are associated with the HA and/or NA protein segments while maintaining the TRIG constellation, which comprised swine (M, NP, and NS gene segments), avian (PB2 and PA gene segments), and human IVs (PB1) [55,69,72,73,74].
The H3N1 viruses were sometimes recognized as bounded outbreaks but were not extensively circulating [75,76]. Furthermore, in 2006, TRIG was demonstrated to have taken the avian HA H2 and N3 variants, resulting in a new triple-reassortant H2N3 SIVs [77]. Yassine et al. [78] showed that North American swine TRIG-containing viruses can infect turkeys, which plays a certain role in IAV epidemiology in swine hosts and human hosts. Several authors reported that genetically related TRIG-containing swine viruses were also detected in Korea, Vietnam, and China [79,80,81].
There are reports on the introduction of IAV with the HA H1 gene of human H1N2 (hu-like H1) seasonal viruses, which are both molecularly and genetically different from the SIVs cH1N1 in Canada [82]. Since 2005, TRIG-containing hu-like viruses H1N1 and H1N2 have appeared in U.S. swine populations [83]. However, their TRIG genes are similar to those found in contemporary triple-reassortant SI viruses.
To better represent the evolution of H1 IAV circulating in North America, a cluster classification was proposed. cH1N1 IAVs, circulating among swine since 1918, evolved, based on the genetic composition of the HA gene, into contemporary a-, b-, and c-clusters, while strains with the H1 HA subtype most similar to seasonal human H1 IAVs form a d-cluster [83]. All four HA gene clusters (a, b, c, and d) are found with the NA genes of the N1 or N2 subtype. The HA genes from the d-cluster of viruses presumably originated from two different introductions of human seasonal viruses, with HA from the H1N2 and H1N1 viruses phylogenetically differentiated into two subclusters of swine origin, d1 and d2, respectively [69,84]. In strains isolated in the U.S. during 2008–2010, the d-cluster HA genes paired with either the N1 or the N2 IAV gene of human origin [85,86]. Until 2009, the H1 IAV evolved as a result of the reassortant formation while maintaining TRIG as the basis and generating genetically and antigenically different viruses [25,69]. Since then, H1N1pdm09 has become firmly established in the U.S. swine population with the emergence of subsequent second-generation reassortants [87,88,89].
A(H1N1)pdm09 IV has also become established in swine in North Carolina and co-circulates with previously enzootic SI virus strains, including avian H1N1 and human-like H1N2 [90]. In 2010, a reassortant IAV (H1N2r) was isolated that caused mild clinical disease in swine in North Carolina. This reassortant virus has a gene constellation, including the internal gene cassette of the A(H1N1)pdm09 viruses and HA and NA genes of the swine-origin H1N2 virus. This reassortant virus has been shown to have both zoonotic and reverse zoonotic potential and no apparent increased virulence or transmissibility compared to A(H1N1)pdm09 viruses [91,92].
During the swine influenza surveillance in 2012, a reassortant IAV strain containing HA and NA from human seasonal IAV was detected. These viruses subsequently reassorted, maintaining only the human-origin H3, which resulted in a novel lineage of viruses that became the most frequently revealed H3 branch in U.S. swine (2010.1 HA clade) [64,93,94]. Powell et al. [95] found that compared to the 2010.1 viruses from 2012 and 2014, the 2010.1 strains from the 2015–2017 period led to equivalent macroscopic lung lesions in swine. A single mutation in amino acid residue 145 in a previously identified HA antigenic motif was associated with a change in antigenic phenotype, potentially reducing vaccine effectiveness. The 2010.1 viruses circulating in swine after 2012 significantly differed from both the H3N2 viruses that existed prior to 2012 in swine and human seasonal influenza H3N2 viruses and showed continued evolution within this lineage [95].
In 2017, the Iowa State University Veterinary Diagnostic Laboratory detected a reverse zoonotic transmission of seasonal human IAV to swine in Oklahoma. A pairwise comparison between the human H3 IAV (H3.2010.2) HA sequences detected in swine and the most similar 2016–2017 seasonal human H3 sequences revealed 99.9% nucleotide identity [96]. H3.2010.2 is a phylogenetic clade of H3N2 circulating in swine. This clade became established following the spillover of a seasonal human H3N2 virus during the 2016–2017 influenza season. H3.2010.2 transmitted and adapted to the swine host and demonstrated reassortment with the internal genes from strains endemic to swine but maintained the human HA and NA. It is genetically and antigenically distinct from H3.2010.1 H3N2, detected earlier in 2010. It was shown that seasonal human-to-swine transmission of IAV became established in the population through adaptation and contributed to the genetic and antigenic diversity of IAV circulating in swine [96].
Exposure to IAVs and infectious individual swine assists in the introduction, transmission, and spread of diverse IAVs. Influenza strains circulating among swine can cause outbreaks in humans. In October 2020, a swine-origin H1N2 IAV variant was detected in a person in Alberta, Canada [97]. The source of the infection was a local swine farm, where a contact person worked in the household. A phylogenetic analysis showed that the isolate was very similar to the strains found on this farm in 2017.
Agricultural fairs also bring humans into close contact with swine and provide a unique interface for the zoonotic transmission of IAVs. Agricultural fairs are held every summer in the United States, where people are exposed to genetically diverse IAVs circulating among exhibition swine. As a result of such fairs, more than 450 laboratory-confirmed cases of zoonotic infections have been recorded since 2010 [98].
An understanding of the IAV transmission dynamics through exhibition swine is therefore critical to reducing the high incidence of various IAV variants registered in connection with agricultural fairs.
3.2. Circulation of Swine Influenza Viruses in South America
IAVs have been found in South American swine populations. Several IAV lineages are circulating among swine populations in Latin America, which apparently existed but remained undetected for many years; however, these lineages were established after the emergence of the seasonal human virus and the global movement of swine. The emergence of IAV with H1 HA from the classical clade of swine lineage has been reported in Mexico, including viruses with HA that share a common ancestor with the H1N1pdm2009 virus, as well as viruses with HA and NA belonging to the H1N1 lineage of Eurasian swine [99,100,101]. Two lineages of H3N2 IAVs were also detected in Mexico: one probably resulted from C-IV H3 importation from North America, and the second resulted from a unique seasonal introduction to humans since the mid-1990s [99].
A report was received from Argentina in 2009 in which clinical disease and the isolation of H1N1pdm IAV were recorded on a commercial swine farm. Later, in 2011, the reassortant H1N1, H1N2, and H3N2 IAVs with internal genes from the H1N1pdm virus were detected. Clinical, pathological, and virological evidence suggested that IV infection spread widely among swine farms in Argentina [102,103,104,105,106].
Since the emergence of H1N1pdm in 2009, IAV outbreaks have also been detected in Brazilian swine herds [99,100,107,108,109,110,111,112,113,114,115,116,117]. A total of 107 strains of H1N1, H1N2, and H3N2 IAVs were characterized during the surveillance of swine in Brazil from 2011 to 2020 [118,119]. A phylogenetic analysis based on surface protein segments (HA and NA) showed that since the mid-1980s, seasonal human IAVs have been introduced at least eight times into the swine population in Brazil. The analysis identified three H1 genetic clades within the 1B lineage and an H3 lineage that has diversified into three genetic clades. The N2 segment from seasonal human H1N2 and H3N2 IAVs was detected six times in swine, and the N1 segment from the human H1N1 virus was also identified once. An additional analysis revealed further reassortment with the H1N1pdm09 viruses, which confirmed the significant contribution of human IAVs to the genetic diversity of IAV in swine and the need for IAV surveillance in swine [118].
Swine H1N2 IAVs, which co-circulate with A(H1N1)pdm09-like viruses, have been recognized in Chile [120,121]. A genetic analysis based on the HA1 domain and an antigenic analysis by the HI assay demonstrated the presence of three antigenic clusters: Chilean H1A (ChH1A), Chilean H1B (ChH1B), and A(H1N1)pdm09-like. It was established that the antigenic sites of the ChH1A and ChH1B strains differed from those of commercial vaccine strains by 10–60% at the amino acid sequence level. These findings confirmed the importance of antigenic analyses and genetic testing to evaluate control measures such as vaccination. These results emphasized the need for ongoing monitoring to determine further transmission of antigenic variants in Chilean swine populations [99,122,123].
3.3. Circulation of Swine Influenza Viruses in Europe
SIV is one of the most significant primary causative agents of swine respiratory diseases in Europe. Three subtypes of IAVs, H1N1, H1N2, and H3N2, have been circulating in the European swine population for decades. These viruses remain endemic in the European swine populations, but they differ in antigenic characteristics from viruses circulating among swine elsewhere.
Since the mid-twentieth century, cH1N1 IAVs have been widely spread in European swine populations and North American swine herds. However, in the late 1970s, cH1N1 viruses were replaced by avian H1avN1 IAVs, which were transmitted from wild ducks to swine and later spread to Asia [124]. In early 1979, the emergence of avian H1avN1 IAVs was reported in Belgium [124] and, later, in Germany and France [125,126,127]. Subsequently, during a long-term 13-year SI surveillance in Germany, the persistent spread of the Eurasian H1avN1 virus was established, as well as the emergence of a novel H1N1 reassortant with human HA and avian NA genes. There has also been a report of a zoonotic infection caused by H1avN1 1C.2.2 IAV and detected in a German child [128]. In addition, the evolution of the A(H1N1)pdm09 antigenic drift variant has been observed [129].
In Serbia, in 2016–2018, during the monitoring of IAV circulation on 13 swine farms, 255 samples were obtained from animals. The IAV genome was detected in 24 samples. As a result of virological studies, eight strains of IAV were isolated. Sequence studies of the HA and NA genes revealed the circulation of H1N1 and H3N2 IAVs. IAVs with the subtype H1 were classified as 1C.2.1 and 1A.3.3.2. clades. The nucleotide sequences of the genes HA H1 showed that three viruses belong to the H1N1pdm09 lineage, and four belong to the Eurasian “avian-like” lineage H1avN1. Based on their NA gene sequences, these seven viruses belong to the Eurasian avian lineage H1avN1. The HA and NA genes of the H3N2 subtype virus belong to the A/swine/Gent/1/1984-like H3N2 lineage [130,131,132].
The genetic diversity of SIVs was investigated in northern Italy from 2017 to 2020. High genotypic diversity was found among the H1N1 and H1N2 strains, while the H3N2 strains showed a stable genetic pattern. The data obtained confirmed the need for continuous monitoring, since the evolution of IAVs could result in the emergence of isolates with zoonotic potential [133].
H1avN1 IAV has therefore become the dominant lineage on both continents and played a key role in the present-day ecology of endemic viruses, both in these regions and worldwide [134].
The European swine influenza H3N2 viruses of human lineage (A/Hong Kong/1/68) emerged in the 1980s, and they differed from the contemporary H3N2 viruses in North America [40,135,136,137]. They acquired the internal H1avN1 IAV gene cassette as a result of reassortment and had two surface protein genes (HA and NA) from the 1968 Hong Kong IAV and six internal genes (M, PB1, PB2, PA, NP, and NS) from an endemic avian H1N1 virus [138,139]. However, this virus rapidly disappeared from the European swine population and, in 1984, was replaced by another virus with a very similar genetic composition [140]. It quickly spread in the population of European swine. Epizootics have been recorded in Belgium [139,141], France [142], Germany [143,144], England [145], Italy [135,138], Spain [146,147], and the Netherlands [148,149].
H1N2 IAVs, first isolated in the UK, became widespread among European swine in the mid-1990s [150,151,152]. Swine human-like H1N2 (H1huN2) IAVs retained the genotype of reassortant H3N2 viruses but acquired the H1 gene from the seasonal human H1N1 virus of the 1980s. H1huN2 IAVs later spread in France (1997 and 2000–2018), Italy (1998), Belgium and the Netherlands (1999 and 2014–2019), Germany (2000, 2009, 2014–2015), and Denmark (2011–2014) [129,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166].
These three subtypes (H1avN1, H3N2, and H1huN2) therefore share internal genes but have clearly distinguishable HAs. All three IAV subtypes became established in the European swine populations and gradually replaced previously circulating cH1N1 IAV [48,134].
H1N1pdm09 IAV has also been prevalent in European swine since the 2009 pandemic [167,168,169,170]. The first reports of H1N1pdm09 IAV came from England and Northern Ireland in 2009 [167]. H1N1pdm09 IAVs were later detected in Finland [171,172] and Germany [169]. A swine reassortant, H1N2r IAV, was isolated in 2010, which caused mild clinical disease in swine in the United Kingdom. This reassortant virus had a novel gene constellation, including the internal gene cassette of the H1N1pdm09 viruses and HA and NA genes from swine-origin H1N2 [92]. In 2016, reports came from Greece about the detection of H1N1pdm09, H1N1, H1N2, and H3N2 IAVs in swine serums collected during 2002–2004 and 2010–2012 [173]. Serological surveillance in Norway from 2009 to 2017 also detected the circulation of H1N1pdm09, H1N1, H1N2, and H3N2 IAVs [174]. Data on the zoonotic transmission of H1N1pdm09 IAV from swine to humans appeared in Germany in 2009–2010 [175] and in France in 2018 [170].
As elsewhere, the introduction of H1N1pdm09 into European swine resulted in an increase in second-generation reassortants between human H1N1pdm09 and established endemic lineages [176,177,178,179]. The circulation of reassortant viruses was reported in the UK [180], Italy [181,182], Germany [157,169,175], France [183], Sweden [184], and Denmark [160,185]. In addition, certain unusual subtypes, such as H3N1 and H1N7 viruses, have been revealed in European swine, but they were apparently unable to survive and spread among swine [186,187,188].
The emergence of H1N1pdm09 and reassortment with endemic swine IAVs therefore significantly diversified the swine IAV ecology in Europe [175,180,181].
3.4. Circulation of Swine Influenza Viruses in Asia
SIV demonstrates a high level of diversity in Asian swine populations due to the introduction of viruses from Europe and North America [81,189]. It has been established that the first isolation of IAVs in Asia was in 1974 (swine/Hong Kong/1/74), although the first detection of cH1N1 was recorded in Chinese swine during the Spanish flu pandemic in 1918. SIV observations conducted in Hong Kong and Japan since the mid-1970s have shown that cH1N1 is widely spread in numerous Asian regions and countries [190,191]. cH1N1s have been discovered in Hong Kong, Japan, India, Taiwan, Singapore, Iran, Thailand, Korea, Vietnam, and Malaysia [181,192,193,194,195,196,197,198,199,200,201,202].
The stock of IAV genetic diversity in swine herds from China, South Korea, Vietnam, and other Asian countries has increased due to the intercontinental movement of swine and their viruses [45,203]. This led to the spread of the European H3N2 and H1avN1 viruses, along with North American TRIG viruses introduced in the late 1990s to early 2000s.
The findings of evolutionary studies have shown that cH1N1 SIV and the 1918 pandemic influenza H1N1 virus had a general ancestor or were very closely related [204,205,206]. The relationship between human and swine H1N1 viruses may be similar to that between human and H1N1pdm09 viruses. Since its appearance, H1N1pdm09 has been constantly transmitted from humans to swine [102,207]. In addition, the sporadic isolation of human-like H1N1 viruses has been observed in swine in Japan [208] and, later, in China [209,210].
The H1N1pdm09 virus was introduced to swine and reassorted with the endemic subtypes in China [57], Japan [211,212], and Thailand [213]. In many Asian countries, H1N1, H1N2, and H3N2 viruses included different internal gene segments from the H1N1pdm09 virus [57,195,214,215,216,217,218,219,220,221,222,223,224,225,226,227], resulting in numerous highly complex reassortant viruses [81,217,225,227,228,229].
Avian influenza H1avN1viruses were detected along with cH1N1 in clinically healthy swine in Southern China in 1993 [198,215]. Later, in 2001, H1avN1 viruses that caused the avian influenza Eurasian variant were detected in swine in Hong Kong and Thailand [45,230].
In Asia, the circulation of reassortant H1N1 swine influenza viruses of North American and/or European origin has been recorded among swine. Reports of H1N1 swine reassortant virus circulation came from Korea [79,231], Thailand [230,232], and Malaysia [233]. There is serological evidence for the prevalence of H1N1 SIVs in Cambodian swine in 2006–2010 [234]. H1N1 SIVs were recorded in Bhutan in 2011–2012 [235]. H1N1 SIVs were detected in Russia in 2016 [236]. A genetic analysis of the H1N1 virus isolate revealed 90% identity similarity between HA and NA genes and those found in U.S. humans in the 1980s.
The first outbreak of influenza in swine caused by reassortant H1N2 viruses determined from the human-like variant of swine H3N2 and cH1N1s viruses was observed in Japan in 1978; later, outbreaks were recorded during 1989–1990 [237].
