Single-Nucleus and Spatial Transcriptomics Revealing Host Response Differences Triggered by Mutated Virus in Severe Dengue
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Cells
2.3. Animals
2.4. Animal Experiment
2.5. RNA Quantification
2.6. Histological Analysis
2.7. Multiplex Cytokine Analysis
2.8. Western Blotting Analysis
2.9. Sequence Alignment
2.10. 10× Genomics Chromium snRNA-Seq Data of the Mouse Liver
2.11. 10× Visium Spatial RNA Sequencing Library Preparation and Data Analysis
2.12. Statistics Analysis and Graphs
3. Results
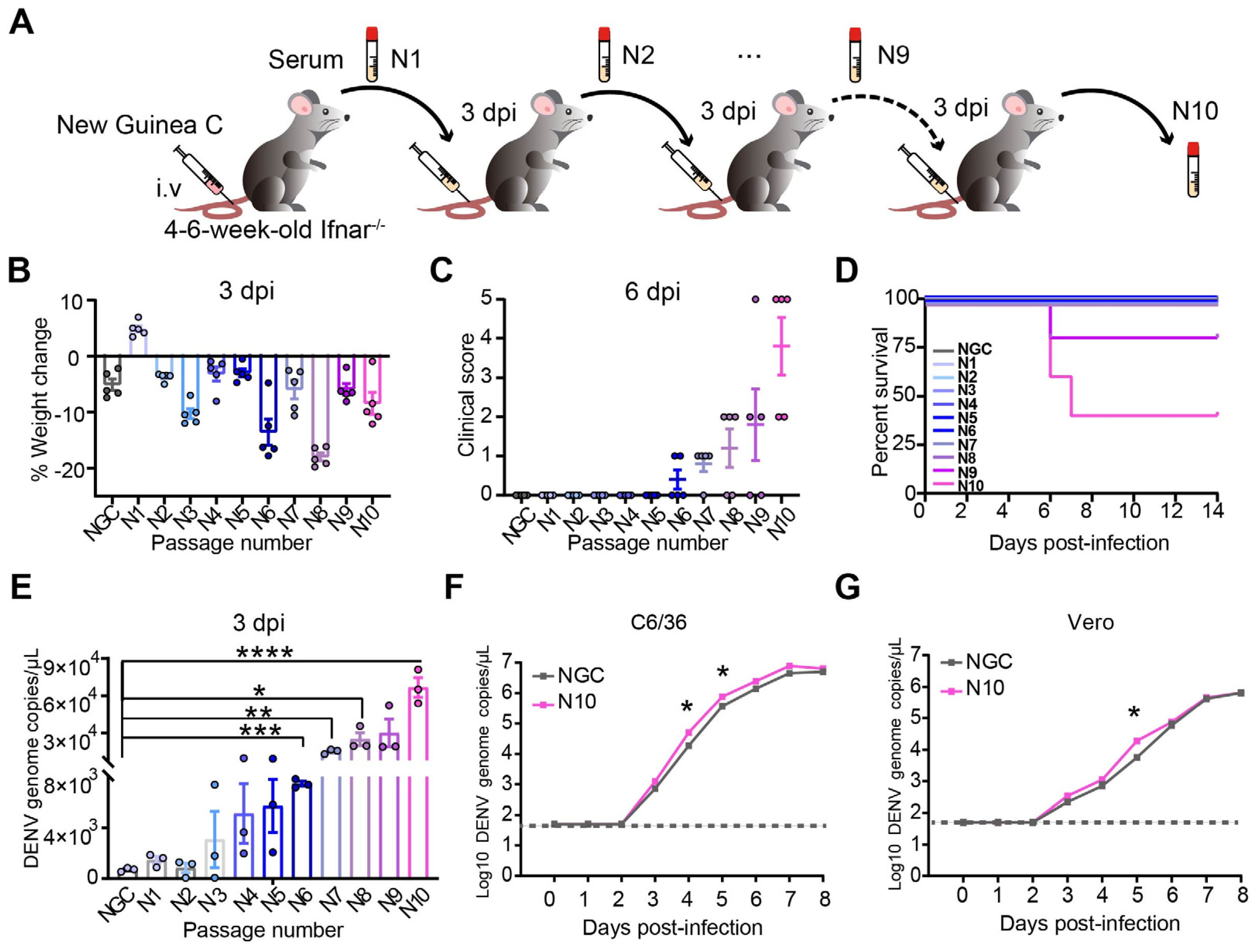
3.1. The Evolution of DENV After Passing Through Ifnar1−/− Mice Increases Viral Pathogenicity and Replicability
3.2. Cumulative Mutations Occur in Adaptive DENV with Enhanced Replication and Pathogenicity
3.3. Cumulative Non-Synonymous Mutations in N10 Cause Severe Liver Damage in Mice
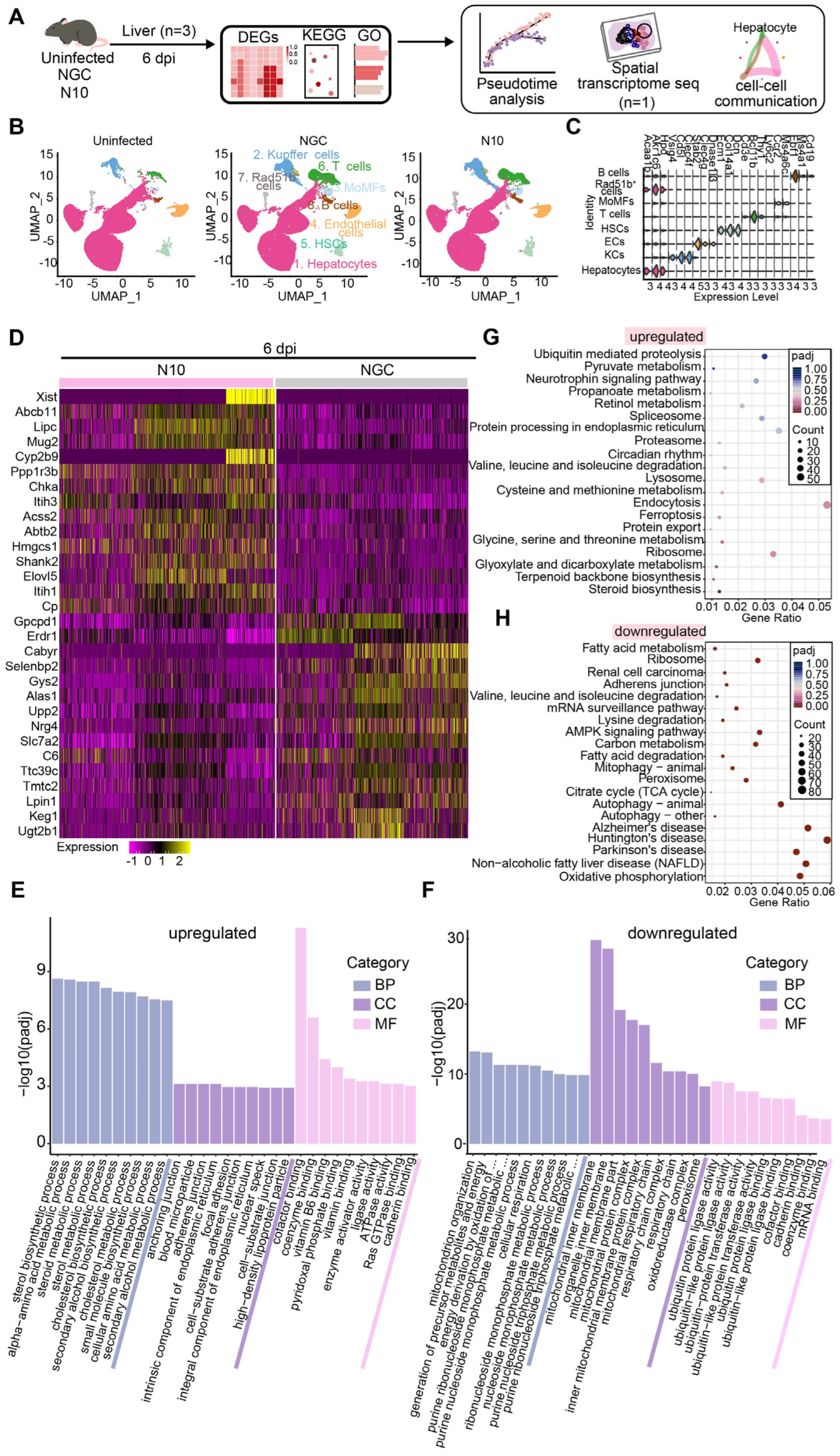
3.4. N10 Strain Virus Induces Aberrant Host Responses in Mouse Livers
3.5. Trajectory of Hepatocellular Differentiation upon N10 Infection
3.6. Spatial Distribution of the Key Host Genes in the Process of N10 Infection
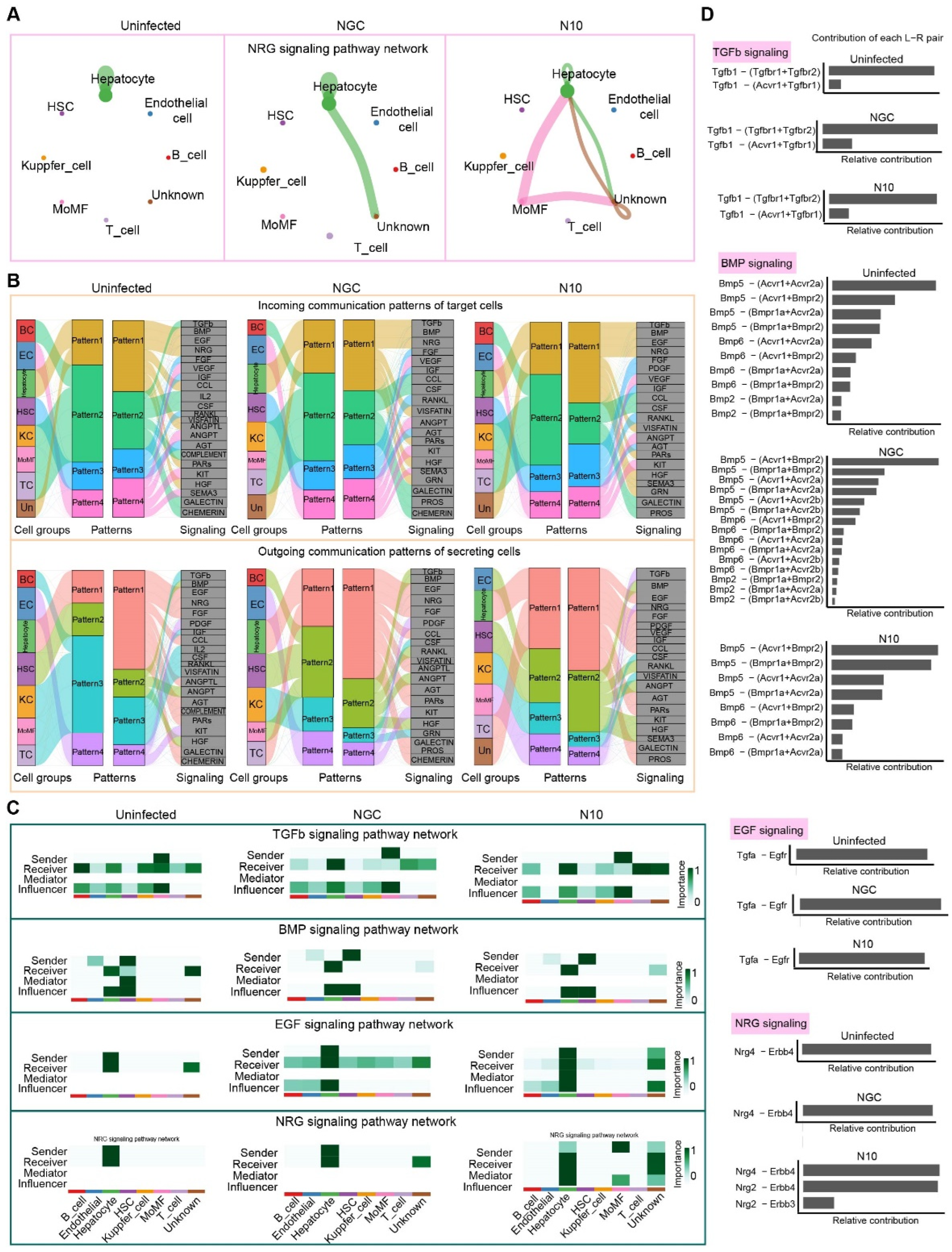
3.7. Cell–Cell Communication in the Process of N10 Infection
3.8. Nrg4 Responds to the Accumulative Mutated N10 Dengue Virus Strain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilder-Smith, A.; Ooi, E.E.; Horstick, O.; Wills, B. Dengue. Lancet 2019, 393, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.D.; Pandey, K.; Dumre, S.P.; Morita, K.; Costello, A. Struggling with a new dengue epidemic in Nepal. Lancet Infect. Dis. 2023, 23, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Burki, T. Bangladesh faces record dengue outbreak. Lancet 2023, 402, 439. [Google Scholar] [CrossRef] [PubMed]
- Sanjeet, B. Dengue outbreak in Peru affects adults and children. Lancet Infect. Dis. 2023, 23, e339. [Google Scholar] [CrossRef]
- Abid, M.A.; Abid, M.B. Climate change and the increased burden of dengue fever in Pakistan. Lancet Infect. Dis. 2023, 23, 17–18. [Google Scholar] [CrossRef]
