Low gH/gL (Sub)Species-Specific Antibody Levels Indicate Elephants at Risk of Fatal Elephant Endotheliotropic Herpesvirus Hemorrhagic Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Serum Samples
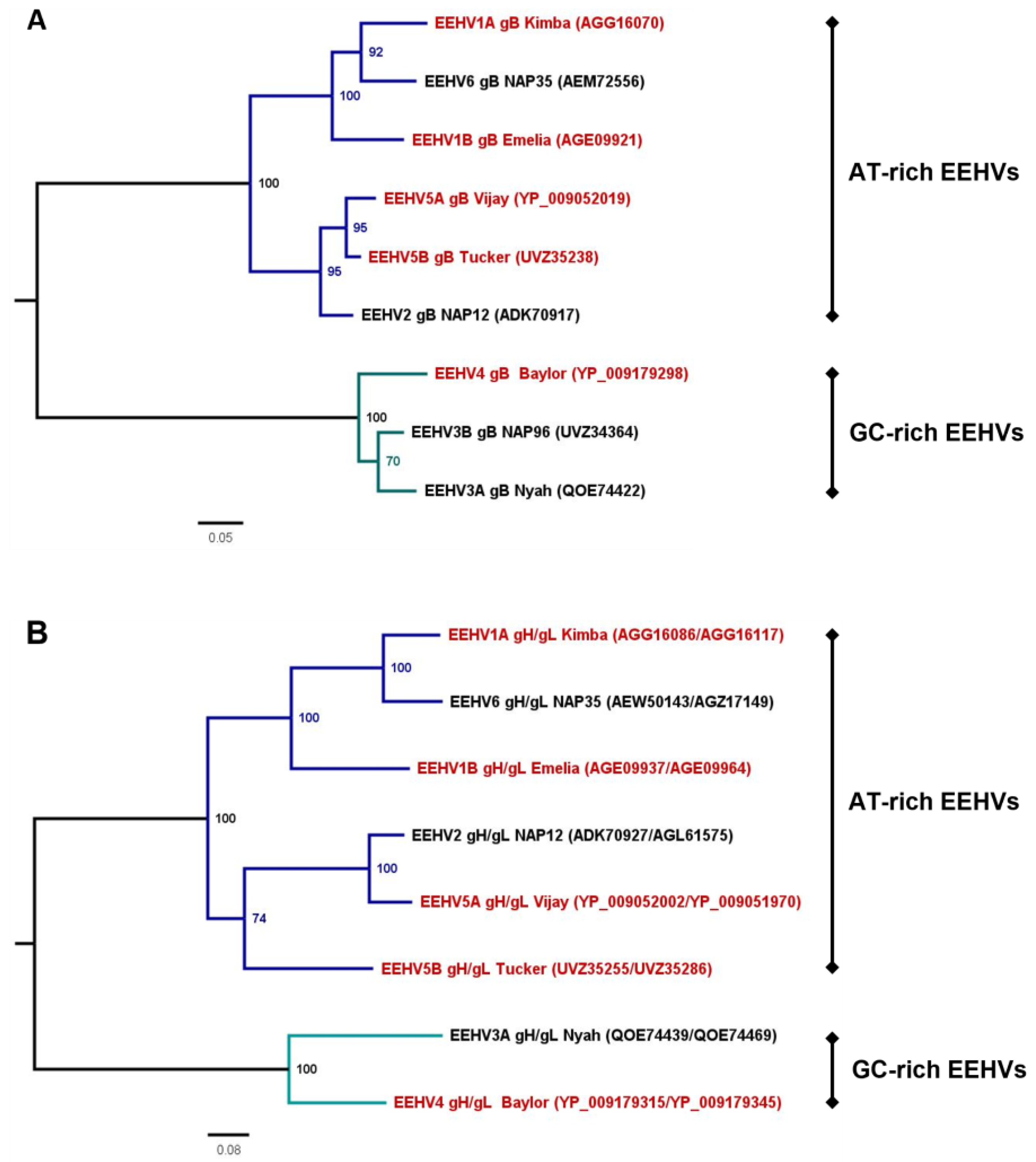
2.2. Phylogenetic Analyses