These viruses, endemic in Japanese swine populations, established a genetically stable lineage [238]. A similar reassortant H1N2 IAV gene cassette was detected in swine in Taiwan [239], China [45,240], and India [241].
Since 2002, a genetically stable line of North American reassortant H1N2 viruses containing HA cH1N1s has become widespread in Korea [79].
Since the 1970s, in Asia, the H3N2 influenza virus has been transmitted from humans to swine on multiple occasions, and the first report of the virus being isolated from swine in Taiwan came a year after the human pandemic in Hong Kong [242]. Outbreaks of viral infection caused by H3N2 IAVs in swine have also been reported in Hong Kong, Japan, Korea, India, and Thailand [189,243,244,245,246,247,248,249,250], but these H3N2 viruses could not become established in swine herds [214,251].
In the 1980s, swine H3N2 viruses were isolated in China, containing human-like HA and NA and the cH1N1 internal gene segments. At about the same time, two triple-reassortant H3N2 viruses were isolated from swine in China that were not genetically related to the North American or European reassortant viruses. In 1999, SIVs appeared in China: the European reassortants of human-like H3N2 SIVs. Further reassortment of these viruses with circulating cH1N1 influenza viruses subsequently resulted in triple-reassortant strains [214]. In addition, the European reassortant human-like H3N2 SIVs were genetically reassorted with circulating human H1 or H3 strains in Thailand [230,232]. The circulation of European viruses in swine was confirmed by serological studies in Malaysia [233]. Since 1998, SIV strains have been isolated in Korea, which are triple-reassortant H3N2 IAVs of the North American swine lineage with three different human-like HAs [79,252]. The swine showed signs of respiratory disease, but no deaths were recorded. There are also several reports on the prevalence of the H3N2 virus in swine in Sri Lanka during 2004–2005 [253], in Laos during 2009 [254], and in Cambodia during 2011–2012 [255].
Influenza epizootics among swine represent a serious problem in the Republic of Kazakhstan. In 1984, due to a virological examination of piglets with clinical signs of respiratory diseases on swine farms situated in East Kazakhstan, three strains of the A/H1N1 influenza virus were isolated [256]. Due to the nature of the interaction with a panel of monoclonal antibodies, the Kazakhstan isolates showed similarities with the A/England/333/80 (H1N1) virus. The performed immunological analysis made it possible to classify the strains isolated from swine as drift variants of the human influenza A(H1N1) virus.
Reassortant A/H3N6 strains were also isolated from swine in Kazakhstan in 1986. The viruses of this complex were characterized by significant antigenic heterogeneity. The populations of several strains contained virions whose enzymatic activity was suppressed by immune serum against NA N6 [256].
In 2008–2009, nine isolates collected from uneven-aged swine belonging to peasant farms of the Republic of Kazakhstan were identified, of which four had A(H1N1), one had A(HswN1), and four had the A/H3N2 antigenic formula. A serological analysis performed to retrospectively confirm the etiological role of influenza viruses showed the presence of antibodies against influenza A/H1N1, A/HswN1, and A/H3N2 viruses in animals, which completely coincided with the results obtained from the isolation of viruses in chicken embryos [256].
In 2013–2014, a serological analysis of 492 blood serums collected from swine belonging to peasant farms in various regions of Kazakhstan was conducted. Data obtained in the study indicated the co-circulation of influenza A/H1N1 and A/H3N2 viruses among animals during the specified period [256].
There are also data on the circulation of H1N1 and H3N2 SIVs among the swine population in Kazakhstan during 2017–2022 [256,257,258,259].
A genetic analysis of the H1N1 IAV strain, isolated in Kazakhstan in 2020, showed that it belongs to clade 1A.3.2.2, lineage 1A. This clade has a wide global distribution, which includes the 2009 H1N1 pandemic strains [18]. The findings emphasized the importance of ongoing SIV monitoring to identify the possibility of the cross-species transmission of this infectious agent.
Thus, the IVs that infect swine most frequently belong to the H1N1 subtype, but at the same time, the H1N2, H3N1, and H3N2 variants are circulating among them. Sometimes, swine are infected with more than one virus simultaneously, resulting in a reassortant strain containing genes from different sources. Reassortment plays a significant role in the emergence of novel virus variants, particularly in the origin of pandemic strains.
3.5. Circulation of Swine Influenza Viruses in Africa and Australia
H1N1pdm09, H1N1, and H3N2 IAVs have been reported in various African countries [260,261], such as Egypt [262,263], Nigeria [264,265,266,267,268], Ghana [269,270], Cameroon [271,272,273], Kenya [274,275,276], Réunion Island [277], Uganda [278], Togo [279], and Burkina Faso [280]. In addition to H1N1pdm09, the circulation of avian influenza viruses such as H5N1, H5N2, and H9N2 has been recorded in Nigeria and Egypt [262,267].
Swine influenza in Australia was first reported in 2009, when a herd of 300 sows was infected with the H1N1pdm09 virus. The outbreak occurred as a result of human-to-swine virus transmission [281,282]. The outbreaks of influenza H1N1pdm were detected on three swine farms in three different states of Australia. Further analysis of the outbreak in Queensland resulted in the identification of two distinct IAV strains in swine. Two employees working in the same piggery were also infected with the same two strains found in swine. Although no reassortment was identified in the above cases, the ability of these viruses to be transmitted between swine and humans was evident, emphasizing the importance of monitoring swine to identify novel influenza infections [282].
An outbreak of SI occurred in Western Australia in 2012 [283], and the infection spread among both swine and farm workers. A molecular analysis of the isolates revealed the circulation of H1N2, H3N2, and H1N1pdm09 IAVs. H1N2, H3N2, and H1N1 SIVs later continued to circulate in Western Australia and Southern Queensland right up to 2016 [284]. A public health investigation identified an unrecognized zoonotic risk and the potential threat of a pandemic in the future [283].
In 2018, there was a report of a 15-year-old teenage girl from Australia infected with A(H3N2) SIV. The virus contained the HA and NA genes derived from the 1990s-like seasonal human viruses and internal protein genes from H1N1pdm09 IAV, emphasizing the potential risk to human health in Australia [285].
4. Circulation of Avian Influenza Viruses in Swine
H1N1 and H3N2 IAVs mainly circulate among swine populations in North America, Europe, and Asia. However, there have been no cases of transmission of the 1957 Asian pandemic H2N2 virus to swine. There is also cross-species transmission of avian viruses to swine on these continents. The reassortant SIVs presently circulating have at least one component of the avian virus gene. Although natural infections of swine with avian influenza viruses have been reported in numerous parts of the world, only the European avian H1avN1 virus has adapted to swine [166,286,287,288]. Other subtypes of avian influenza virus have been found sporadically in swine in North America and Asia. There are reports of sporadic isolation of atypical H1N7 and H3N1 IAVs from swine in Europe [186,287] and the H4N6, H3N3, H3N1, and H2N3 subtypes in North America [75,77,289]. Numerous interspecies transmissions of H9N2 viruses from host birds to swine occurred in Asia [290]. H9N2 IAVs isolated from swine in Hong Kong in 1998 were fully avian IAV variants [249,290,291]. However, since 2003, reassortant avian strains have emerged with certain genes closely associated with the highly pathogenic avian influenza (HPAI) H5N1virus [292,293,294,295,296]. Infection with the avian H5N1 virus was also detected in swine in Indonesia [297] and Vietnam [231]. The avian H5N2 virus has been transmitted from wild migratory birds to Korean swine [298].
H3N1 SIV, which is a reassortant virus with genes of the human-like H3N2 isolate and circulating cH1N1SIVs, has been isolated from swine in Taiwan [239]. There are data on the circulation of H3N1 SIVs in Indonesia and Korea [299]. H3N1 SIVs circulating in Korea represented a reassortment of co-circulating North American reassortant H1N1 and H3N2 viruses [299].
Equine-like H3N8 SIVs were also isolated from swine in Central China [300].
Thus, some avian IAVs, such as H9N2, H5N1, H5N2, H4N8, and H6N6, as well as equine H3N8 IAV, have been sporadically detected in swine from certain Asian countries [81,189,300,301,302]. The simultaneous circulation of so many genetically diverse IAVs in swine explains the very complex ecology of swine IAVs in Asia.
5. Conclusions
Swine play an important role in the formation of present-day influenza ecology. Swine have specific receptors for avian, porcine, and human IAV in the tracheal epithelium, which may contribute to the emergence of new reassortant variants that could potentially pose a public health threat [8,249,303]. The complex interaction of IAVs of human, avian, and porcine evolutionary origin in swine is supported by the detection of multiple IAV subtypes in swine populations, including H1, H2, H3, H4, H5, H7, and H9 [304]. Swine play an important role in the epidemiology of IV, which is dangerous to humans [305]. For example, influenza A(H1N1)pdm09 is thought to have been present in swine herds for several months before it became a pandemic strain in humans [35].
Human-to-swine transmission of IV occurs much more frequently than swine-to-human transmission [30]. Despite the genetic diversity of SIVs found in the swine population, only H1 and H3 IAVs formed stable lineages similar to human IVs. The nature of H1 and H3 viruses nevertheless differs between the two host populations. In addition, the distribution of subtypes and genotypes of endemic IAVs varies greatly by geographic region due to the repeated introduction of avian and human influenza viruses. An exception is H1N1pdm09, which is mainly widespread due to multiple human-to-swine reintroductions [57,102,167,176,306,307,308,309,310,311,312]. New cases of infection with avian influenza H5N1 virus have been confirmed in dairy cattle and pigs [313,314]. However, no information has been found on the presence of swine-origin IVs in dairy cows.
The continuous monitoring and genome-wide genetic characterization of SIVs make it possible to obtain a complete picture of the infectious disease process, predict the epidemiological and epizootic situation, and choose the right strategy and tactics for preventive and anti-epidemic measures. Understanding the transmission modes is of decisive importance for developing effective control strategies and making reasonable surveillance recommendations.
This study may provide the information needed to develop a more effective method for monitoring SIVs and selecting countermeasures to prevent interspecies transmission, which could become a potential pre-pandemic situation.
Author Contributions
Conceptualization, N.K. and R.W.; Methodology, N.K. and R.W.; Software, G.L. and N.S.; Validation, T.G., N.S. and N.O.; Formal Analysis, R.W., N.K., T.G. and G.L.; Investigation, N.K., R.W. and N.O.; Resources, N.S., T.G., N.O. and G.L.; Data Curation, N.K. and R.W.; Writing—Original Draft Preparation, N.K.; Writing—Review and Editing, R.W. and N.K.; Visualization, R.W.; Supervision, R.W.; Project Administration, N.K.; Funding Acquisition, N.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of the Republic of Kazakhstan under the following project: “Detection of transmission of influenza viruses at the human-animal interface in Kazakhstan” (Grant No. AP19677931).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Knipe, D.; Howley, P. Orthomyxoviridae: The Viruses and Their Replication. Fields Virol. 2020, 1, 596–648. [Google Scholar]
- Encinas, P.; Del Real, G.; Dutta, J.; Khan, Z.; van Bakel, H.; Del Burgo, M.Á.M.; García-Sastre, A.; Nelson, M.I. Evolution of influenza A virus in intensive and free-range swine farms in Spain. Virus Evol. 2021, 7, veab099. [Google Scholar] [CrossRef] [PubMed]
- Simon-Grifé, M.; Martín-Valls, G.E.; Vilar, M.J.; García-Bocanegra, I.; Mora, M.; Martín, M.; Mateu, E.; Casal, J. Seroprevalence and risk factors of swine influenza in Spain. Vet. Microbiol. 2011, 149, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Parys, A.; Vereecke, N.; Vandoorn, E.; Theuns, S.; Van Reeth, K. Surveillance and Genomic Characterization of Influenza A and D Viruses in Swine, Belgium and the Netherlands, 2019–2021. Emerg. Infect. Dis. 2023, 29, 1459–1464. [Google Scholar] [CrossRef]
- Nakatsu, S.; Murakami, S.; Shindo, K.; Horimoto, T.; Sagara, H.; Noda, T.; Kawaoka, Y. Influenza C and D Viruses Package Eight Organized Ribonucleoprotein Complexes. J. Virol. 2018, 92, e02084-17. [Google Scholar] [CrossRef]