- Mondal, N. The resurgence of dengue epidemic and climate change in India. Lancet 2023, 401, 727–728. [Google Scholar] [CrossRef]
- Walsh, M.R.; Alam, M.S.; Pierce, K.K.; Carmolli, M.; Alam, M.; Dickson, D.M.; Bak, D.M.; Afreen, S.; Nazib, F.; Golam, K.; et al. Safety and durable immunogenicity of the TV005 tetravalent dengue vaccine, across serotypes and age groups, in dengue-endemic Bangladesh: A randomised, controlled trial. Lancet Infect. Dis. 2023, 24, p150–p160. [Google Scholar] [CrossRef]
- Dengue emergency in the Americas: Time for a new continental eradication plan. Lancet Reg. Health Am. 2023, 22, 100539. [CrossRef]
- Lenharo, M. Dengue is breaking records in the Americas—What’s behind the surge? Nature 2023. online ahead of print. [Google Scholar] [CrossRef]
- Ainsworth, C. Tropical diseases move north. Nature 2023. online ahead of print. [Google Scholar] [CrossRef]
- Osawa, T.; Aoki, M.; Ehara, H.; Sekine, S.I. Structures of dengue virus RNA replicase complexes. Mol. Cell 2023, 83, 2781–2791.e2784. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.M.; Katzelnick, L.; Bedford, T. Dengue genetic divergence generates within-serotype antigenic variation, but serotypes dominate evolutionary dynamics. eLife 2019, 8, e42496. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, E.; Muñoz, M.L.; David, R.E.; Pérez-Ramírez, G.; Navarrete-Espinosa, J.; Díaz-Badillo, Á.; Domínguez-de-la-Cruz, E.; Moreno-Galeana, M.; Brito-Carreón, C.A. Epidemiological implications of the genetic diversification of dengue virus (DENV) serotypes and genotypes in Mexico. Infect. Genet. Evol. 2020, 84, 104391. [Google Scholar] [CrossRef] [PubMed]
- Yenamandra, S.P.; Koo, C.; Chiang, S.; Lim, H.S.J.; Yeo, Z.Y.; Ng, L.C.; Hapuarachchi, H.C. Evolution, heterogeneity and global dispersal of cosmopolitan genotype of Dengue virus type 2. Sci. Rep. 2021, 11, 13496. [Google Scholar] [CrossRef]