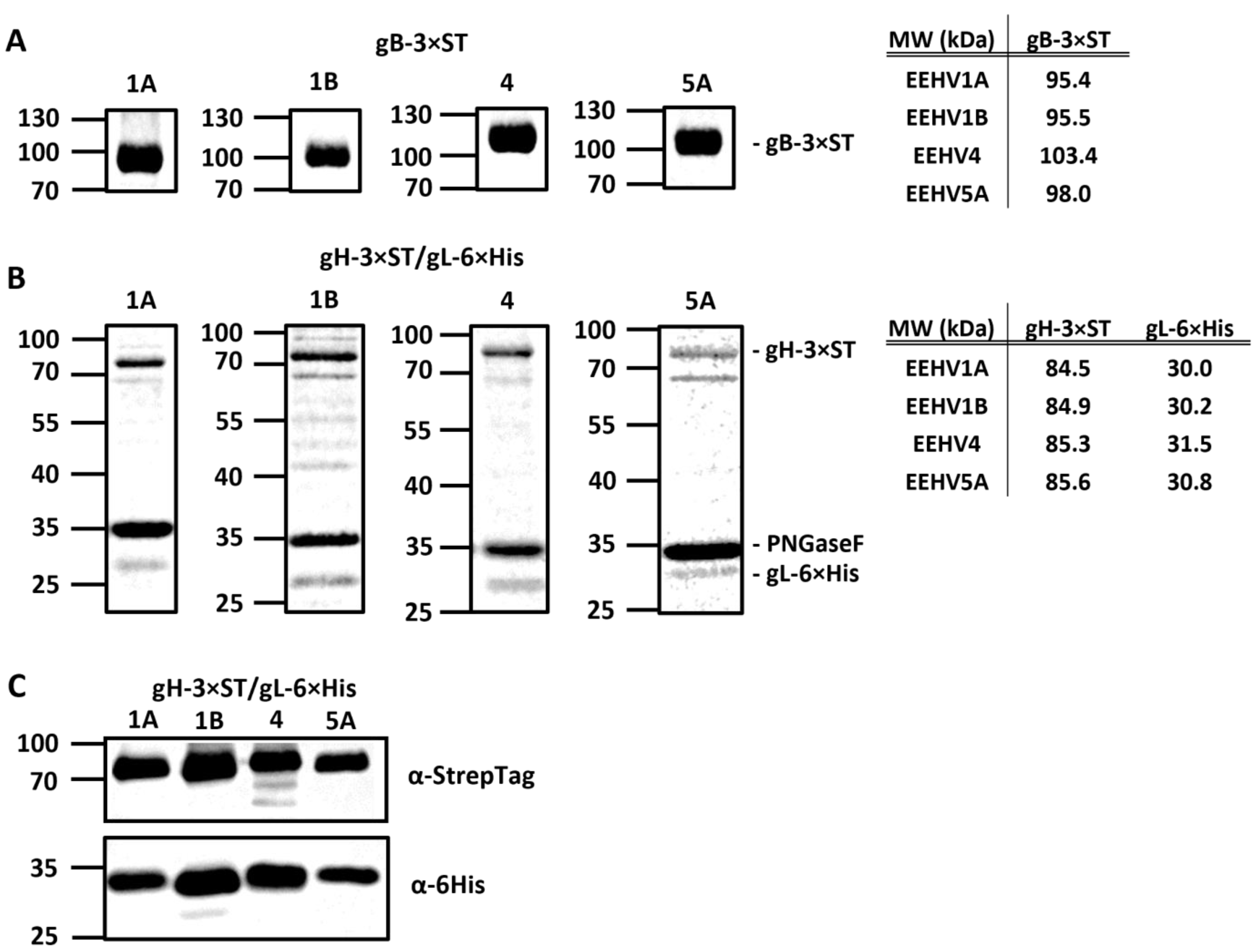
2.3. Expression of Recombinant EEHV Proteins
2.4. ELISAs
2.5. Statistical Analyses
3. Results