- Fouchier, R.A.M.; Munster, V.; Wallensten, A.; Bestebroer, T.M.; Herfst, S.; Smith, D.; Rimmelzwaan, G.F.; Olsen, B.; Osterhaus, A.D.M.E. Characterization of a Novel Influenza A Virus Hemagglutinin Subtype (H16) Obtained from Black-Headed Gulls. J. Virol. 2005, 79, 2814–2822. [Google Scholar] [CrossRef]
- Röhm, C.; Zhou, N.; Süss, J.; Mackenzie, J.; Webster, R.G. Characterization of a Novel Influenza Hemagglutinin, H15: Criteria for Determination of Influenza A Subtypes. Virology 1996, 217, 508–516. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and Ecology of Influenza A Viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, Y.; Tefsen, B.; Shi, Y.; Gao, G.F. Bat-Derived Influenza-like Viruses H17N10 and H18N11. Trends Microbiol. 2014, 22, 183–191. [Google Scholar] [CrossRef]
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.A.; Chen, L.-M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A Distinct Lineage of Influenza A Virus from Bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New World Bats Harbor Diverse Influenza A Viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shen, H.; McDowell, C.; Liu, Q.; Duff, M.; Lee, J.; Lang, Y.; Hesse, D.; Richt, J.A.; Ma, W. Virus Survival and Fitness When Multiple Genotypes and Subtypes of Influenza A Viruses Exist and Circulate in Swine. Virology 2019, 532, 30–38. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Gordon, M.L. A Systematic Review of Influenza A Virus Prevalence and Transmission Dynamics in Backyard Swine Populations Globally. Porc. Health Manag. 2022, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.P.; Gordon, M.L. A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide. Pathogens 2020, 9, 355. [Google Scholar] [CrossRef] [PubMed]
- Borland, S.; Gracieux, P.; Jones, M.; Mallet, F.; Yugueros-Marcos, J. Influenza A Virus Infection in Cats and Dogs: A Literature Review in the Light of the “One Health” Concept. Front. Public Health 2020, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Klivleyeva, N.G.; Glebova, T.I.; Shamenova, M.G.; Saktaganov, N.T. Influenza A Viruses Circulating in Dogs: A Review of the Scientific Literature. Open Vet. J. 2022, 12, 676–687. [Google Scholar] [CrossRef]
- Klivleyeva, N.G.; Ongarbayeva, N.S.; Baimukhametova, A.M.; Saktaganov, N.T.; Lukmanova, G.V.; Glebova, T.I.; Sayatov, M.K.; Berezin, V.E.; Nusupbaeva, G.E.; Aikimbayev, A.M. Detection of Influenza Virus and Pathogens of Acute Respiratory Viral Infections in Population of Kazakhstan during 2018-2019 Epidemic Season. Russ. J. Infect. Immun. 2021, 11, 137–147. [Google Scholar] [CrossRef]
- Klivleyeva, N.G.; Ongarbayeva, N.S.; Korotetskiy, I.S.; Glebova, T.I.; Saktaganov, N.T.; Shamenova, M.G.; Baimakhanova, B.B.; Shevtsov, A.B.; Amirgazin, A.; Berezin, V.E.; et al. Coding-Complete Genome Sequence of Swine Influenza Virus Isolate A/Swine/Karaganda/04/2020 (H1N1) from Kazakhstan. Microbiol. Resour. Announc. 2021, 10, e0078621. [Google Scholar] [CrossRef]
- Glebova, T.; Klivleyeva, N.; Saktaganov, N.; Shamenova, M.; Lukmanova, G.; Baimukhametova, A.; Baiseiit, S.; Ongarbayeva, N.; Orynkhanov, K.; Ametova, A.; et al. Circulation of Influenza Viruses in the Dog Population in Kazakhstan (2023–2024). Open Vet. J. 2024, 14, 1896–1904. [Google Scholar] [CrossRef]
- Ma, W.; Kahn, R.E.; Richt, J.A. The Pig as a Mixing Vessel for Influenza Viruses: Human and Veterinary Implications. J. Mol. Genet. Med. 2008, 3, 158–166. [Google Scholar] [CrossRef]
- Tao, H.; Li, L.; White, M.C.; Steel, J.; Lowen, A.C. Influenza A Virus Coinfection through Transmission Can Support High Levels of Reassortment. J. Virol. 2015, 89, 8453–8461. [Google Scholar] [CrossRef] [PubMed]
- Canini, L.; Holzer, B.; Morgan, S.; Hemmink, J.D.; Clark, B.; Woolhouse, M.E.J.; Tchilian, E.; Charleston, B. Timelines of Infection and Transmission Dynamics of H1N1pdm09 in Swine. PLoS Pathog. 2020, 16, e1008628. [Google Scholar] [CrossRef] [PubMed]
- Salomon, R.; Webster, R.G. The Influenza Virus Enigma. Cell 2009, 136, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Lager, K.M.; Vincent, A.L.; Janke, B.H.; Gramer, M.R.; Richt, J.A. The Role of Swine in the Generation of Novel Influenza Viruses. Zoonoses Public Health 2009, 56, 326–337. [Google Scholar] [CrossRef]
- Vincent, A.; Awada, L.; Brown, I.; Chen, H.; Claes, F.; Dauphin, G.; Donis, R.; Culhane, M.; Hamilton, K.; Lewis, N.; et al. Review of Influenza A Virus in Swine Worldwide: A Call for Increased Surveillance and Research. Zoonoses Public Health 2014, 61, 4–17. [Google Scholar] [CrossRef]
- Lindstrom, S.; Garten, R.; Balish, A.; Shu, B.; Emery, S.; Berman, L.; Barnes, N.; Sleeman, K.; Gubareva, L.; Villanueva, J.; et al. Human Infections with Novel Reassortant Influenza A(H3N2)v Viruses, United States, 2011. Emerg. Infect. Dis. 2012, 18, 834–837. [Google Scholar] [CrossRef]
- Nelson, M.I.; Gramer, M.R.; Vincent, A.L.; Holmes, E.C. Global Transmission of Influenza Viruses from Humans to Swine. J. Gen. Virol. 2012, 93, 2195–2203. [Google Scholar] [CrossRef]
- Nelson, M.I.; Vincent, A.L.; Kitikoon, P.; Holmes, E.C.; Gramer, M.R. Evolution of Novel Reassortant A/H3N2 Influenza Viruses in North American Swine and Humans, 2009–2011. J. Virol. 2012, 86, 8872–8878. [Google Scholar] [CrossRef]
- Nelson, M.I.; Wentworth, D.E.; Culhane, M.R.; Vincent, A.L.; Viboud, C.; LaPointe, M.P.; Lin, X.; Holmes, E.C.; Detmer, S.E. Introductions and Evolution of Human-Origin Seasonal Influenza A Viruses in Multinational Swine Populations. J. Virol. 2014, 88, 10110–10119. [Google Scholar] [CrossRef]
- Nelson, M.I.; Vincent, A.L. Reverse Zoonosis of Influenza to Swine: New Perspectives on the Human–Animal Interface. Trends Microbiol. 2015, 23, 142–153. [Google Scholar] [CrossRef]
- Kasowski, E.J.; Garten, R.J.; Bridges, C.B. Influenza Pandemic Epidemiologic and Virologic Diversity: Reminding Ourselves of the Possibilities. Clin. Infect. Dis. 2011, 52, S44–S49. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.H. History and Epidemiology of Swine Influenza in Europe. Curr. Top. Microbiol. Immunol. 2013, 370, 133–146. [Google Scholar] [CrossRef]
- Mostafa, A.; Abdelwhab, E.; Mettenleiter, T.; Pleschka, S. Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview. Viruses 2018, 10, 497. [Google Scholar] [CrossRef] [PubMed]
- Ma, W. Swine Influenza Virus: Current Status and Challenge. Virus Res. 2020, 288, 198118. [Google Scholar] [CrossRef]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef]
- Smith, G.J.D.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and Evolutionary Genomics of the 2009 Swine-Origin H1N1 Influenza A Epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef]
- Ito, T.; Couceiro, J.N.S.S.; Kelm, S.; Baum, L.G.; Krauss, S.; Castrucci, M.R.; Donatelli, I.; Kida, H.; Paulson, J.C.; Webster, R.G.; et al. Molecular Basis for the Generation in Pigs of Influenza A Viruses with Pandemic Potential. J. Virol. 1998, 72, 7367–7373. [Google Scholar] [CrossRef]
- Scholtissek, C.; Bürger, H.; Bachmann, P.A.; Hannoun, C. Genetic Relatedness of Hemagglutinins of the H1 Subtype of Influenza a Viruses Isolated from Swine and Birds. Virology 1983, 129, 521–523. [Google Scholar] [CrossRef]
- Scholtissek, C. Pigs as “Mixing Vessels” for the Creation of New Pandemic Influenza A Viruses. Med. Princ. Pract. 1990, 2, 65–71. [Google Scholar] [CrossRef]
- Van Reeth, K.; Brown, I.; Olsen, C. Influenza Virus. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; John Wiley & Sons, Inc.: Ames, IA, USA, 2012; pp. 557–571. [Google Scholar]
- Myers, K.P.; Olsen, C.W.; Gray, G.C. Cases of Swine Influenza in Humans: A Review of the Literature. Clin. Infect. Dis. 2007, 44, 1084–1088. [Google Scholar] [CrossRef]
- Nelson, M.I.; Stratton, J.; Killian, M.L.; Janas-Martindale, A.; Vincent, A.L. Continual Reintroduction of Human Pandemic H1N1 Influenza A Viruses into Swine in the United States, 2009 to 2014. J. Virol. 2015, 89, 6218–6226. [Google Scholar] [CrossRef] [PubMed]
- Löndt, B.Z.; Brookes, S.M.; Nash, B.J.; Núñez, A.; Stagg, D.A.; Brown, I.H. The Infectivity of Pandemic 2009 H1N1 and Avian Influenza Viruses for Pigs: An Assessment by Ex Vivo Respiratory Tract Organ Culture*. Influenza Other Respir. Viruses 2013, 7, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Detmer, S.E.; Wentworth, D.E.; Tan, Y.; Schwartzbard, A.; Halpin, R.A.; Stockwell, T.B.; Lin, X.; Vincent, A.L.; Gramer, M.R.; et al. Genomic Reassortment of Influenza A Virus in North American Swine, 1998–2011. J. Gen. Virol. 2012, 93, 2584–2589. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Smith, G.J.D.; Pybus, O.G.; Zhu, H.; Bhatt, S.; Poon, L.L.M.; Riley, S.; Bahl, J.; Ma, S.K.; Cheung, C.L.; et al. Long-Term Evolution and Transmission Dynamics of Swine Influenza A Virus. Nature 2011, 473, 519–522. [Google Scholar] [CrossRef]
- Kitikoon, P.; Nelson, M.I.; Killian, M.L.; Anderson, T.K.; Koster, L.; Culhane, M.R.; Vincent, A.L. Genotype Patterns of Contemporary Reassorted H3N2 Virus in US Swine. J. Gen. Virol. 2013, 94, 1236–1241. [Google Scholar] [CrossRef]
- Anderson, T.K.; Nelson, M.I.; Kitikoon, P.; Swenson, S.L.; Korslund, J.A.; Vincent, A.L. Population Dynamics of Cocirculating Swine Influenza A Viruses in the United States from 2009 to 2012. Influenza Other Respir. Viruses 2013, 7, 42–51. [Google Scholar] [CrossRef]
- Simon, G.; Larsen, L.E.; Dürrwald, R.; Foni, E.; Harder, T.; Van Reeth, K.; Markowska-Daniel, I.; Reid, S.M.; Dan, A.; Maldonado, J.; et al. European Surveillance Network for Influenza in Pigs: Surveillance Programs, Diagnostic Tools and Swine Influenza Virus Subtypes Identified in 14 European Countries from 2010 to 2013. PLoS ONE 2014, 9, e115815. [Google Scholar] [CrossRef]
- Koen, J.S. A Practical Method for Field Diagnosis of Swine Diseases. Am. J. Vet. Med. 1919, 14, 468–470. [Google Scholar]
- Shope, R.E. Swine Influenza: III. Filtration Experiments and Etiology. J. Exp. Med. 1931, 54, 373–385. [Google Scholar] [CrossRef]
- Detmer, S.E.; Patnayak, D.P.; Jiang, Y.; Gramer, M.R.; Goyal, S.M. Detection of Influenza a Virus in Porcine Oral Fluid Samples. J. Vet. Diagn. Investig. 2011, 23, 241–247. [Google Scholar] [CrossRef]
- Easterday, B.; van Reeth, K. Swine Influenza. In Diseases of Swine; Straw, B., D’Allaire, S., Mengeling, W., Taylor, D., Eds.; Blackwell Sciences: Ames, IA, USA, 1999; pp. 277–290. [Google Scholar]
- Leuwerke, B.; Kitikoon, P.; Evans, R.; Thacker, E. Comparison of Three Serological Assays to Determine the Cross-Reactivity of Antibodies from Eight Genetically Diverse U.S. Swine Influenza Viruses. J. Vet. Diagn. Investig. 2008, 20, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.N.; Senne, D.A.; Landgraf, J.S.; Swenson, S.L.; Erickson, G.; Rossow, K.; Liu, L.; Yoon, K.; Krauss, S.; Webster, R.G. Genetic Reassortment of Avian, Swine, and Human Influenza A Viruses in American Pigs. J. Virol. 1999, 73, 8851–8856. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Richt, J.A. Chapter 3 Swine Influenza Viruses: A North American Perspective. Adv. Virus Res. 2008, 72, 127–154. [Google Scholar]
- Bastien, N.; Antonishyn, N.A.; Brandt, K.; Wong, C.E.; Chokani, K.; Vegh, N.; Horsman, G.B.; Tyler, S.; Graham, M.R.; Plummer, F.A.; et al. Human Infection with a Triple Reassortant Swine Influenza A(H1N1) Virus Containing the Hemagglutinin and Neuraminidase Genes of Seasonal Influenza Virus. J. Infect. Dis. 2010, 201, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Vijaykrishna, D.; Poon, L.L.M.; Zhu, H.C.; Ma, S.K.; Li, O.T.W.; Cheung, C.L.; Smith, G.J.D.; Peiris, J.S.M.; Guan, Y. Reassortment of Pandemic H1N1/2009 Influenza A Virus in Swine. Science 2010, 328, 1529. [Google Scholar] [CrossRef]
- Webby, R.J.; Swenson, S.L.; Krauss, S.L.; Gerrish, P.J.; Goyal, S.M.; Webster, R.G. Evolution of Swine H3N2 Influenza Viruses in the United States. J. Virol. 2000, 74, 8243–8251. [Google Scholar] [CrossRef]
- Richt, J.A.; Lager, K.M.; Janke, B.H.; Woods, R.D.; Webster, R.G.; Webby, R.J. Pathogenic and Antigenic Properties of Phylogenetically Distinct Reassortant H3N2 Swine Influenza Viruses Cocirculating in the United States. J. Clin. Microbiol. 2003, 41, 3198–3205. [Google Scholar] [CrossRef]
- Webby, R.J.; Rossow, K.; Erickson, G.; Sims, Y.; Webster, R. Multiple Lineages of Antigenically and Genetically Diverse Influenza A Virus Co-Circulate in the United States Swine Population. Virus Res. 2004, 103, 67–73. [Google Scholar] [CrossRef]
- Vincent, A.L.; Anderson, T.K.; Lager, K.M. A Brief Introduction to Influenza A Virus in Swine. In Animal Influenza Virus; Humana Press: New York, NY, USA, 2020; pp. 249–271. [Google Scholar]
- Gramer, M.R.; Lee, J.H.; Choi, Y.K.; Goyal, S.M.; Joo, H.S. Serologic and Genetic Characterization of North American H3N2 Swine Influenza A Viruses. Can. J. Vet. Res. 2007, 71, 201–206. [Google Scholar]
- Olsen, C.W.; Karasin, A.I.; Carman, S.; Li, Y.; Bastien, N.; Ojkic, D.; Alves, D.; Charbonneau, G.; Henning, B.M.; Low, D.E.; et al. Triple Reassortant H3N2 Influenza A Viruses, Canada, 2005. Emerg. Infect. Dis. 2006, 12, 1132–1135. [Google Scholar] [CrossRef]