- Nwe, K.M.; Ngwe Tun, M.M.; Muthugala, R.; Nabeshima, T.; Balingit, J.C.; Rajamanthri, L.; Jayawardana, D.; Attanayake, S.; Inoue, S.; Takamatsu, Y.; et al. Clinical, Virological, and Immunological Features in Cosmopolitan Genotype DENV-2-Infected Patients during a Large Dengue Outbreak in Sri Lanka in 2017. Am. J. Trop. Med Hyg. 2023, 109, 917–925. [Google Scholar] [CrossRef]
- Jiang, L.; Ma, D.; Ye, C.; Li, L.; Li, X.; Yang, J.; Zhao, Y.; Xi, J.; Wang, X.; Chen, J.; et al. Molecular Characterization of Dengue Virus Serotype 2 Cosmospolitan Genotype From 2015 Dengue Outbreak in Yunnan, China. Front. Cell. Infect. Microbiol. 2018, 8, 219. [Google Scholar] [CrossRef]
- Ngwe Tun, M.M.; Muthugala, R.; Nabeshima, T.; Rajamanthri, L.; Jayawardana, D.; Attanayake, S.; Soe, A.M.; Dumre, S.P.; Ando, T.; Hayasaka, D.; et al. Unusual, neurological and severe dengue manifestations during the outbreak in Sri Lanka, 2017. J. Clin. Virol. 2020, 125, 104304. [Google Scholar] [CrossRef]
- Chan, K.W.K.; Watanabe, S.; Jin, J.Y.; Pompon, J.; Teng, D.; Alonso, S.; Vijaykrishna, D.; Halstead, S.B.; Marzinek, J.K.; Bond, P.J.; et al. A T164S mutation in the dengue virus NS1 protein is associated with greater disease severity in mice. Sci. Transl. Med. 2019, 11, eaat7726. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, X.; Guo, X.; Peng, W.; Zhu, Y.; Wang, Z.; Yu, X.; Shi, H.; Li, Y.; Zhang, L.; et al. Neighboring mutation-mediated enhancement of dengue virus infectivity and spread. EMBO Rep. 2022, 23, e55671. [Google Scholar] [CrossRef]
- Tan, G.K.; Ng, J.K.; Trasti, S.L.; Schul, W.; Yip, G.; Alonso, S. A non mouse-adapted dengue virus strain as a new model of severe dengue infection in AG129 mice. PLoS Negl. Trop. Dis. 2010, 4, e672. [Google Scholar] [CrossRef]
- Yang, J.; Lin, Y.; Jiang, L.; Xi, J.; Wang, X.; Guan, J.; Chen, J.; Pan, Y.; Luo, J.; Ye, C.; et al. Comparison analysis of microRNAs in response to dengue virus type 2 infection between the Vero cell-adapted strain and its source, the clinical C6/36 isolated strain. Virus Res. 2018, 250, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Zhao, L.; Zhang, J.; Wang, Y.; Liu, M.; Hua, D.; Ding, X.; Zhou, X.; Zeng, J.; Yan, H.; et al. Effective Infection with Dengue Virus in Experimental Neonate and Adult Mice through the Intranasal Route. Viruses 2022, 14, 1394. [Google Scholar] [CrossRef] [PubMed]