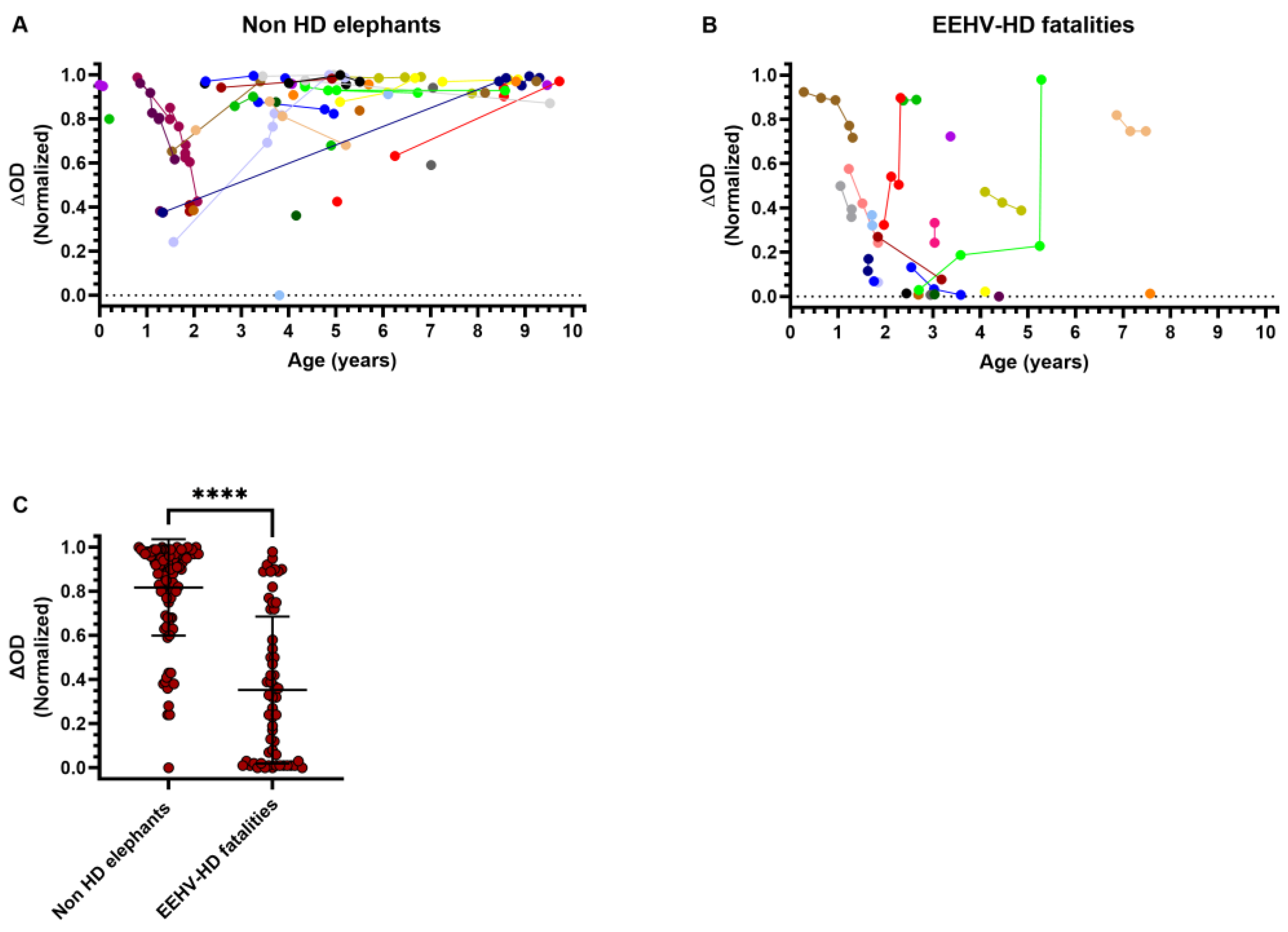
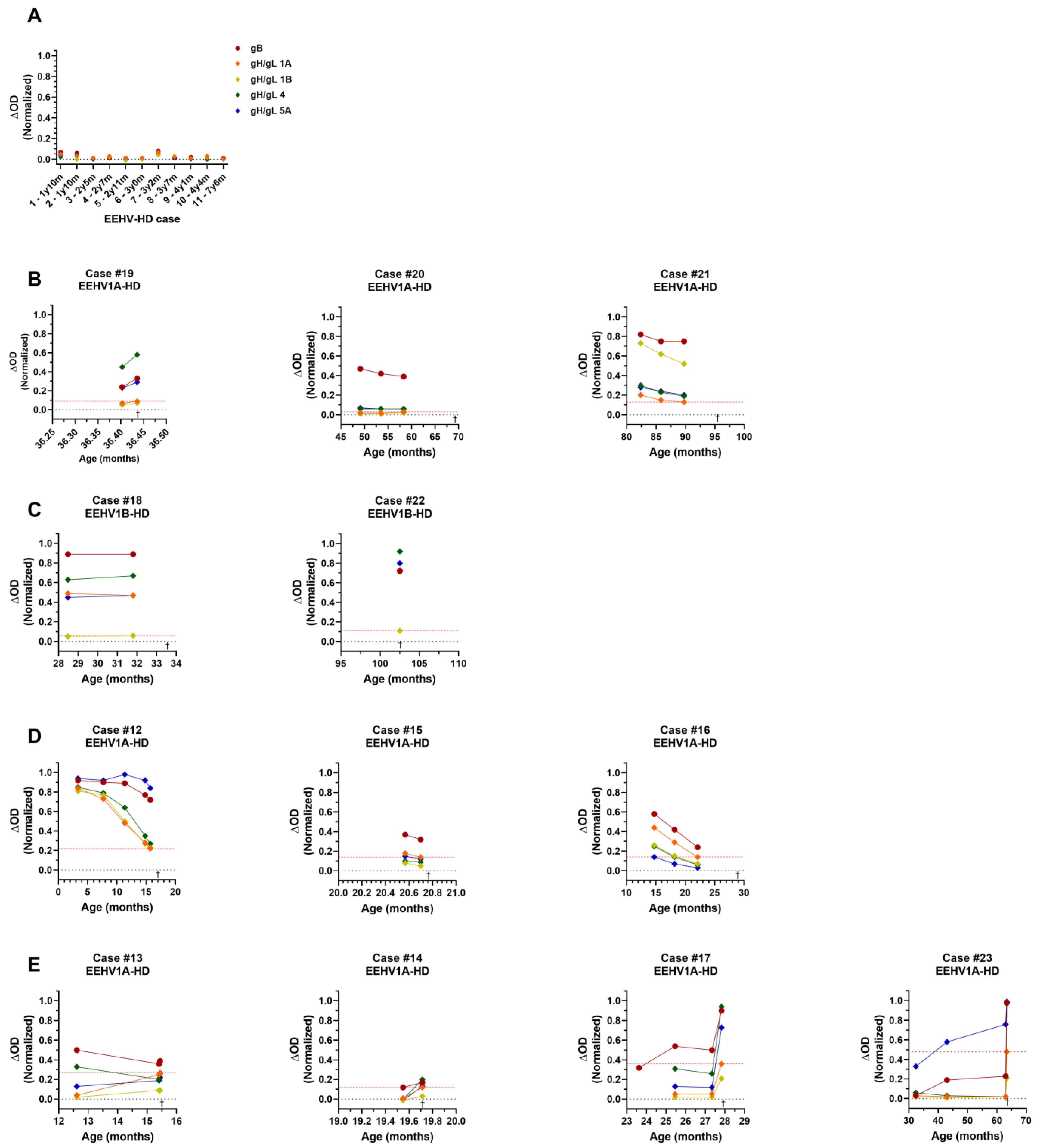
3.1. High Levels of EEHV gB-Specific Antibodies Found in Sera