- Walia, R.R.; Anderson, T.K.; Vincent, A.L. Regional Patterns of Genetic Diversity in Swine Influenza A Viruses in the United States from 2010 to 2016. Influenza Other Respir. Viruses 2019, 13, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Karasin, A.I.; Landgraf, J.; Swenson, S.; Erickson, G.; Goyal, S.; Woodruff, M.; Scherba, G.; Anderson, G.; Olsen, C.W. Genetic Characterization of H1N2 Influenza A Viruses Isolated from Pigs throughout the United States. J. Clin. Microbiol. 2002, 40, 1073–1079. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Karasin, A.I.; Schutten, M.M.; Cooper, L.A.; Smith, C.B.; Subbarao, K.; Anderson, G.A.; Carman, S.; Olsen, C.W. Genetic Characterization of H3N2 Influenza Viruses Isolated from Pigs in North America, 1977-1999: Evidence for Wholly Human and Reassortant Virus Genotypes. Virus Res. 2000, 68, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Goyal, S.M.; Farnham, M.W.; Joo, H.S. Phylogenetic Analysis of H1N2 Isolates of Influenza A Virus from Pigs in the United States. Virus Res. 2002, 87, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Evseenko, V.A.; Boon, A.C.M.; Brockwell-Staats, C.; Franks, J.; Rubrum, A.; Daniels, C.S.; Gramer, M.R.; Webby, R.J. Genetic Composition of Contemporary Swine Influenza Viruses in the West Central Region of the United States of America. Influenza Other Respir. Viruses 2011, 5, 188–197. [Google Scholar] [CrossRef]
- Lorusso, A.; Vincent, A.L.; Harland, M.L.; Alt, D.; Bayles, D.O.; Swenson, S.L.; Gramer, M.R.; Russell, C.A.; Smith, D.J.; Lager, K.M.; et al. Genetic and Antigenic Characterization of H1 Influenza Viruses from United States Swine from 2008. J. Gen. Virol. 2011, 92, 919–930. [Google Scholar] [CrossRef]
- Nfon, C.K.; Berhane, Y.; Hisanaga, T.; Zhang, S.; Handel, K.; Kehler, H.; Labrecque, O.; Lewis, N.S.; Vincent, A.L.; Copps, J.; et al. Characterization of H1N1 Swine Influenza Viruses Circulating in Canadian Pigs in 2009. J. Virol. 2011, 85, 8667–8679. [Google Scholar] [CrossRef]
- Nfon, C.; Berhane, Y.; Zhang, S.; Handel, K.; Labrecque, O.; Pasick, J. Molecular and Antigenic Characterization of Triple-Reassortant H3N2 Swine Influenza Viruses Isolated from Pigs, Turkey and Quail in Canada. Transbound. Emerg. Dis. 2011, 58, 394–401. [Google Scholar] [CrossRef]
- De Jong, J.C.; Van Nieuwstadt, A.P.; Kimman, T.G.; Loeffen, W.L.A.; Bestebroer, T.M.; Bijlsma, K.; Verweij, C.; Osterhaus, A.D.M.E.; Claas, E.C.J. Antigenic Drift in Swine Influenza H3 Haemagglutinins with Implications for Vaccination Policy. Vaccine 1999, 17, 1321–1328. [Google Scholar] [CrossRef]
- Lewis, N.S.; Anderson, T.K.; Kitikoon, P.; Skepner, E.; Burke, D.F.; Vincent, A.L. Substitutions near the Hemagglutinin Receptor-Binding Site Determine the Antigenic Evolution of Influenza A H3N2 Viruses in U.S. Swine. J. Virol. 2014, 88, 4752–4763. [Google Scholar] [CrossRef]
- Abente, E.J.; Santos, J.; Lewis, N.S.; Gauger, P.C.; Stratton, J.; Skepner, E.; Anderson, T.K.; Rajao, D.S.; Perez, D.R.; Vincent, A.L. The Molecular Determinants of Antibody Recognition and Antigenic Drift in the H3 Hemagglutinin of Swine Influenza A Virus. J. Virol. 2016, 90, 8266–8280. [Google Scholar] [CrossRef] [PubMed]
- Lekcharoensuk, P.; Lager, K.M.; Vemulapalli, R.; Woodruff, M.; Vincent, A.L.; Richt, J.A. Novel Swine Influenza Virus Subtype H3N1, United States. Emerg. Infect. Dis. 2006, 12, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Gramer, M.; Rossow, K.; Yoon, K.-J. Isolation and Genetic Characterization of New Reassortant H3N1 Swine Influenza Virus from Pigs in the Midwestern United States. J. Virol. 2006, 80, 5092–5096. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Vincent, A.L.; Gramer, M.R.; Brockwell, C.B.; Lager, K.M.; Janke, B.H.; Gauger, P.C.; Patnayak, D.P.; Webby, R.J.; Richt, J.A. Identification of H2N3 Influenza A Viruses from Swine in the United States. Proc. Natl. Acad. Sci. USA 2007, 104, 20949–20954. [Google Scholar] [CrossRef]
- Yassine, H.M.; Al-Natour, M.Q.; Lee, C.W.; Saif, Y.M. Interspecies and Intraspecies Transmission of Triple Reassortant H3N2 Influenza A Viruses. Virol. J. 2007, 4, 129. [Google Scholar] [CrossRef]
- Pascua, P.N.Q.; Song, M.-S.; Lee, J.H.; Choi, H.-W.; Han, J.H.; Kim, J.-H.; Yoo, G.-J.; Kim, C.-J.; Choi, Y.-K. Seroprevalence and Genetic Evolutions of Swine Influenza Viruses under Vaccination Pressure in Korean Swine Herds. Virus Res. 2008, 138, 43–49. [Google Scholar] [CrossRef]
- Ngo, L.T.; Hiromoto, Y.; Pham, V.P.; Le, H.T.H.; Nguyen, H.T.; Le, V.T.; Takemae, N.; Saito, T. Isolation of Novel Triple-reassortant Swine H3N2 Influenza Viruses Possessing the Hemagglutinin and Neuraminidase Genes of a Seasonal Influenza Virus in Vietnam in 2010. Influenza Other Respir. Viruses 2012, 6, 6–10. [Google Scholar] [CrossRef]
- Zhu, H.; Webby, R.; Lam, T.T.Y.; Smith, D.K.; Peiris, J.S.M.; Guan, Y. History of Swine Influenza Viruses in Asia. In Swine Influenza; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2011; Volume 370, pp. 57–68. [Google Scholar]
- Karasin, A.I.; West, K.; Carman, S.; Olsen, C.W. Characterization of Avian H3N3 and H1N1 Influenza A Viruses Isolated from Pigs in Canada. J. Clin. Microbiol. 2004, 42, 4349–4354. [Google Scholar] [CrossRef]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Gramer, M.R.; Richt, J.A.; Janke, B.H. Characterization of a Newly Emerged Genetic Cluster of H1N1 and H1N2 Swine Influenza Virus in the United States. Virus Genes 2009, 39, 176–185. [Google Scholar] [CrossRef]
- Nelson, M.I.; Lemey, P.; Tan, Y.; Vincent, A.; Lam, T.T.-Y.; Detmer, S.; Viboud, C.; Suchard, M.A.; Rambaut, A.; Holmes, E.C.; et al. Spatial Dynamics of Human-Origin H1 Influenza A Virus in North American Swine. PLoS Pathog. 2011, 7, e1002077. [Google Scholar] [CrossRef]
- Ma, W.; Lager, K.M.; Lekcharoensuk, P.; Ulery, E.S.; Janke, B.H.; Solorzano, A.; Webby, R.J.; Garcia-Sastre, A.; Richt, J.A. Viral Reassortment and Transmission after Co-Infection of Pigs with Classical H1N1 and Triple-Reassortant H3N2 Swine Influenza Viruses. J. Gen. Virol. 2010, 91, 2314–2321. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.K.; Macken, C.A.; Lewis, N.S.; Scheuermann, R.H.; Van Reeth, K.; Brown, I.H.; Swenson, S.L.; Simon, G.; Saito, T.; Berhane, Y.; et al. A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza A Viruses. mSphere 2016, 1, e00275-16. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Hause, B.; Stigger-Rosser, E.; Darnell, D.; Corzo, C.; Juleen, K.; Simonson, R.; Brockwell-Staats, C.; Rubrum, A.; Wang, D.; et al. Multiple Reassortment between Pandemic (H1N1) 2009 and Endemic Influenza Viruses in Pigs, United States. Emerg. Infect. Dis. 2011, 17, 1624–1629. [Google Scholar] [CrossRef] [PubMed]
- Kitikoon, P.; Vincent, A.L.; Gauger, P.C.; Schlink, S.N.; Bayles, D.O.; Gramer, M.R.; Darnell, D.; Webby, R.J.; Lager, K.M.; Swenson, S.L.; et al. Pathogenicity and Transmission in Pigs of the Novel A(H3N2)v Influenza Virus Isolated from Humans and Characterization of Swine H3N2 Viruses Isolated in 2010-2011. J. Virol. 2012, 86, 6804–6814. [Google Scholar] [CrossRef]
- Rajao, D.S.; Vincent, A.L.; Perez, D.R. Adaptation of Human Influenza Viruses to Swine. Front. Vet. Sci. 2019, 5, 347. [Google Scholar] [CrossRef]
- Bakre, A.A.; Jones, L.P.; Kyriakis, C.S.; Hanson, J.M.; Bobbitt, D.E.; Bennett, H.K.; Todd, K.V.; Orr-Burks, N.; Murray, J.; Zhang, M.; et al. Molecular Epidemiology and Glycomics of Swine Influenza Viruses Circulating in Commercial Swine Farms in the Southeastern and Midwest United States. Vet. Microbiol. 2020, 251, 108914. [Google Scholar] [CrossRef]
- Everett, H.E.; Nash, B.; Londt, B.Z.; Kelly, M.D.; Coward, V.; Nunez, A.; Van Diemen, P.M.; Brown, I.H.; Brookes, S.M. Interspecies Transmission of Reassortant Swine Influenza a Virus Containing Genes from Swine Influenza A(H1N1)Pdm09 and A(H1N2) Viruses. Emerg. Infect. Dis. 2020, 26, 273–281. [Google Scholar] [CrossRef]
- Cook, P.W.; Stark, T.; Jones, J.; Kondor, R.; Zanders, N.; Benfer, J.; Scott, S.; Jang, Y.; Janas-Martindale, A.; Lindstrom, S.; et al. Detection and Characterization of Swine Origin Influenza A(H1N1) Pandemic 2009 Viruses in Humans Following Zoonotic Transmission. J. Virol. 2020, 95. [Google Scholar] [CrossRef]
- Rajão, D.S.; Gauger, P.C.; Anderson, T.K.; Lewis, N.S.; Abente, E.J.; Killian, M.L.; Perez, D.R.; Sutton, T.C.; Zhang, J.; Vincent, A.L. Novel Reassortant Human-Like H3N2 and H3N1 Influenza A Viruses Detected in Pigs Are Virulent and Antigenically Distinct from Swine Viruses Endemic to the United States. J. Virol. 2015, 89, 11213–11222. [Google Scholar] [CrossRef]
- Zeller, M.A.; Anderson, T.K.; Walia, R.W.; Vincent, A.L.; Gauger, P.C. ISU FLUture: A Veterinary Diagnostic Laboratory Web-Based Platform to Monitor the Temporal Genetic Patterns of Influenza A Virus in Swine. BMC Bioinform. 2018, 19, 397. [Google Scholar] [CrossRef]
- Powell, J.D.; Abente, E.J.; Chang, J.; Souza, C.K.; Rajao, D.S.; Anderson, T.K.; Zeller, M.A.; Gauger, P.C.; Lewis, N.S.; Vincent, A.L. Characterization of Contemporary 2010.1 H3N2 Swine Influenza A Viruses Circulating in United States Pigs. Virology 2021, 553, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Zeller, M.A.; Souza, C.K.; Anderson, T.K.; Vincent, A.L.; Harmon, K.; Li, G.; Zhang, J.; Gauger, P.C. Characterization of a 2016-2017 Human Seasonal H3 Influenza A Virus Spillover Now Endemic to U.S. Swine. mSphere 2022, 7, e0080921. [Google Scholar] [CrossRef] [PubMed]
- Kanji, J.N.; Pabbaraju, K.; Croxen, M.; Detmer, S.; Bastien, N.; Li, Y.; Majer, A.; Keshwani, H.; Zelyas, N.; Achebe, I.; et al. Characterization of Swine Influenza A(H1N2) Variant, Alberta, Canada, 2020. Emerg. Infect. Dis. 2021, 27, 3045–3051. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Perofsky, A.; McBride, D.S.; Rambo-Martin, B.L.; Wilson, M.M.; Barnes, J.R.; van Bakel, H.; Khan, Z.; Dutta, J.; Nolting, J.M.; et al. A Heterogeneous Swine Show Circuit Drives Zoonotic Transmission of Influenza A Viruses in the United States. J. Virol. 2020, 94, e01453-20. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.; Culhane, M.R.; Rovira, A.; Torremorell, M.; Guerrero, P.; Norambuena, J. Novel Human-like Influenza A Viruses Circulate in Swine in Mexico and Chile. PLoS Curr. 2015, 7. [Google Scholar] [CrossRef]
- Nelson, M.I.; Schaefer, R.; Gava, D.; Cantão, M.E.; Ciacci-Zanella, J.R. Influenza A Viruses of Human Origin in Swine, Brazil. Emerg. Infect. Dis. 2015, 21, 1339–1347. [Google Scholar] [CrossRef]
- Mena, I.; Nelson, M.I.; Quezada-Monroy, F.; Dutta, J.; Cortes-Fernández, R.; Lara-Puente, J.H.; Castro-Peralta, F.; Cunha, L.F.; Trovão, N.S.; Lozano-Dubernard, B.; et al. Origins of the 2009 H1N1 Influenza Pandemic in Swine in Mexico. eLife 2016, 5, e16777. [Google Scholar] [CrossRef]
- Pereda, A.; Cappuccio, J.; Quiroga, M.A.; Baumeister, E.; Insarralde, L.; Ibar, M.; Sanguinetti, R.; Cannilla, M.L.; Franzese, D.; Cabrera, O.E.E.; et al. Pandemic (H1N1) 2009 Outbreak on Pig Farm, Argentina. Emerg. Infect. Dis. 2010, 16, 304–307. [Google Scholar] [CrossRef]
- Pereda, A.; Rimondi, A.; Cappuccio, J.; Sanguinetti, R.; Angel, M.; Ye, J.; Sutton, T.; Dibárbora, M.; Olivera, V.; Craig, M.I.; et al. Evidence of Reassortment of Pandemic H1N1 Influenza Virus in Swine in Argentina: Are We Facing the Expansion of Potential Epicenters of Influenza Emergence? Influenza Other Respir. Viruses 2011, 5, 409–412. [Google Scholar] [CrossRef]
- Cappuccio, J.A.; Pena, L.; Dibárbora, M.; Rimondi, A.; Piñeyro, P.; Insarralde, L.; Quiroga, M.A.; Machuca, M.; Craig, M.I.; Olivera, V.; et al. Outbreak of Swine Influenza in Argentina Reveals a Non-Contemporary Human H3N2 Virus Highly Transmissible among Pigs. J. Gen. Virol. 2011, 92, 2871–2878. [Google Scholar] [CrossRef]
- Cappuccio, J.; Dibarbora, M.; Lozada, I.; Quiroga, A.; Olivera, V.; Dángelo, M.; Pérez, E.; Barrales, H.; Perfumo, C.; Pereda, A.; et al. Two Years of Surveillance of Influenza a Virus Infection in a Swine Herd. Results of Virological, Serological and Pathological Studies. Comp. Immunol. Microbiol. Infect. Dis. 2017, 50, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Dibárbora, M.; Cappuccio, J.; Olivera, V.; Quiroga, M.; Machuca, M.; Perfumo, C.; Pérez, D.; Pereda, A. Swine Influenza: Clinical, Serological, Pathological, and Virological Cross-Sectional Studies in Nine Farms in Argentina. Influenza Other Respir. Viruses 2013, 7, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, R.; Zanella, J.R.C.; Brentano, L.; Vincent, A.L.; Ritterbusch, G.A.; Silveira, S.; Caron, L.; Mores, N. Isolamento e Caracterização Do Vírus Da Influenza Pandêmico H1N1 Em Suínos No Brasil. Pesqui. Vet. Bras. 2011, 31, 761–767. [Google Scholar] [CrossRef]
- Rajão, D.S.; Alves, F.; Del Puerto, H.L.; Braz, G.F.; Oliveira, F.G.; Ciacci-Zanella, J.R.; Schaefer, R.; dos Reis, J.K.P.; Guedes, R.M.C.; Lobato, Z.I.P.; et al. Serological Evidence of Swine Influenza in Brazil. Influenza Other Respir. Viruses 2013, 7, 109–112. [Google Scholar] [CrossRef]
- Rajão, D.S.; Couto, D.H.; Gasparini, M.R.; Costa, A.T.R.; Reis, J.K.P.; Lobato, Z.I.P.; Guedes, R.M.C.; Leite, R.C. Diagnosis and Clinic-Pathological Findings of Influenza Virus Infection in Brazilian Pigs. Pesqui. Vet. Bras. 2013, 33, 30–36. [Google Scholar] [CrossRef]