- Boonyasuppayakorn, S.; Reichert, E.D.; Manzano, M.; Nagarajan, K.; Padmanabhan, R. Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antivir. Res. 2014, 106, 125–134. [Google Scholar] [CrossRef]
- Krishnaswami, S.R.; Grindberg, R.V.; Novotny, M.; Venepally, P.; Lacar, B.; Bhutani, K.; Linker, S.B.; Pham, S.; Erwin, J.A.; Miller, J.A. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 2016, 11, 499–524. [Google Scholar] [CrossRef]
- Dong, Y.; Jain, R.W.; Lozinski, B.M.; D’Mello, C.; Visser, F.; Ghorbani, S.; Zandee, S.; Brown, D.I.; Prat, A.; Xue, M. Single-cell and spatial RNA sequencing identify perturbators of microglial functions with aging. Nat. Aging 2022, 2, 508–525. [Google Scholar] [CrossRef]
- Wang, S.; Zha, L.; Cui, X.; Yeh, Y.T.; Liu, R.; Jing, J.; Shi, H.; Chen, W.; Hanover, J.; Yin, J. Epigenetic regulation of hepatic lipid metabolism by DNA methylation. Adv. Sci. 2023, 10, 2206068. [Google Scholar] [CrossRef]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 2024, 19, 3292–3320. [Google Scholar] [CrossRef]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.-H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Hudson, W.H.; Sudmeier, L.J. Localization of T cell clonotypes using the Visium spatial transcriptomics platform. STAR Protoc. 2022, 3, 101391. [Google Scholar] [CrossRef]
- Tamura, T.; Zhang, J.; Madan, V.; Biswas, A.; Schwoerer, M.P.; Cafiero, T.R.; Heller, B.L.; Wang, W.; Ploss, A. Generation and characterization of genetically and antigenically diverse infectious clones of dengue virus serotypes 1-4. Emerg. Microbes Infect. 2022, 11, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Kudlacek, S.T.; Metz, S.; Thiono, D.; Payne, A.M.; Phan, T.T.N.; Tian, S.; Forsberg, L.J.; Maguire, J.; Seim, I.; Zhang, S.; et al. Designed, highly expressing, thermostable dengue virus 2 envelope protein dimers elicit quaternary epitope antibodies. Sci. Adv. 2021, 7, eabg4084. [Google Scholar] [CrossRef] [PubMed]