- Rajão, D.S.; Costa, A.T.R.; Brasil, B.S.A.F.; Del Puerto, H.L.; Oliveira, F.G.; Alves, F.; Braz, G.F.; Reis, J.K.P.; Guedes, R.M.C.; Lobato, Z.I.P.; et al. Genetic Characterization of Influenza Virus Circulating in Brazilian Pigs during 2009 and 2010 Reveals a High Prevalence of the Pandemic H1N1 Subtype. Influenza Other Respir. Viruses 2013, 7, 783–790. [Google Scholar] [CrossRef]
- Ribeiro Amorim, A.; Garcete Fornells, L.A.M.; da Costa Reis, F.; Jacinto Rezende, D.; da Silva Mendes, G.; Couceiro, J.N.d.S.S.; de Oliveira Santos, N.S. Influenza A Virus Infection of Healthy Piglets in an Abattoir in Brazil: Animal-Human Interface and Risk for Interspecies Transmission. Mem. Inst. Oswaldo Cruz 2013, 108, 548–553. [Google Scholar] [CrossRef]
- Schmidt, C.; Cibulski, S.P.; Andrade, C.P.; Teixeira, T.F.; Varela, A.P.M.; Scheffer, C.M.; Franco, A.C.; de Almeida, L.L.; Roehe, P.M. Swine Influenza Virus and Association with the Porcine Respiratory Disease Complex in Pig Farms in Southern Brazil. Zoonoses Public Health 2016, 63, 234–240. [Google Scholar] [CrossRef]
- Schmidt, C.; Cibulski, S.P.; Muterle Varela, A.P.; Mengue Scheffer, C.; Wendlant, A.; Quoos Mayer, F.; Lopes de Almeida, L.; Franco, A.C.; Roehe, P.M. Full-Genome Sequence of a Reassortant H1N2 Influenza A Virus Isolated from Pigs in Brazil. Genome Announc. 2014, 2, e01319-14. [Google Scholar] [CrossRef]
- Schaefer, R.; Rech, R.R.; Gava, D.; Cantão, M.E.; da Silva, M.C.; Silveira, S.; Zanella, J.R.C. A Human-like H1N2 Influenza Virus Detected during an Outbreak of Acute Respiratory Disease in Swine in Brazil. Arch. Virol. 2015, 160, 29–38. [Google Scholar] [CrossRef]
- Souza, C.K.; Oldiges, D.P.; Poeta, A.P.S.; Vaz, I.d.S.; Schaefer, R.; Gava, D.; Ciacci-Zanella, J.R.; Canal, C.W.; Corbellini, L.G. Serological Surveillance and Factors Associated with Influenza A Virus in Backyard Pigs in Southern Brazil. Zoonoses Public Health 2019, 66, 125–132. [Google Scholar] [CrossRef]
- Serafini Poeta Silva, A.P.; de Freitas Costa, E.; Sousa e Silva, G.; Souza, C.K.; Schaefer, R.; da Silva Vaz, I.; Corbellini, L.G. Biosecurity Practices Associated with Influenza A Virus Seroprevalence in Sows from Southern Brazilian Breeding Herds. Prev. Vet. Med. 2019, 166, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ciacci-Zanella, J.R.; Schaefer, R.; Gava, D.; Haach, V.; Cantão, M.E.; Coldebella, A. Influenza A Virus Infection in Brazilian Swine Herds Following the Introduction of Pandemic 2009 H1N1. Vet. Microbiol. 2015, 180, 118–122. [Google Scholar] [CrossRef]
- Tochetto, C.; Junqueira, D.M.; Anderson, T.K.; Gava, D.; Haach, V.; Cantão, M.E.; Vincent Baker, A.L.; Schaefer, R. Introductions of Human-Origin Seasonal H3N2, H1N2 and Pre-2009 H1N1 Influenza Viruses to Swine in Brazil. Viruses 2023, 15, 576. [Google Scholar] [CrossRef]
- Haach, V.; Gava, D.; Cantão, M.E.; Schaefer, R. Evaluation of Two Multiplex RT-PCR Assays for Detection and Subtype Differentiation of Brazilian Swine Influenza Viruses. Braz. J. Microbiol. 2020, 51, 1447–1451. [Google Scholar] [CrossRef]
- Bravo-Vasquez, N.; Di Pillo, F.; Lazo, A.; Jiménez-Bluhm, P.; Schultz-Cherry, S.; Hamilton-West, C. Presence of Influenza Viruses in Backyard Poultry and Swine in El Yali Wetland, Chile. Prev. Vet. Med. 2016, 134, 211–215. [Google Scholar] [CrossRef]
- Jimenez-Bluhm, P.; Di Pillo, F.; Bahl, J.; Osorio, J.; Schultz-Cherry, S.; Hamilton-West, C. Circulation of Influenza in Backyard Productive Systems in Central Chile and Evidence of Spillover from Wild Birds. Prev. Vet. Med. 2018, 153, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Vasquez, N.; Baumberger, C.; Jimenez-Bluhm, P.; Di Pillo, F.; Lazo, A.; Sanhueza, J.; Schultz-Cherry, S.; Hamilton-West, C. Risk Factors and Spatial Relative Risk Assessment for Influenza A Virus in Poultry and Swine in Backyard Production Systems of Central Chile. Vet. Med. Sci. 2020, 6, 518–526. [Google Scholar] [CrossRef]
- Tapia, R.; Torremorell, M.; Culhane, M.; Medina, R.A.; Neira, V. Antigenic Characterization of Novel H1 Influenza A Viruses in Swine. Sci. Rep. 2020, 10, 4510. [Google Scholar] [CrossRef]
- Pensaert, M.; Ottis, K.; Vandeputte, J.; Kaplan, M.M.; Bachmann, P.A. Evidence for the Natural Transmission of Influenza A Virus from Wild Ducts to Swine and Its Potential Importance for Man. Bull. World Health Organ. 1981, 59, 75–78. [Google Scholar]
- Ottis, K.; Bachmann, P.A. Occurrence of Hsw1N1 Subtype Influenza A Viruses in Wild Ducks in Europe. Arch. Virol. 1980, 63, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Gourreau, J.M.; Hannoun, C.; Kaiser, C. Diffusion Du Virus de La Grippe Du Porc (H1N1=Hsw1N1) En France. Ann. Inst. Pasteur Virol. 1981, 132, 287–294. [Google Scholar] [CrossRef]
- Witte, K.H.; Nienhoff, H.; Ernst, H.; Schmidt, U.; Prager, D. Erstmaliges Auftreten Einer Durch Das Schweineinfluenzavirus Verursachten Epizootie in Schweinebestanden Der Bundesrepublik Deutschland. Tierärztliche Umsch. 1981, 36, 591–606. [Google Scholar]
- Dürrwald, R.; Wedde, M.; Biere, B.; Oh, D.Y.; Heßler-Klee, M.; Geidel, C.; Volmer, R.; Hauri, A.M.; Gerst, K.; Thürmer, A.; et al. Zoonotic Infection with Swine A/H1avN1 Influenza Virus in a Child, Germany, June 2020. Eurosurveillance 2020, 25, 2001638. [Google Scholar] [CrossRef] [PubMed]
- Zell, R.; Groth, M.; Krumbholz, A.; Lange, J.; Philipps, A.; Dürrwald, R. Cocirculation of Swine H1N1 Influenza A Virus Lineages in Germany. Viruses 2020, 12, 762. [Google Scholar] [CrossRef] [PubMed]
- Maksimović Zorić, J.; Milićević, V.; Veljović, L.; Radosavljević, V.; Kureljušić, B.; Stevančević, O.; Chiapponi, C. Genetic Analysis of Influenza A Viruses in Pigs from Commercial Farms in Serbia. Vet. Ital. 2023, 59, 179–189. [Google Scholar] [CrossRef]
- Zorić, J.M.; Veljović, L.; Radosavljević, V.; Glišić, D.; Kureljušić, J.; Maletić, J.; Savić, B. Protein Sequence Features of H1N1 Swine Influenza A Viruses Detected on Commercial Swine Farms in Serbia. J. Vet. Res. 2023, 67, 147–154. [Google Scholar] [CrossRef]
- Maksimović Zorić, J.; Milićević, V.; Stevančević, O.; Chiapponi, C.; Potkonjak, A.; Stojanac, N.; Kureljušić, B.; Veljović, L.; Radosavljević, V.; Savić, B. Phylogenetic Analysis of HA and NA Genes of Swine Influenza Viruses in Serbia in 2016–2018. Acta Vet. 2020, 70, 110–125. [Google Scholar] [CrossRef]
- Chiapponi, C.; Prosperi, A.; Moreno, A.; Baioni, L.; Faccini, S.; Manfredi, R.; Zanni, I.; Gabbi, V.; Calanchi, I.; Fusaro, A.; et al. Genetic Variability among Swine Influenza Viruses in Italy: Data Analysis of the Period 2017–2020. Viruses 2022, 14, 47. [Google Scholar] [CrossRef]
- Zell, R.; Scholtissek, C.; Ludwig, S. Genetics, Evolution, and the Zoonotic Capacity of European Swine Influenza Viruses. In Swine Influenza; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 370, pp. 29–55. [Google Scholar]
- Castrucci, M.R.; Donatelli, I.; Sidoli, L.; Barigazzi, G.; Kawaoka, Y.; Webster, R.G. Genetic Reassortment between Avian and Human Influenza a Viruses in Italian Pigs. Virology 1993, 193, 503–506. [Google Scholar] [CrossRef]
- Campitelli, L.; Donatelli, I.; Foni, E.; Castrucci, M.R.; Fabiani, C.; Kawaoka, Y.; Krauss, S.; Webster, R.G. Continued Evolution of H1N1 and H3N2 Influenza Viruses in Pigs in Italy. Virology 1997, 232, 310–318. [Google Scholar] [CrossRef]
- Schrader, C.; Süss, J. Molecular Epidemiology of Porcine H3N2 Influenza A Viruses Isolated in Germany between 1982 and 2001. Intervirology 2004, 47, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Castrucci, M.R.; Campitelli, L.; Ruggieri, A.; Barigazzi, G.; Sidoli, L.; Daniels, R.; Oxford, J.S.; Donatelli, I. Antigenic and Sequence Analysis of H3 Influenza Virus Haemagglutinins from Pigs in Italy. J. Gen. Virol. 1994, 75, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Haesebrouck, F.; Biront, P.; Pensaert, M.B.; Leunen, J. Epizootics of Respiratory Tract Disease in Swine in Belgium Due to H3N2 Influenza Virus and Experimental Reproduction of Disease. Am. J. Vet. Res. 1985, 46, 1926–1928. [Google Scholar] [PubMed]
- Zell, R.; Scholtissek, C.; Ludwig, S. Emergence of Human-Like H3N2 in European Pigs. In Swine Influenza; Richt, J., Webby, R., Eds.; Springer: New York, NY, USA, 2013; pp. 39–41. [Google Scholar]
- Haesebrouck, F.; Pensaert, M. Influenza in Swine in Belgium (1969–1986) Epizootiologic Aspects. Comp. Immunol. Microbiol. Infect. Dis. 1988, 11, 215–222. [Google Scholar] [CrossRef]
- Madec, F.; Gourreau, J.M.; Kaiser, C.; Aymard, M. Apparition de Manifestations Grippales Chezles Porcs En Association Avec Un Virus A/H3N2. Bull. Acad. Vet. Fr. 1984, 57, 513–522. [Google Scholar]
- Zhang, X.M.; Herbst, W.; Lange-Herbst, H.; Schliesser, T. Seroprevalence of Porcine and Human Influenza A Virus Antibodies in Pigs between 1986 and 1988 in Hassia. J. Vet. Med. Ser. B 1989, 36, 765–770. [Google Scholar] [CrossRef]
- Zell, R.; Groth, M.; Krumbholz, A.; Lange, J.; Philipps, A.; Dürrwald, R. Novel Reassortant Swine H3N2 Influenza A Viruses in Germany. Sci. Rep. 2020, 10, 14296. [Google Scholar] [CrossRef]
- Brown, I.H.; Harris, P.A.; Alexander, D.J. Serological Studies of Influenza Viruses in Pigs in Great Britain 1991–2. Epidemiol. Infect. 1995, 114, 511–520. [Google Scholar] [CrossRef]
- Castro, J.M.; del Pozo, M.; Simarro, I. Identification of H3N2 Influenza Virus Isolated from Pigs with Respiratory Problems in Spain. Vet. Rec. 1988, 122, 418–419. [Google Scholar] [CrossRef]
- Yus, E.; Sanjuan, M.L.; Garcia, F.; Castro, J.M.; Simarro, I. Influenza A Viruses: Epidemiologic Study in Fatteners in Spain (1987–89). J. Vet. Med. Ser. B 1992, 39, 113–118. [Google Scholar] [CrossRef]
- Loeffen, W.L.A.; Hunneman, W.A.; Quak, J.; Verheijden, J.H.M.; Stegeman, J.A. Population Dynamics of Swine Influenza Virus in Farrow-to-Finish and Specialised Finishing Herds in the Netherlands. Vet. Microbiol. 2009, 137, 45–50. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Loeffen, W.L.A.; Kamp, E.M.; Stockhofe-Zurwieden, N.; Van Nieuwstadt, A.P.K.M.I.; Bongers, J.H.; Hunneman, W.A.; Elbers, A.R.W.; Baars, J.; Nell, T.; Van Zijderveld, F.G. Survey of Infectious Agents Involved in Acute Respiratory Disease in Finishing Pigs. Vet. Rec. 1999, 145, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.H.; Chakraverty, P.; Harris, P.A.; Alexander, D.J. Disease Outbreaks in Pigs in Great Britain Due to an Influenza A Virus of H1N2 Subtype. Vet. Rec. 1995, 136, 328–329. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.H.; Harris, P.A.; McCauley, J.W.; Alexander, D.J. Multiple Genetic Reassortment of Avian and Human Influenza A Viruses in European Pigs, Resulting in the Emergence of an H1N2 Virus of Novel Genotype. J. Gen. Virol. 1998, 79, 2947–2955. [Google Scholar] [CrossRef]
- Marozin, S.; Gregory, V.; Cameron, K.; Bennett, M.; Valette, M.; Aymard, M.; Foni, E.; Barigazzi, G.; Lin, Y.; Hay, A. Antigenic and Genetic Diversity among Swine Influenza A H1N1 and H1N2 Viruses in Europe. J. Gen. Virol. 2002, 83, 735–745. [Google Scholar] [CrossRef]
- Gourreau, J.M.; Kaiser, C.; Valette, M.; Douglas, A.R.; Labie, J.; Aymard, M. Isolation of Two H1N2 Influenza Viruses from Swine in France. Arch. Virol. 1994, 135, 365–382. [Google Scholar] [CrossRef]
- Van Reeth, K.; Pensaert, M.; Brown, I.H. Isolations of H1N2 Influenza A Virus from Pigs in Belgium. Vet. Rec. 2000, 146, 588–589. [Google Scholar] [CrossRef]
- Van Reeth, K.; Brown, I.; Essen, S.; Pensaert, M. Genetic Relationships, Serological Cross-Reaction and Cross-Protection between H1N2 and Other Influenza a Virus Subtypes Endemic in European Pigs. Virus Res. 2004, 103, 115–124. [Google Scholar] [CrossRef]
- Schrader, C.; Süss, J. Genetic Characterization of a Porcine H1N2 Influenza Virus Strain Isolated in Germany. Intervirology 2003, 46, 66–70. [Google Scholar] [CrossRef]
- Zell, R.; Motzke, S.; Krumbholz, A.; Wutzler, P.; Herwig, V.; Dürrwald, R. Novel Reassortant of Swine Influenza H1N2 Virus in Germany. J. Gen. Virol. 2008, 89, 271–276. [Google Scholar] [CrossRef]
- Breum, S.Ø.; Hjulsager, C.K.; Trebbien, R.; Larsen, L.E. Influenza A Virus with a Human-Like N2 Gene Is Circulating in Pigs. Genome Announc. 2013, 1, e00712-13. [Google Scholar] [CrossRef]
- Ma, J.; Shen, H.; Liu, Q.; Bawa, B.; Qi, W.; Duff, M.; Lang, Y.; Lee, J.; Yu, H.; Bai, J.; et al. Pathogenicity and Transmissibility of Novel Reassortant H3N2 Influenza Viruses with 2009 Pandemic H1N1 Genes in Pigs. J. Virol. 2015, 89, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Krog, J.S.; Hjulsager, C.K.; Larsen, M.A.; Larsen, L.E. Triple-Reassortant Influenza A Virus with H3 of Human Seasonal Origin, NA of Swine Origin, and Internal A(H1N1) Pandemic 2009 Genes Is Established in Danish Pigs. Influenza Other Respir. Viruses 2017, 11, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Pippig, J.; Ritzmann, M.; Büttner, M.; Neubauer-Juric, A. Influenza A Viruses Detected in Swine in Southern Germany after the H1N1 Pandemic in 2009. Zoonoses Public Health 2016, 63, 555–568. [Google Scholar] [CrossRef]
- Ryt-Hansen, P.; Larsen, I.; Kristensen, C.S.; Krog, J.S.; Wacheck, S.; Larsen, L.E. Longitudinal Field Studies Reveal Early Infection and Persistence of Influenza A Virus in Piglets despite the Presence of Maternally Derived Antibodies. Vet. Res. 2019, 50, 36. [Google Scholar] [CrossRef] [PubMed]
- Chastagner, A.; Hervé, S.; Quéguiner, S.; Hirchaud, E.; Lucas, P.; Gorin, S.; Béven, V.; Barbier, N.; Deblanc, C.; Blanchard, Y.; et al. Genetic and Antigenic Evolution of European Swine Influenza a Viruses of Ha-1c (Avian-like) and Ha-1b (Human-like) Lineages in France from 2000 to 2018. Viruses 2020, 12, 1304. [Google Scholar] [CrossRef]
- Bhatta, T.R.; Ryt-Hansen, P.; Nielsen, J.P.; Larsen, L.E.; Larsen, I.; Chamings, A.; Goecke, N.B.; Alexandersen, S. Infection Dynamics of Swine Influenza Virus in a Danish Pig Herd Reveals Recurrent Infections with Different Variants of the H1n2 Swine Influenza a Virus Subtype. Viruses 2020, 12, 1013. [Google Scholar] [CrossRef]
- Chepkwony, S.; Parys, A.; Vandoorn, E.; Stadejek, W.; Xie, J.; King, J.; Graaf, A.; Pohlmann, A.; Beer, M.; Harder, T.; et al. Genetic and Antigenic Evolution of H1 Swine Influenza A Viruses Isolated in Belgium and the Netherlands from 2014 through 2019. Sci. Rep. 2021, 11, 11276. [Google Scholar] [CrossRef]
- Van Reeth, K.; Brown, I.H.; Dürrwald, R.; Foni, E.; Labarque, G.; Lenihan, P.; Maldonado, J.; Markowska-Daniel, I.; Pensaert, M.; Pospisil, Z.; et al. Seroprevalence of H1N1, H3N2 and H1N2 Influenza Viruses in Pigs in Seven European Countries in 2002–2003. Influenza Other Respir. Viruses 2008, 2, 99–105. [Google Scholar] [CrossRef]
- Welsh, M.D.; Baird, P.M.; Guelbenzu-Gonzalo, M.P.; Hanna, A.; Reid, S.M.; Essen, S.; Russell, C.; Thomas, S.; Barrass, L.; McNeilly, F.; et al. Initial Incursion of Pandemic (H1N1) 2009 Influenza A Virus into European Pigs. Vet. Rec. 2010, 166, 642–645. [Google Scholar] [CrossRef]
- Williamson, S.M.; Tucker, A.W.; McCrone, I.S.; Bidewell, C.A.; Brons, N.; Habernoll, H.; Essen, S.C.; Brown, I.H.; Wood, J.L.N. Descriptive Clinical and Epidemiological Characteristics of Influenza A H1N1 2009 Virus Infections in Pigs in England. Vet. Rec. 2012, 171, 271. [Google Scholar] [CrossRef]
- Lange, J.; Groth, M.; Schlegel, M.; Krumbholz, A.; Wieczorek, K.; Ulrich, R.; Köppen, S.; Schulz, K.; Appl, D.; Selbitz, H.J.; et al. Reassortants of the Pandemic (H1N1) 2009 Virus and Establishment of a Novel Porcine H1N2 Influenza Virus, Lineage in Germany. Vet. Microbiol. 2013, 167, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Chastagner, A.; Enouf, V.; Peroz, D.; Hervé, S.; Lucas, P.; Quéguiner, S.; Gorin, S.; Beven, V.; Behillil, S.; Leneveu, P.; et al. Bidirectional Human-Swine Transmission of Seasonal Influenza A(H1N1)Pdm09 Virus in Pig Herd, France, 2018. Emerg. Infect. Dis. 2019, 25, 1940–1943. [Google Scholar] [CrossRef] [PubMed]
- Nokireki, T.; Laine, T.; London, L.; Ikonen, N.; Huovilainen, A. The First Detection of Influenza in the Finnish Pig Population: A Retrospective Study. Acta Vet. Scand. 2013, 55, 69. [Google Scholar] [CrossRef][Green Version]
- Haimi-Hakala, M.; Hälli, O.; Laurila, T.; Raunio-Saarnisto, M.; Nokireki, T.; Laine, T.; Nykäsenoja, S.; Pelkola, K.; Segales, J.; Sibila, M.; et al. Etiology of Acute Respiratory Disease in Fattening Pigs in Finland. Porc. Health Manag. 2017, 3, 19. [Google Scholar] [CrossRef]
- Kyriakis, C.S.; Papatsiros, V.G.; Athanasiou, L.V.; Valiakos, G.; Brown, I.H.; Simon, G.; Van Reeth, K.; Tsiodras, S.; Spyrou, V.; Billinis, C. Serological Evidence of Pandemic H1N1 Influenza Virus Infections in Greek Swine. Zoonoses Public Health 2016, 63, 370–373. [Google Scholar] [CrossRef]
- Er, J.C.; Lium, B.; Framstad, T. Antibodies of Influenza A(H1N1)Pdm09 Virus in Pigs’ Sera Cross-React with Other Influenza A Virus Subtypes. A Retrospective Epidemiological Interpretation of Norway’s Serosurveillance Data from 2009–2017. Epidemiol. Infect. 2020, 148, e73. [Google Scholar] [CrossRef]
- Starick, E.; Lange, E.; Fereidouni, S.; Bunzenthal, C.; Höveler, R.; Kuczka, A.; grosse Beilage, E.; Hamann, H.-P.; Klingelhöfer, I.; Steinhauer, D.; et al. Reassorted Pandemic (H1N1) 2009 Influenza A Virus Discovered from Pigs in Germany. J. Gen. Virol. 2011, 92, 1184–1188. [Google Scholar] [CrossRef]
- Hofshagen, M.; Gjerset, B.; Er, C.; Tarpai, A.; Brun, E.; Dannevig, B.; Bruheim, T.; Fostad, I.G.; Iversen, B.; Hungnes, O.; et al. Pandemic Influenza A(H1N1)v: Human to Pig Transmission in Norway? Eurosurveillance 2009, 14, 19406. [Google Scholar] [CrossRef][Green Version]
- Watson, S.J.; Langat, P.; Reid, S.M.; Lam, T.T.-Y.; Cotten, M.; Kelly, M.; Van Reeth, K.; Qiu, Y.; Simon, G.; Bonin, E.; et al. Molecular Epidemiology and Evolution of Influenza Viruses Circulating within European Swine between 2009 and 2013. J. Virol. 2015, 89, 9920–9931. [Google Scholar] [CrossRef]
- Chiapponi, C.; Ebranati, E.; Pariani, E.; Faccini, S.; Luppi, A.; Baioni, L.; Manfredi, R.; Carta, V.; Merenda, M.; Affanni, P.; et al. Genetic Analysis of Human and Swine Influenza A Viruses Isolated in Northern Italy during 2010–2015. Zoonoses Public Health 2018, 65, 114–123. [Google Scholar] [CrossRef]
- Henritzi, D.; Petric, P.P.; Lewis, N.S.; Graaf, A.; Pessia, A.; Starick, E.; Breithaupt, A.; Strebelow, G.; Luttermann, C.; Parker, L.M.K.; et al. Surveillance of European Domestic Pig Populations Identifies an Emerging Reservoir of Potentially Zoonotic Swine Influenza A Viruses. Cell Host Microbe 2020, 28, 614–627.e6. [Google Scholar] [CrossRef]
- Howard, W.A.; Essen, S.C.; Strugnell, B.W.; Russell, C.; Barrass, L.; Reid, S.M.; Brown, I.H. Reassortant Pandemic (H1N1) 2009 Virus in Pigs, United Kingdom. Emerg. Infect. Dis. 2011, 17, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Di Trani, L.; Faccini, S.; Vaccari, G.; Nigrelli, D.; Boniotti, M.B.; Falcone, E.; Boni, A.; Chiapponi, C.; Sozzi, E.; et al. Novel H1N2 Swine Influenza Reassortant Strain in Pigs Derived from the Pandemic H1N1/2009 Virus. Vet. Microbiol. 2011, 149, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Chiapponi, C.; Barbieri, I.; Manfredi, R.; Zanni, I.; Barigazzi, G.; Foni, E. Genetic Diversity among H1N1 and H1N2 Swine Influenza Viruses in Italy: Preliminary Results. In Proceedings of the 5th International Symposium on Emerging and Re-Emerging Pig Diseases, Krakow, Poland, 24–27 June 2007; Markowska-Daniel, I., Ed.; p. 261. [Google Scholar]
- Franck, N.; Queguiner, S.; Gorin, S.; Eveno, E.; Fablet, C.; Madec, F.; Kuntz-Simon, G. Molecular Epidemiology of Swine Influenza Virus in France: Identification of Novel H1N1 Reassortants. In Proceedings of the 5th International Symposium on Emerging and Re-Emerging Pig Diseases, Krakow, Poland, 24–27 June 2007; Markowska-Daniel, I., Ed.; pp. 24–27. [Google Scholar]
- Bálint, Á.; Metreveli, G.; Widén, F.; Zohari, S.; Berg, M.; Isaksson, M.; Renström, L.H.; Wallgren, P.; Belák, S.; Segall, T.; et al. The First Swedish H1N2 Swine Influenza Virus Isolate Represents an Uncommon Reassortant. Virol. J. 2009, 6, 180. [Google Scholar] [CrossRef]
- Hjulsager, C.; Bragstad, K.; Botner, A.; Nielsen, E.; Vigre, H.; Enoe, C.; Larsen, L. New Swine Influenza A H1N2 Reassortment Found in Danish Swine. In Proceedings of the 19th IPVS Congress; Copenhagen, Denmark, 16–19 July 2006, p. 265.
- Moreno, A.; Barbieri, I.; Sozzi, E.; Luppi, A.; Lelli, D.; Lombardi, G.; Zanoni, M.G.; Cordioli, P. Novel Swine Influenza Virus Subtype H3N1 in Italy. Vet. Microbiol. 2009, 138, 361–367. [Google Scholar] [CrossRef]
- Brown, I.H.; Alexander, D.J.; Chakraverty, P.; Harris, P.A.; Manvell, R.J. Isolation of an Influenza A Virus of Unusual Subtype (H1N7) from Pigs in England, and the Subsequent Experimental Transmission from Pig to Pig. Vet. Microbiol. 1994, 39, 125–134. [Google Scholar] [CrossRef]
- Brown, I.H.; Hill, M.L.; Harris, P.A.; Alexander, D.J.; McCauley, J.W. Genetic Characterisation of an Influenza A Virus of Unusual Subtype (H1N7) Isolated from Pigs in England. Arch. Virol. 1997, 142, 1045–1050. [Google Scholar] [CrossRef]
- Choi, Y.K.; Pascua, P.N.Q.; Song, M.S. Swine Influenza Viruses: An Asian Perspective. In Swine Influenza; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2013; Volume 370, pp. 147–172. [Google Scholar] [CrossRef]
- Arikawa, J.; Yamane, N.; Odagiri, T.; Ishida, N. Serological Evidence of H1 Influenza Virus Infection among Japanese Hogs. Acta Virol. 1979, 23, 508–511. [Google Scholar]
- Nerome, K.; Ishida, M.; Oya, A.; Oda, K. The Possible Origin of H1N1 (Hsw 1N1) Virus in the Swine Population of Japan and Antigenic Analysis of the Isolates. J. Gen. Virol. 1982, 62, 171–175. [Google Scholar] [CrossRef]
- Samadieh, B.; Shakeri, F. Swine Influenza in Iran and Its Serological Relationship with Human Influenza. In Proceedings of the 4th International Pig Veterinary Society Congress, Ames, IA, USA, 22–24 June 1976. [Google Scholar]
- Yamane, N.; Arikawa, J.; Odagiri, T.; Ishida, N.; Kumasaka, M. Distribution of Antibodies Against Swine and Hong Kong Influenza Viruses Among Pigs in 1977. Tohoku J. Exp. Med. 1978, 126, 199–200. [Google Scholar] [CrossRef]
- Shortridge, K.F.; Webster, R.G. Geographical Distribution of Swine (Hswlnl) and Hong Kong (H3n2) Influenza Virus Variants in Pigs in Southeast Asia. Intervirology 1979, 11, 9–15. [Google Scholar] [CrossRef]
- Nerome, K.; Ishida, M.; Oya, A.; Kanai, C.; Suwicha, K. Isolation of an Influenza H1N1 Virus from a Pig. Virology 1982, 117, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Kupradinun, S.; Peanpijit, P.; Bhodhikosoom, C.; Yoshioka, Y.; Endo, A.; Nerome, K. The First Isolation of Swine H1N1 Influenza Viruses from Pigs in Thailand. Arch. Virol. 1991, 118, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Mukherjee, K.K.; Mondal, M.C.; Chakravarti, S.K.; Chakraborty, M.S. A Serological Survey of Influenza a Antibody in Human and Pig Sera in Calcutta. Folia Microbiol. 1995, 40, 345–348. [Google Scholar] [CrossRef]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Li, P.H.; Kawaoka, Y.; Webster, R.G. Emergence of Avian H1N1 Influenza Viruses in Pigs in China. J. Virol. 1996, 70, 8041–8046. [Google Scholar] [CrossRef] [PubMed]
- Song, D.S.; Lee, C.S.; Jung, K.; Kang, B.K.; Oh, J.S.; Yoon, Y.D.; Lee, J.H.; Park, B.K. Isolation and Phylogenetic Analysis of H1N1 Swine Influenza Virus Isolated in Korea. Virus Res. 2007, 125, 98–103. [Google Scholar] [CrossRef]
- Lee, C.S.; Kang, B.K.; Kim, H.K.; Park, S.J.; Park, B.K.; Jung, K.; Song, D.S. Phylogenetic Analysis of Swine Influenza Viruses Recently Isolated in Korea. Virus Genes 2008, 37, 168–176. [Google Scholar] [CrossRef]
- Malik Peiris, J.S.; Poon, L.L.M.; Guan, Y. Emergence of a Novel Swine-Origin Influenza A Virus (S-OIV) H1N1 Virus in Humans. J. Clin. Virol. 2009, 45, 169–173. [Google Scholar] [CrossRef]
- Qi, X.; Pang, B.; Lu, C.P. Genetic Characterization of H1N1 Swine Influenza A Viruses Isolated in Eastern China. Virus Genes 2009, 39, 193–199. [Google Scholar] [CrossRef]
- Nelson, M.I.; Viboud, C.; Vincent, A.L.; Culhane, M.R.; Detmer, S.E.; Wentworth, D.E.; Rambaut, A.; Suchard, M.A.; Holmes, E.C.; Lemey, P. Global Migration of Influenza A Viruses in Swine. Nat. Commun. 2015, 6, 6696. [Google Scholar] [CrossRef]
- Smith, G.J.D.; Bahl, J.; Vijaykrishna, D.; Zhang, J.; Poon, L.L.M.; Chen, H.; Webster, R.G.; Peiris, J.S.M.; Guan, Y. Dating the Emergence of Pandemic Influenza Viruses. Proc. Natl. Acad. Sci. USA 2009, 106, 11709–11712. [Google Scholar] [CrossRef]
- Kanegae, Y.; Sugita, S.; Shortridge, K.F.; Yoshioka, Y.; Nerome, K. Origin and Evolutionary Pathways of the H1 Hemagglutinin Gene of Avian, Swine and Human Influenza Viruses: Cocirculation of Two Distinct Lineages of Swine Virus. Arch. Virol. 1994, 134, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Gorman, O.T.; Bean, W.J.; Kawaoka, Y.; Donatelli, I.; Guo, Y.J.; Webster, R.G. Evolution of Influenza A Virus Nucleoprotein Genes: Implications for the Origins of H1N1 Human and Classical Swine Viruses. J. Virol. 1991, 65, 3704–3714. [Google Scholar] [CrossRef] [PubMed]
- Weingartl, H.M.; Berhane, Y.; Hisanaga, T.; Neufeld, J.; Kehler, H.; Emburry-Hyatt, C.; Hooper-McGreevy, K.; Kasloff, S.; Dalman, B.; Bystrom, J.; et al. Genetic and Pathobiologic Characterization of Pandemic H1N1 2009 Influenza Viruses from a Naturally Infected Swine Herd. J. Virol. 2010, 84, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Katsuda, K.; Sato, S.; Shirahata, T.; Lindstrom, S.; Nerome, R.; Ishida, M.; Nerome, K.; Goto, H. Antigenic and Genetic Characteristics of H1N1 Human Influenza Virus Isolated from Pigs in Japan. J. Gen. Virol. 1995, 76, 1247–1249. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, G.-H.; Hua, R.-H.; Zhang, Q.; Liu, T.-Q.; Liao, M.; Tong, G.-Z. Isolation and Genetic Analysis of Human Origin H1N1 and H3N2 Influenza Viruses from Pigs in China. Biochem. Biophys. Res. Commun. 2007, 356, 91–96. [Google Scholar] [CrossRef]
- Yu, H.; Zhou, Y.-J.; Li, G.-X.; Zhang, G.-H.; Liu, H.-L.; Yan, L.-P.; Liao, M.; Tong, G.-Z. Further Evidence for Infection of Pigs with Human-like H1N1 Influenza Viruses in China. Virus Res. 2009, 140, 85–90. [Google Scholar] [CrossRef]
- Matsuu, A.; Uchida, Y.; Takemae, N.; Mawatari, T.; Kasai Yoneyama, S.; Kasai, T.; Nakamura, R.; Eto, M.; Saito, T. Genetic Characterization of Swine Influenza Viruses Isolated in Japan between 2009 and 2012. Microbiol. Immunol. 2012, 56, 792–803. [Google Scholar] [CrossRef]
- Mine, J.; Uchida, Y.; Takemae, N.; Saito, T. Genetic Characterization of Influenza A Viruses in Japanese Swine in 2015 to 2019. J. Virol. 2020, 94, e02169-19. [Google Scholar] [CrossRef]