- Munt, J.E.; Henein, S.; Adams, C.; Young, E.; Hou, Y.J.; Conrad, H.; Zhu, D.; Dong, S.; Kose, N.; Yount, B.; et al. Homotypic antibodies target novel E glycoprotein domains after natural DENV 3 infection/vaccination. Cell Host Microbe 2023, 31, 1850–1865.e1855. [Google Scholar] [CrossRef] [PubMed]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med. 2015, 7, 304ra142. [Google Scholar] [CrossRef]
- Prestwood, T.R.; Prigozhin, D.M.; Sharar, K.L.; Zellweger, R.M.; Shresta, S. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J. Virol. 2008, 82, 8411–8421. [Google Scholar] [CrossRef]
- Prasad, D.; Bhriguvanshi, A. Clinical Profile, Liver Dysfunction and Outcome of Dengue Infection in Children: A Prospective Observational Study. Pediatr. Infect. Dis. J. 2020, 39, 97–101. [Google Scholar] [CrossRef]
- Devarbhavi, H.; Ganga, D.; Menon, M.; Kothari, K.; Singh, R. Dengue hepatitis with acute liver failure: Clinical, biochemical, histopathological characteristics and predictors of outcome. J. Gastroenterol. Hepatol. 2020, 35, 1223–1228. [Google Scholar] [CrossRef]
- Cunha, M.S.; de Moura Coletti, T.; Guerra, J.M.; Ponce, C.C.; Fernandes, N.; Résio, R.A.; Claro, I.M.; Salles, F.; Lima Neto, D.F.; Sabino, E. A fatal case of dengue hemorrhagic fever associated with dengue virus 4 (DENV-4) in Brazil: Genomic and histopathological findings. Braz. J. Microbiol. 2022, 53, 1305–1312. [Google Scholar] [CrossRef]
- Robinson, M.W.; Harmon, C.; O’Farrelly, C. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 2016, 13, 267–276. [Google Scholar] [CrossRef]
- Dang, T.T.; Pham, M.H.; Bui, H.V.; Van Le, D. Whole genome sequencing and genetic variations in several dengue virus type 1 strains from unusual dengue epidemic of 2017 in Vietnam. Virol. J. 2020, 17, 7. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Coello Escoto, A.; Huang, A.T.; Garcia-Carreras, B.; Chowdhury, N.; Maljkovic Berry, I.; Chavez, C.; Buchy, P.; Duong, V.; Dussart, P.; et al. Antigenic evolution of dengue viruses over 20 years. Science 2021, 374, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Waickman, A.T.; Lu, J.Q.; Fang, H.; Waldran, M.J.; Gebo, C.; Currier, J.R.; Ware, L.; Van Wesenbeeck, L.; Verpoorten, N.; Lenz, O.; et al. Evolution of inflammation and immunity in a dengue virus 1 human infection model. Sci. Transl. Med. 2022, 14, eabo5019. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.C.; Lima de Mendonça, M.C.; Damasceno Dos Santos Rodrigues, C.; Fonseca, V.; Ribeiro, M.S.; Brandão, A.P.; Venâncio da Cunha, R.; Dias, A.I.; Santos Vilas Boas, L.; Felix, A.C.; et al. Dengue Virus Serotype 2 Intrahost Diversity in Patients with Different Clinical Outcomes. Viruses 2021, 13, 349. [Google Scholar] [CrossRef]
- Alen, M.M.; Schols, D. Dengue virus entry as target for antiviral therapy. J. Trop. Med. 2012, 2012, 628475. [Google Scholar] [CrossRef]
- Talarico, L.B.; Damonte, E.B. Characterization of in vitro dengue virus resistance to carrageenan. J. Med. Virol. 2016, 88, 1120–1129. [Google Scholar] [CrossRef]
- Laiton-Donato, K.; Alvarez, D.A.; Peláez-Carvajal, D.; Mercado, M.; Ajami, N.J.; Bosch, I.; Usme-Ciro, J.A. Molecular characterization of dengue virus reveals regional diversification of serotype 2 in Colombia. Virol. J. 2019, 16, 62. [Google Scholar] [CrossRef]
- Fibriansah, G.; Lim, X.N.; Lok, S.M. Morphological Diversity and Dynamics of Dengue Virus Affecting Antigenicity. Viruses 2021, 13, 1446. [Google Scholar] [CrossRef]
- Deval, H.; Behera, S.P.; Agrawal, A.; Singh, R.; Misra, B.; Janardhan, V.; Patil, G.; Sah, K.; Kumar, N.; Singh, R.; et al. Genetic characterization of dengue virus serotype 2 isolated from dengue fever outbreaks in eastern Uttar Pradesh and western Bihar, India. J. Med. Virol. 2021, 93, 3322–3329. [Google Scholar] [CrossRef]
- Gualano, R.C.; Pryor, M.J.; Cauchi, M.R.; Wright, P.J.; Davidson, A.D. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J. Gen. Virol. 1998, 79 Pt 3, 437–446. [Google Scholar] [CrossRef]
- Davidson, A.D. Development and application of dengue virus reverse genetic systems. Methods Mol. Biol. 2014, 1138, 113–130. [Google Scholar] [CrossRef]
- VanBlargan, L.A.; Mukherjee, S.; Dowd, K.A.; Durbin, A.P.; Whitehead, S.S.; Pierson, T.C. The type-specific neutralizing antibody response elicited by a dengue vaccine candidate is focused on two amino acids of the envelope protein. PLoS Pathog. 2013, 9, e1003761. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, L.; Jiang, T.; Li, X.; Fan, H.; Hong, W.; Zhang, Y.; Zhu, Q.; Ye, Q.; Tong, Y.; et al. Isolation and characterization of dengue virus serotype 2 from the large dengue outbreak in Guangdong, China in 2014. Sci. China Life Sci. 2014, 57, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Mittal, V.; Chhabra, M.; Kumari, R.; Singh, P.; Venkatesh, S. Molecular epidemiology and evolutionary analysis of dengue virus type 2, circulating in Delhi, India. Virusdisease 2016, 27, 400–404. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Yuan, Y.; Cai, F.; Li, Z.; Wei, Q.; Wang, W. Single-Nucleus and Spatial Transcriptomics Revealing Host Response Differences Triggered by Mutated Virus in Severe Dengue. Viruses 2024, 16, 1779. https://doi.org/10.3390/v16111779
Chen Q, Yuan Y, Cai F, Li Z, Wei Q, Wang W. Single-Nucleus and Spatial Transcriptomics Revealing Host Response Differences Triggered by Mutated Virus in Severe Dengue. Viruses. 2024; 16(11):1779. https://doi.org/10.3390/v16111779
Chicago/Turabian StyleChen, Qian, Yizhen Yuan, Fangzhou Cai, Zhe Li, Qiang Wei, and Wei Wang. 2024. "Single-Nucleus and Spatial Transcriptomics Revealing Host Response Differences Triggered by Mutated Virus in Severe Dengue" Viruses 16, no. 11: 1779. https://doi.org/10.3390/v16111779
APA StyleChen, Q., Yuan, Y., Cai, F., Li, Z., Wei, Q., & Wang, W. (2024). Single-Nucleus and Spatial Transcriptomics Revealing Host Response Differences Triggered by Mutated Virus in Severe Dengue. Viruses, 16(11), 1779. https://doi.org/10.3390/v16111779