- Kitikoon, P.; Sreta, D.; Nuntawan Na Ayudhya, S.; Wongphatcharachai, M.; Lapkuntod, J.; Prakairungnamthip, D.; Bunpapong, N.; Suradhat, S.; Thanawongnuwech, R.; Amonsin, A. Brief Report: Molecular Characterization of a Novel Reassorted Pandemic H1N1 2009 in Thai Pigs. Virus Genes 2011, 43, 1–5. [Google Scholar] [CrossRef]
- Yu, H.; Hua, R.-H.; Zhang, Q.; Liu, T.-Q.; Liu, H.-L.; Li, G.-X.; Tong, G.-Z. Genetic Evolution of Swine Influenza A (H3N2) Viruses in China from 1970 to 2006. J. Clin. Microbiol. 2008, 46, 1067–1075. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, P.-C.; Zhou, Y.-J.; Li, G.-X.; Pan, J.; Yan, L.-P.; Shi, X.-X.; Liu, H.-L.; Tong, G.-Z. Isolation and Genetic Characterization of Avian-like H1N1 and Novel Ressortant H1N2 Influenza Viruses from Pigs in China. Biochem. Biophys. Res. Commun. 2009, 386, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, K.; Saikumar, G.; Arya, R.S.; Gupta, A.; Somvanshi, R.; Pattnaik, B. Influenza A H1N1 Virus in Indian Pigs & Its Genetic Relatedness with Pandemic Human Influenza A 2009 H1N1. Indian J. Med. Res. 2010, 132, 160–167. [Google Scholar]
- Song, M.-S.; Lee, J.H.; Pascua, P.N.Q.; Baek, Y.H.; Kwon, H.; Park, K.J.; Choi, H.-W.; Shin, Y.-K.; Song, J.-Y.; Kim, C.-J.; et al. Evidence of Human-to-Swine Transmission of the Pandemic (H1N1) 2009 Influenza Virus in South Korea. J. Clin. Microbiol. 2010, 48, 3204–3211. [Google Scholar] [CrossRef]
- Zhao, G.; Fan, Q.; Zhong, L.; Li, Y.; Liu, W.; Liu, X.; Gao, S.; Peng, D.; Liu, X. Isolation and Phylogenetic Analysis of Pandemic H1N1/09 Influenza Virus from Swine in Jiangsu Province of China. Res. Vet. Sci. 2012, 93, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Trevennec, K.; Leger, L.; Lyazrhi, F.; Baudon, E.; Cheung, C.Y.; Roger, F.; Peiris, M.; Garcia, J. Transmission of Pandemic Influenza H1N1 (2009) in Vietnamese Swine in 2009–2010. Influenza Other Respir. Viruses 2012, 6, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, J.; Qiao, C.; Yang, H.; Zhang, Y.; Xin, X.; Chen, H. Co-Circulation of Pandemic 2009 H1N1, Classical Swine H1N1 and Avian-like Swine H1N1 Influenza Viruses in Pigs in China. Infect. Genet. Evol. 2013, 13, 331–338. [Google Scholar] [CrossRef]
- Hiromoto, Y.; Parchariyanon, S.; Ketusing, N.; Netrabukkana, P.; Hayashi, T.; Kobayashi, T.; Takemae, N.; Saito, T. Isolation of the Pandemic (H1N1) 2009 Virus and Its Reassortant with an H3N2 Swine Influenza Virus from Healthy Weaning Pigs in Thailand in 2011. Virus Res. 2012, 169, 175–181. [Google Scholar] [CrossRef]
- Charoenvisal, N.; Keawcharoen, J.; Sreta, D.; Chaiyawong, S.; Nonthabenjawan, N.; Tantawet, S.; Jittimanee, S.; Arunorat, J.; Amonsin, A.; Thanawongnuwech, R. Genetic Characterization of Thai Swine Influenza Viruses after the Introduction of Pandemic H1N1 2009. Virus Genes 2013, 47, 75–85. [Google Scholar] [CrossRef]
- Poonsuk, S.; Sangthong, P.; Petcharat, N.; Lekcharoensuk, P. Genesis and Genetic Constellations of Swine Influenza Viruses in Thailand. Vet. Microbiol. 2013, 167, 314–326. [Google Scholar] [CrossRef]
- Kirisawa, R.; Ogasawara, Y.; Yoshitake, H.; Koda, A.; Furuya, T. Genomic Reassortants of Pandemic a (H1N1) 2009 Virus and Endemic Porcine H1 and H3 Viruses in Swine in Japan. J. Vet. Med. Sci. 2014, 76, 1457–1470. [Google Scholar] [CrossRef]
- Mine, J.; Abe, H.; Parchariyanon, S.; Boonpornprasert, P.; Ubonyaem, N.; Nuansrichay, B.; Takemae, N.; Tanikawa, T.; Tsunekuni, R.; Uchida, Y.; et al. Genetic and Antigenic Dynamics of Influenza A Viruses of Swine on Pig Farms in Thailand. Arch. Virol. 2019, 164, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xiao, Y.; Liu, J.; Wang, D.; Li, F.; Wang, C.; Li, C.; Zhu, J.; Song, J.; Sun, H.; et al. Prevalent Eurasian Avian-like H1N1 Swine Influenza Virus with 2009 Pandemic Viral Genes Facilitating Human Infection. Proc. Natl. Acad. Sci. USA 2020, 117, 17204–17210. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-F.; Wang, X.-H.; Li, X.-L.; Zhang, L.; Li, H.-H.; Lu, C.; Yang, C.-L.; Feng, J.; Han, W.; Ren, W.-K.; et al. Novel Triple-Reassortant H1N1 Swine Influenza Viruses in Pigs in Tianjin, Northern China. Vet. Microbiol. 2016, 183, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, H.J.; Jin, Y.H.; Yeoul, J.J.; Lee, K.K.; Oem, J.K.; Lee, M.H.; Park, C.K. Isolation of Influenza A(H3N2)v Virus from Pigs and Characterization of Its Biological Properties in Pigs and Mice. Arch. Virol. 2013, 158, 2351–2357. [Google Scholar] [CrossRef] [PubMed]
- Baudon, E.; Poon, L.L.; Dao, T.D.; Pham, N.T.; Cowling, B.J.; Peyre, M.; Nguyen, K.V.; Peiris, M. Detection of Novel Reassortant Influenza A (H3N2) and H1N1 2009 Pandemic Viruses in Swine in Hanoi, Vietnam. Zoonoses Public Health 2015, 62, 429–434. [Google Scholar] [CrossRef]
- Takemae, N.; Parchariyanon, S.; Damrongwatanapokin, S.; Uchida, Y.; Ruttanapumma, R.; Watanabe, C.; Yamaguchi, S.; Saito, T. Genetic Diversity of Swine Influenza Viruses Isolated from Pigs during 2000 to 2005 in Thailand. Influenza Other Respir. Viruses 2008, 2, 181–189. [Google Scholar] [CrossRef]
- Choi, C.; Ha, S.K.; Chae, C. Detection and Isolation of H1N1 Influenza Virus from Pigs in Korea. Vet. Rec. 2004, 154, 274–275. [Google Scholar] [CrossRef]
- Chutinimitkul, S.; Thippamom, N.; Damrongwatanapokin, S.; Payungporn, S.; Thanawongnuwech, R.; Amonsin, A.; Boonsuk, P.; Sreta, D.; Bunpong, N.; Tantilertcharoen, R.; et al. Genetic Characterization of H1N1, H1N2 and H3N2 Swine Influenza Virus in Thailand. Arch. Virol. 2008, 153, 1049–1056. [Google Scholar] [CrossRef]
- Suriya, R.; Hassan, L.; Omar, A.R.; Aini, I.; Tan, C.G.; Lim, Y.S.; Kamaruddin, M.I. Seroprevalence and Risk Factors for Influenza A Viruses in Pigs in Peninsular Malaysia. Zoonoses Public Health 2008, 55, 342–351. [Google Scholar] [CrossRef]
- Rith, S.; Netrabukkana, P.; Sorn, S.; Mumford, E.; Mey, C.; Holl, D.; Goutard, F.; Bunthin, Y.; Fenwick, S.; Robertson, I.; et al. Serologic Evidence of Human Influenza Virus Infections in Swine Populations, Cambodia. Influenza Other Respir. Viruses 2013, 7, 271–279. [Google Scholar] [CrossRef]
- Monger, V.R.; Stegeman, J.A.; Koop, G.; Dukpa, K.; Tenzin, T.; Loeffen, W.L.A. Seroprevalence and Associated Risk Factors of Important Pig Viral Diseases in Bhutan. Prev. Vet. Med. 2014, 117, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, I.; Kurskaya, O.; Murashkina, T.; Leonov, S.; Sharshov, K.; Kabilov, M.; Alikina, T.; Tolstykh, N.; Gorodov, V.; Alekseev, A.; et al. Genome Sequence of an Unusual Reassortant H1N1 Swine Influenza Virus Isolated from a Pig in Russia, 2016. Genome Announc. 2017, 5, e00747-17. [Google Scholar] [CrossRef]
- Sugimura, T.; Yonemochi, H.; Ogawa, T.; Tanaka, Y.; Kumagai, T. Isolation of a Recombinant Influenza Virus (Hsw1N2) from Swine in Japan. Arch. Virol. 1980, 66, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, S.; Hayashi, T.; Kojima, H.; Usami, Y.; Kubo, M.; Takemae, N.; Uchida, Y.; Saito, T. Occurrence of a Pig Respiratory Disease Associated with Swine Influenza a (H1N2) Virus in Tochigi Prefecture, Japan. J. Vet. Med. Sci. 2010, 72, 481–488. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsai, C.-P.; Pan, M.-J. New H1N2 and H3N1 Influenza Viruses in Taiwanese Pig Herds. Vet. Rec. 2003, 153, 408. [Google Scholar] [PubMed]
- Qi, X.; Lu, C.P. Genetic Characterization of Novel Reassortant H1N2 Influenza A Viruses Isolated from Pigs in Southeastern China. Arch. Virol. 2006, 151, 2289–2299. [Google Scholar] [CrossRef]
- Prakash, N.; Devangi, P.; Madhuuri, K.; Khushbu, P.; Deepali, P. Phylogenetic Analysis of H1N1 Swine Flu Virus Isolated In India. J. Antivir. Antiretrovir. 2011, 3, 11–13. [Google Scholar] [CrossRef]
- Kundin, W.D. Hong Kong A-2 Influenza Virus Infection among Swine during a Human Epidemic in Taiwan. Nature 1970, 228, 857. [Google Scholar] [CrossRef]
- Shortridge, K.F.; Webster, R.G.; Butterfield, W.K.; Campbell, C.H. Persistence of Hong Kong Influenza Virus Variants in Pigs. Science 1977, 196, 1454–1455. [Google Scholar] [CrossRef]
- Singh, P.B.; Rodrigues, F.M.; Gupta, N.P. Haemagglutination-Inhibiting Antibodies against Influenza Type A Viruses in Swine Sera in India. Indian J. Med. Res. 1978, 68, 187–191. [Google Scholar]
- Nerome, K.; Ishida, M.; Nakayama, M.; Oya, A.; Kanai, C.; Suwicha, K. Antigenic and Genetic Analysis of A/Hong Kong (H3N2) Influenza Viruses Isolated from Swine and Man. J. Gen. Virol. 1981, 56, 441–445. [Google Scholar] [CrossRef]
- Nerome, K.; Kanegae, Y.; Shortridge, K.F.; Sugita, S.; Ishida, M. Genetic Analysis of Porcine H3N2 Viruses Originating in Southern China. J. Gen. Virol. 1995, 76, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Arikawa, J.; Yamane, N.; Totsukawa, K.; Ishida, N. The Follow-Up Study of Swine and Hong Kong Influenza Virus Infection among Japanese Hogs. Tohoku J. Exp. Med. 1982, 136, 353–358. [Google Scholar] [CrossRef]
- Kida, H.; Shortridge, K.F.; Webster, R.G. Origin of the Hemagglutinin Gene of H3N2 Influenza Viruses from Pigs in China. Virology 1988, 162, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.M.; Guan, Y.; Markwell, D.; Ghose, P.; Webster, R.G.; Shortridge, K.F. Cocirculation of Avian H9N2 and Contemporary “Human” H3N2 Influenza A Viruses in Pigs in Southeastern China: Potential for Genetic Reassortment? J. Virol. 2001, 75, 9679–9686. [Google Scholar] [CrossRef] [PubMed]
- Lyoo, Y.; Kim, L. Sero-Epidemiology and Genetic Characterization of Swine Influenza Virus. Korean J. Vet. Res. 1998, 38, 53–63. [Google Scholar]
- Zhu, H.; Zhou, B.; Fan, X.; Lam, T.T.Y.; Wang, J.; Chen, A.; Chen, X.; Chen, H.; Webster, R.G.; Webby, R.; et al. Novel Reassortment of Eurasian Avian-Like and Pandemic/2009 Influenza Viruses in Swine: Infectious Potential for Humans. J. Virol. 2011, 85, 10432–10439. [Google Scholar] [CrossRef] [PubMed]
- Song, D.S.; Lee, J.Y.; Oh, J.S.; Lyoo, K.S.; Yoon, K.J.; Park, Y.H.; Park, B.K. Isolation of H3N2 Swine Influenza Virus in South Korea. J. Vet. Diagn. Investig. 2003, 15, 30–34. [Google Scholar] [CrossRef]
- Perera, H.K.K.; Wickramasinghe, G.; Cheung, C.L.; Nishiura, H.; Smith, D.K.; Poon, L.L.M.; Perera, A.K.C.; Ma, S.K.; Sunil-Chandra, N.P.; Guan, Y.; et al. Swine Influenza in Sri Lanka. Emerg. Infect. Dis. 2013, 19, 481–484. [Google Scholar] [CrossRef]
- Conlan, J.V.; Vongxay, K.; Jarman, R.G.; Gibbons, R.V.; Lunt, R.A.; Fenwick, S.; Thompson, R.C.A.; Blacksell, S.D. Serologic Study of Pig-Associated Viral Zoonoses in Laos. Am. J. Trop. Med. Hyg. 2012, 86, 1077–1084. [Google Scholar] [CrossRef]
- Osbjer, K.; Berg, M.; Sokerya, S.; Chheng, K.; San, S.; Davun, H.; Magnusson, U.; Olsen, B.; Zohari, S. Influenza A Virus in Backyard Pigs and Poultry in Rural Cambodia. Transbound. Emerg. Dis. 2017, 64, 1557–1568. [Google Scholar] [CrossRef]
- Saktaganov, N.T.; Klivleyeva, N.G.; Ongarbayeva, N.S.; Glebova, T.I.; Lukmanova, G.V.; Baimukhametova, A.M. Study on Antigenic Relationships and Biological Properties of Swine Influenza a/H1n1 Virus Strains Isolated in Northern Kazakhstan in 2018. Sel’skokhozyaistvennaya Biol. 2020, 55, 355–363. [Google Scholar] [CrossRef]
- Klivleyeva, N.G.; Ongarbayeva, N.S.; Saktaganov, N.T.; Glebova, T.I.; Lukmanova, G.V.; Shamenova, M.G.; Sayatov, M.K.; Berezin, V.E.; Nusupbaeva, G.E.; Aikimbayev, A.M.; et al. Circulation of Influenza Viruses Among Humans and Swine in the Territory of Kazakhstan During 2017–2018. Bulletin 2019, 2, 6–13. [Google Scholar] [CrossRef]
- Ongarbayeva, N.S.; Saktaganov, N.T.; Glebova, T.I.; Shamenova, M.G.; Klivleyeva, N.G. Isolation and Characteristics of Influenza Viruses Circulating among Swine Populations in Kazakhstan during 2018–2019. Bull. Karaganda Univ. Biol. Med. Geogr. Ser. 2022, 105, 70–77. [Google Scholar] [CrossRef]
- Lukmanova, G.; Klivleyeva, N.; Glebova, T.; Ongarbayeva, N.; Shamenova, M.; Saktaganov, N.; Baimukhametova, A.; Baiseiit, S.; Ismagulova, D.; Ismailov, E.; et al. Influenza A Virus Surveillance in Domestic Pigs in Kazakhstan 2018-2021. Ciênc. Rural 2024, 54, e20230403. [Google Scholar] [CrossRef]
- Couacy-Hymann, E.; Kouakou, V.A.; Aplogan, G.L.; Awoume, F.; Kouakou, C.K.; Kakpo, L.; Sharp, B.R.; McClenaghan, L.; McKenzie, P.; Webster, R.G.; et al. Surveillance for Influenza Viruses in Poultry and Swine, West Africa, 2006-2008. Emerg. Infect. Dis. 2012, 18, 1446–1452. [Google Scholar] [CrossRef]
- El Zowalaty, M.E.; Abdelgadir, A.; Borkenhagen, L.K.; Ducatez, M.F.; Bailey, E.S.; Gray, G.C. Influenza A Viruses Are Likely Highly Prevalent in South African Swine Farms. Transbound. Emerg. Dis. 2022, 69, 2373–2383. [Google Scholar] [CrossRef]
- El-Sayed, A.; Awad, W.; Fayed, A.; Hamann, H.P.; Zschöck, M. Avian Influenza Prevalence in Pigs, Egypt. Emerg. Infect. Dis. 2010, 16, 726–727. [Google Scholar] [CrossRef]
- Gomaa, M.R.; Kandeil, A.; El-Shesheny, R.; Shehata, M.M.; McKenzie, P.P.; Webby, R.J.; Ali, M.A.; Kayali, G. Evidence of Infection with Avian, Human, and Swine Influenza Viruses in Pigs in Cairo, Egypt. Arch. Virol. 2018, 163, 359–364. [Google Scholar] [CrossRef]
- Adeola, O.A.; Adeniji, J.A.; Olugasa, B.O. Detection of Haemagglutination-Inhibiting Antibodies against Human H1 and H3 Strains of Influenza A Viruses in Pigs in Ibadan, Nigeria. Zoonoses Public Health 2010, 57, e89–e94. [Google Scholar] [CrossRef]
- Adeola, O.A.; Olugasa, B.O.; Emikpe, B.O. Molecular Detection of Influenza A(H1N1)Pdm09 Viruses with M Genes from Human Pandemic Strains among Nigerian Pigs, 2013-2015: Implications and Associated Risk Factors. Epidemiol. Infect. 2017, 145, 3345–3360. [Google Scholar] [CrossRef]
- Meseko, C.A.; Odaibo, G.N.; Olaleye, D.O. Detection and Isolation of 2009 Pandemic Influenza A/H1N1 Virus in Commercial Piggery, Lagos Nigeria. Vet. Microbiol. 2014, 168, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Meseko, C.; Globig, A.; Ijomanta, J.; Joannis, T.; Nwosuh, C.; Shamaki, D.; Harder, T.; Hoffman, D.; Pohlmann, A.; Beer, M.; et al. Evidence of Exposure of Domestic Pigs to Highly Pathogenic Avian Influenza H5N1 in Nigeria. Sci. Rep. 2018, 8, 5900. [Google Scholar] [CrossRef] [PubMed]
- Jolaoluwa Awosanya, E.; Ogundipe, G.; Babalobi, O.; Omilabu, S. Prevalence and Correlates of Influenza-a in Piggery Workers and Pigs in Two Communities in Lagos, Nigeria. Pan Afr. Med. J. 2013, 16, 102. [Google Scholar] [CrossRef] [PubMed]
- Adeola, O.A.; Olugasa, B.O.; Emikpe, B.O. Antigenic Detection of Human Strain of Influenza Virus A (H3N2) in Swine Populations at Three Locations in Nigeria and Ghana during the Dry Early Months of 2014. Zoonoses Public Health 2016, 63, 106–111. [Google Scholar] [CrossRef]
- Ayim-Akonor, M.; Mertens, E.; May, J.; Harder, T. Exposure of Domestic Swine to Influenza A Viruses in Ghana Suggests Unidirectional, Reverse Zoonotic Transmission at the Human–Animal Interface. Zoonoses Public Health 2020, 67, 697–707. [Google Scholar] [CrossRef]
- Njabo, K.Y.; Fuller, T.L.; Chasar, A.; Pollinger, J.P.; Cattoli, G.; Terregino, C.; Monne, I.; Reynes, J.-M.; Njouom, R.; Smith, T.B. Pandemic A/H1N1/2009 Influenza Virus in Swine, Cameroon, 2010. Vet. Microbiol. 2012, 156, 189–192. [Google Scholar] [CrossRef]
- Larison, B.; Njabo, K.Y.; Chasar, A.; Fuller, T.; Harrigan, R.J.; Smith, T.B. Spillover of PH1N1 to Swine in Cameroon: An Investigation of Risk Factors. BMC Vet. Res. 2014, 10, 55. [Google Scholar] [CrossRef]
- Snoeck, C.J.; Abiola, O.J.; Sausy, A.; Okwen, M.P.; Olubayo, A.G.; Owoade, A.A.; Muller, C.P. Serological Evidence of Pandemic (H1N1) 2009 Virus in Pigs, West and Central Africa. Vet. Microbiol. 2015, 176, 165–171. [Google Scholar] [CrossRef]
- Munyua, P.; Onyango, C.; Mwasi, L.; Waiboci, L.W.; Arunga, G.; Fields, B.; Mott, J.A.; Cardona, C.J.; Kitala, P.; Nyaga, P.N.; et al. Identification and Characterization of Influenza A Viruses in Selected Domestic Animals in Kenya, 2010–2012. PLoS ONE 2018, 13, e0192721. [Google Scholar] [CrossRef]
- Osoro, E.M.; Lidechi, S.; Nyaundi, J.; Marwanga, D.; Mwatondo, A.; Muturi, M.; Ng’ang’a, Z.; Njenga, K. Detection of Pandemic Influenza A/H1N1/Pdm09 Virus among Pigs but Not in Humans in Slaughterhouses in Kenya, 2013–2014. BMC Res. Notes 2019, 12, 628. [Google Scholar] [CrossRef]
- Osoro, E.M.; Lidechi, S.; Marwanga, D.; Nyaundi, J.; Mwatondo, A.; Muturi, M.; Ng’ang’a, Z.; Njenga, K. Seroprevalence of Influenza A Virus in Pigs and Low Risk of Acute Respiratory Illness among Pig Workers in Kenya. Environ. Health Prev. Med. 2019, 24, 53. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, E.; Pascalis, H.; Temmam, S.; Hervé, S.; Saulnier, A.; Turpin, M.; Barbier, N.; Hoarau, J.; Quéguiner, S.; Gorin, S.; et al. Influenza A(H1N1) Pdm09 Virus in Pigs, Réunion Island. Emerg. Infect. Dis. 2012, 18, 1665–1668. [Google Scholar] [CrossRef] [PubMed]
- Dione, M.; Masembe, C.; Akol, J.; Amia, W.; Kungu, J.; Lee, H.S.; Wieland, B. The Importance of On-Farm Biosecurity: Sero-Prevalence and Risk Factors of Bacterial and Viral Pathogens in Smallholder Pig Systems in Uganda. Acta Trop. 2018, 187, 214–221. [Google Scholar] [CrossRef]
- Ducatez, M.F.; Awoume, F.; Webby, R.J. Influenza A(H1N1)Pdm09 Virus in Pigs, Togo, 2013. Vet. Microbiol. 2015, 177, 201–205. [Google Scholar] [CrossRef]
- Tialla, D.; Sausy, A.; Cissé, A.; Sagna, T.; Ilboudo, A.K.; Ouédraogo, G.A.; Hübschen, J.M.; Tarnagda, Z.; Snoeck, C.J. Serological Evidence of Swine Exposure to Pandemic H1N1/2009 Influenza A Virus in Burkina Faso. Vet. Microbiol. 2020, 241, 108572. [Google Scholar] [CrossRef]
- Holyoake, P.K.; Kirkland, P.D.; Davis, R.J.; Arzey, K.E.; Watson, J.; Lunt, R.A.; Wang, J.; Wong, F.; Moloney, B.J.; Dunn, S.E. The First Identified Case of Pandemic H1N1 Influenza in Pigs in Australia. Aust. Vet. J. 2011, 89, 427–431. [Google Scholar] [CrossRef]
- Deng, Y.M.; Iannello, P.; Smith, I.; Watson, J.; Barr, I.G.; Daniels, P.; Komadina, N.; Harrower, B.; Wong, F.Y.K. Transmission of Influenza A(H1N1) 2009 Pandemic Viruses in Australian Swine. Influenza Other Respir. Viruses 2012, 6, e42–e47. [Google Scholar] [CrossRef]
- Smith, D.W.; Barr, I.G.; Loh, R.; Levy, A.; Tempone, S.; O’Dea, M.; Watson, J.; Wong, F.Y.K.; Effler, P.V. Respiratory Illness in a Piggery Associated with the First Identified Outbreak of Swine Influenza in Australia: Assessing the Risk to Human Health and Zoonotic Potential. Trop. Med. Infect. Dis. 2019, 4, 96. [Google Scholar] [CrossRef]
- Wong, F.Y.K.; Donato, C.; Deng, Y.-M.; Teng, D.; Komadina, N.; Baas, C.; Modak, J.; O’Dea, M.; Smith, D.W.; Effler, P.V.; et al. Divergent Human-Origin Influenza Viruses Detected in Australian Swine Populations. J. Virol. 2018, 92, e00316-18. [Google Scholar] [CrossRef]
- Deng, Y.M.; Wong, F.Y.K.; Spirason, N.; Kaye, M.; Beazley, R.; Grau, M.L.; Shan, S.; Stevens, V.; Subbarao, K.; Sullivan, S.; et al. Locally Acquired Human Infection with Swine-Origin Influenza A(H3N2) Variant Virus, Australia, 2018. Emerg. Infect. Dis. 2020, 26, 143–147. [Google Scholar] [CrossRef]
- Donatelli, I.; Campitelli, L.; Castrucci, M.R.; Ruggieri, A.; Sidoli, L.; Oxford, J.S. Detection of Two Antigenic Subpopulations of A(H1N1) Influenza Viruses from Pigs: Antigenic Drift or Interspecies Transmission? J. Med. Virol. 1991, 34, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.H.; Ludwig, S.; Olsen, C.W.; Hannoun, C.; Scholtissek, C.; Hinshaw, V.S.; Harris, P.A.; McCauley, J.W.; Strong, I.; Alexander, D.J. Antigenic and Genetic Analyses of H1N1 Influenza A Viruses from European Pigs. J. Gen. Virol. 1997, 78, 553–562. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kyriakis, C.S.; Brown, I.H.; Foni, E.; Kuntz-Simon, G.; Maldonado, J.; Madec, F.; Essen, S.C.; Chiapponi, C.; Van Reeth, K. Virological Surveillance and Preliminary Antigenic Characterization of Influenza Viruses in Pigs in Five European Countries from 2006 to 2008. Zoonoses Public Health 2011, 58, 93–101. [Google Scholar] [CrossRef]
- Karasin, A.I.; Brown, I.H.; Carman, S.; Olsen, C.W. Isolation and Characterization of H4N6 Avian Influenza Viruses from Pigs with Pneumonia in Canada. J. Virol. 2000, 74, 9322–9327. [Google Scholar] [CrossRef]
- Cong, Y.L.; Wang, C.F.; Yan, C.M.; Peng, J.S.; Jiang, Z.L.; Liu, J.H. Swine Infection with H9N2 Influenza Viruses in China in 2004. Virus Genes 2008, 36, 461–469. [Google Scholar] [CrossRef]
- Cong, Y.L.; Pu, J.; Liu, Q.F.; Wang, S.; Zhang, G.Z.; Zhang, X.L.; Fan, W.X.; Brown, E.G.; Liu, J.H. Antigenic and Genetic Characterization of H9N2 Swine Influenza Viruses in China. J. Gen. Virol. 2007, 88, 2035–2041. [Google Scholar] [CrossRef]
- Xu, C.; Fan, W.; Wei, R.; Zhao, H. Isolation and Identification of Swine Influenza Recombinant A/Swine/Shandong/1/2003(H9N2) Virus. Microbes Infect. 2004, 6, 919–925. [Google Scholar] [CrossRef]
- Choi, Y.K.; Nguyen, T.D.; Ozaki, H.; Webby, R.J.; Puthavathana, P.; Buranathal, C.; Chaisingh, A.; Auewarakul, P.; Hanh, N.T.H.; Ma, S.K.; et al. Studies of H5N1 Influenza Virus Infection of Pigs by Using Viruses Isolated in Vietnam and Thailand in 2004. J. Virol. 2005, 79, 10821–10825. [Google Scholar] [CrossRef]
- Shi, W.F.; Gibbs, M.J.; Zhang, Y.Z.; Zhang, Z.; Zhao, X.M.; Jin, X.; Zhu, C.D.; Yang, M.F.; Yang, N.N.; Cui, Y.J.; et al. Genetic Analysis of Four Porcine Avian Influenza Viruses Isolated from Shandong, China. Arch. Virol. 2008, 153, 211–217. [Google Scholar] [CrossRef]
- Zhu, Q.; Yang, H.; Chen, W.; Cao, W.; Zhong, G.; Jiao, P.; Deng, G.; Yu, K.; Yang, C.; Bu, Z.; et al. A Naturally Occurring Deletion in Its NS Gene Contributes to the Attenuation of an H5N1 Swine Influenza Virus in Chickens. J. Virol. 2008, 82, 220–228. [Google Scholar] [CrossRef]
- Li, H.; Yu, K.; Yang, H.; Xin, X.; Chen, J.; Zhao, P.; Bi, Y. Isolation and Characterization of H5N1 and H9N2 Influenza Viruses from Pigs in China. Chin. J. Prev. Vet. Med. 2003, 26, 1–6. [Google Scholar]
- Takano, R.; Nidom, C.A.; Kiso, M.; Muramoto, Y.; Yamada, S.; Shinya, K.; Sakai-Tagawa, Y.; Kawaoka, Y. A Comparison of the Pathogenicity of Avian and Swine H5N1 Influenza Viruses in Indonesia. Arch. Virol. 2009, 154, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Pascua, P.N.Q.; Song, M.-S.; Baek, Y.H.; Kim, C.-J.; Choi, H.-W.; Sung, M.-H.; Webby, R.J.; Webster, R.G.; Poo, H.; et al. Isolation and Genetic Characterization of H5N2 Influenza Viruses from Pigs in Korea. J. Virol. 2009, 83, 4205–4215. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.Y.; Song, M.S.; Lee, E.H.; Lee, Y.M.; Kim, S.Y.; Kim, H.K.; Choi, J.K.; Kim, C.J.; Webby, R.J.; Choi, Y.K. Isolation and Characterization of Novel H3N1 Swine Influenza Viruses from Pigs with Respiratory Diseases in Korea. J. Clin. Microbiol. 2006, 44, 3923–3927. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Zhou, H.; Jiang, T.; Li, C.; Zhang, A.; Guo, X.; Zou, W.; Chen, H.; Jin, M. Isolation and Molecular Characterization of Equine H3N8 Influenza Viruses from Pigs in China. Arch. Virol. 2009, 154, 887–890. [Google Scholar] [CrossRef]
- Su, S.; Qi, W.; Chen, J.; Cao, N.; Zhu, W.; Yuan, L.; Wang, H.; Zhang, G. Complete Genome Sequence of an Avian-Like H4N8 Swine Influenza Virus Discovered in Southern China. J. Virol. 2012, 86, 9542. [Google Scholar] [CrossRef]
- Zhang, G.; Kong, W.; Qi, W.; Long, L.-P.; Cao, Z.; Huang, L.; Qi, H.; Cao, N.; Wang, W.; Zhao, F.; et al. Identification of an H6N6 Swine Influenza Virus in Southern China. Infect. Genet. Evol. 2011, 11, 1174–1177. [Google Scholar] [CrossRef]
- Subbarao*, K.; Katz, J. Avian Influenza Viruses Infecting Humans. Cell. Mol. Life Sci. 2000, 57, 1770–1784. [Google Scholar] [CrossRef]
- Brockwell-Staats, C.; Webster, R.G.; Webby, R.J. Diversity of Influenza Viruses in Swine and the Emergence of a Novel Human Pandemic Influenza A (H1N1). Influenza Other Respir. Viruses 2009, 3, 207–213. [Google Scholar] [CrossRef]
- Subbarao, K.; Murphy, B.R.; Fauci, A.S. Development of Effective Vaccines against Pandemic Influenza. Immunity 2006, 24, 5–9. [Google Scholar] [CrossRef]
- Forgie, S.E.; Keenliside, J.; Wilkinson, C.; Webby, R.; Lu, P.; Sorensen, O.; Fonseca, K.; Barman, S.; Rubrum, A.; Stigger, E.; et al. Swine Outbreak of Pandemic Influenza A Virus on a Canadian Research Farm Supports Human-to-Swine Transmission. Clin. Infect. Dis. 2011, 52, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Howden, K.J.; Brockhoff, E.J.; Caya, F.D.; McLeod, L.J.; Lavoie, M.; Ing, J.D.; Bystrom, J.M.; Alexandersen, S.; Pasick, J.M.; Berhane, Y.; et al. Special Report Rapport Spécial: An Investigation into Human Pandemic Influenza Virus (H1N1) 2009 on an Alberta Swine Farm. Can. Vet. J. 2009, 50, 1153–1161. [Google Scholar]
- Pascua, P.N.Q.; Lim, G.J.; Kwon, H.i.; Kim, Y.i.; Kim, E.H.; Park, S.J.; Kim, S.M.; Decano, A.G.; Choi, E.J.; Jeon, H.Y.; et al. Complete Genome Sequences of Novel Reassortant H1N2 Swine Influenza Viruses Isolated from Pigs in the Republic of Korea. Genome Announc. 2013, 1, e00552-13. [Google Scholar] [CrossRef] [PubMed]
- Pascua, P.N.Q.; Lim, G.; Kwon, H.; Park, S.; Kim, E.; Song, M.; Kim, C.J.; Choi, Y. Emergence of H3N2pM-like and Novel Reassortant H3N1 Swine Viruses Possessing Segments Derived from the A (H1N1)Pdm09 Influenza Virus, Korea. Influenza Other Respir. Viruses 2013, 7, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Lam, T.T.-Y.; Fan, X.; Chen, X.; Zeng, Y.; Zhou, J.; Duan, L.; Tse, M.; Chan, C.-H.; Li, L.; et al. Expansion of Genotypic Diversity and Establishment of 2009 H1N1 Pandemic-Origin Internal Genes in Pigs in China. J. Virol. 2014, 88, 10864–10874. [Google Scholar] [CrossRef]
- Kim, S.H.; Roh, I.S.; Lee, K.K.; Park, C.K. A Novel Reassortant H1N2 Virus Related to the Pandemic H1N1 2009 Influenza Virus Isolated from Korean Pigs. Virus Genes 2014, 48, 193–198. [Google Scholar] [CrossRef]
- Kanehira, K.; Takemae, N.; Uchida, Y.; Hikono, H.; Saito, T. Reassortant Swine Influenza Viruses Isolated in Japan Contain Genes from Pandemic A(H1N1) 2009. Microbiol. Immunol. 2014, 58, 327–341. [Google Scholar] [CrossRef]
- Caserta, L.C.; Frye, E.A.; Butt, S.L.; Laverack, M.; Nooruzzaman, M.; Covaleda, L.M.; Thompson, A.C.; Koscielny, M.P.; Cronk, B.; Johnson, A.; et al. Spillover of Highly Pathogenic Avian Influenza H5N1 Virus to Dairy Cattle. Nature 2024, 634, 669–676. [Google Scholar] [CrossRef]
- Lim, E.H.; Lim, S.-I.; Kim, M.J.; Kwon, M.; Kim, M.-J.; Lee, K.-B.; Choe, S.; An, D.-J.; Hyun, B.-H.; Park, J.-Y.; et al. First Detection of Influenza D Virus Infection in Cattle and Pigs in the Republic of Korea. Microorganisms 2023, 11, 1751. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).