of Several EEHV-HD Fatalities
3.2. Development of gB and gH/gL ELISAs for EEHV1A, 1B, 4, and 5A
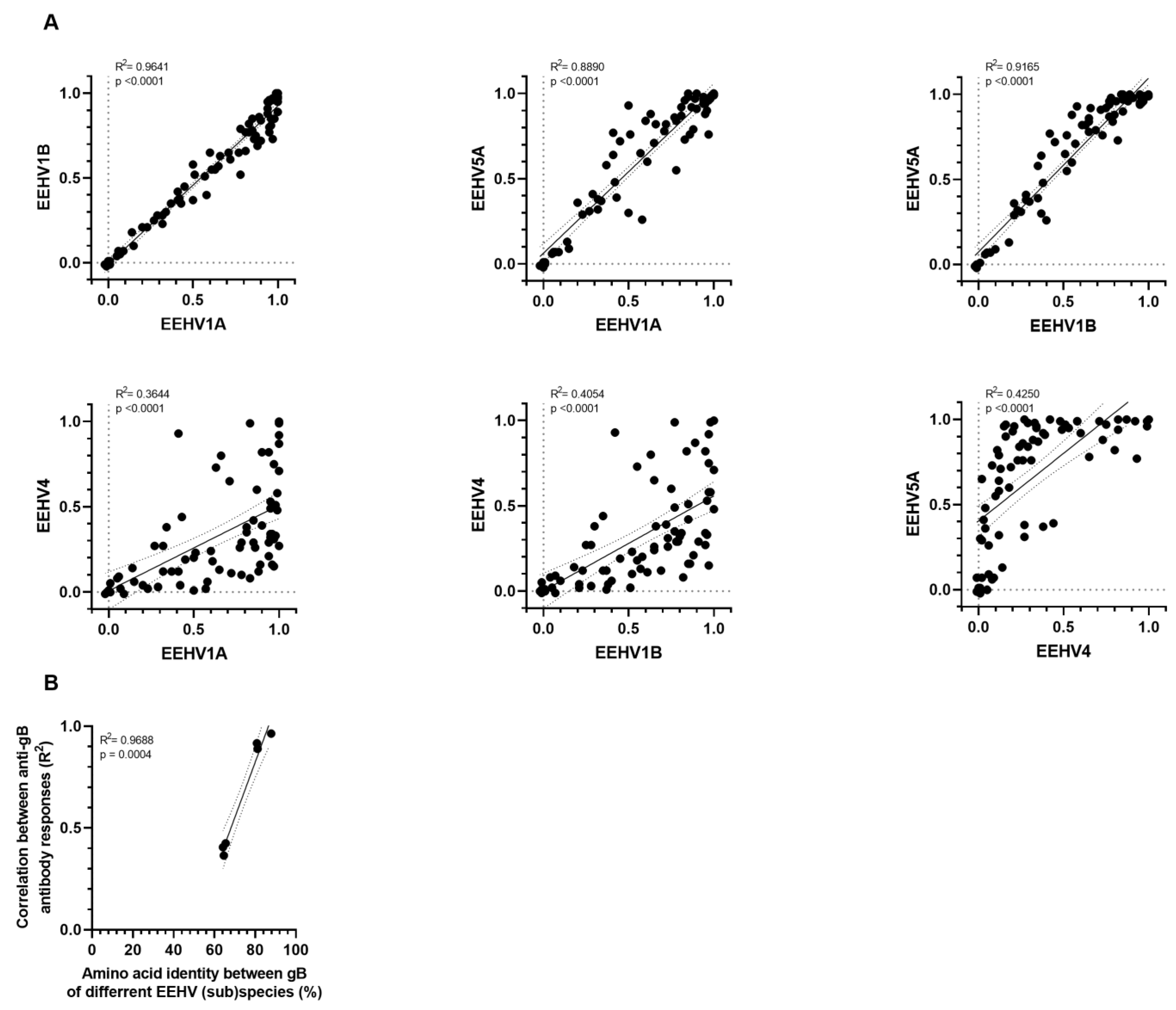
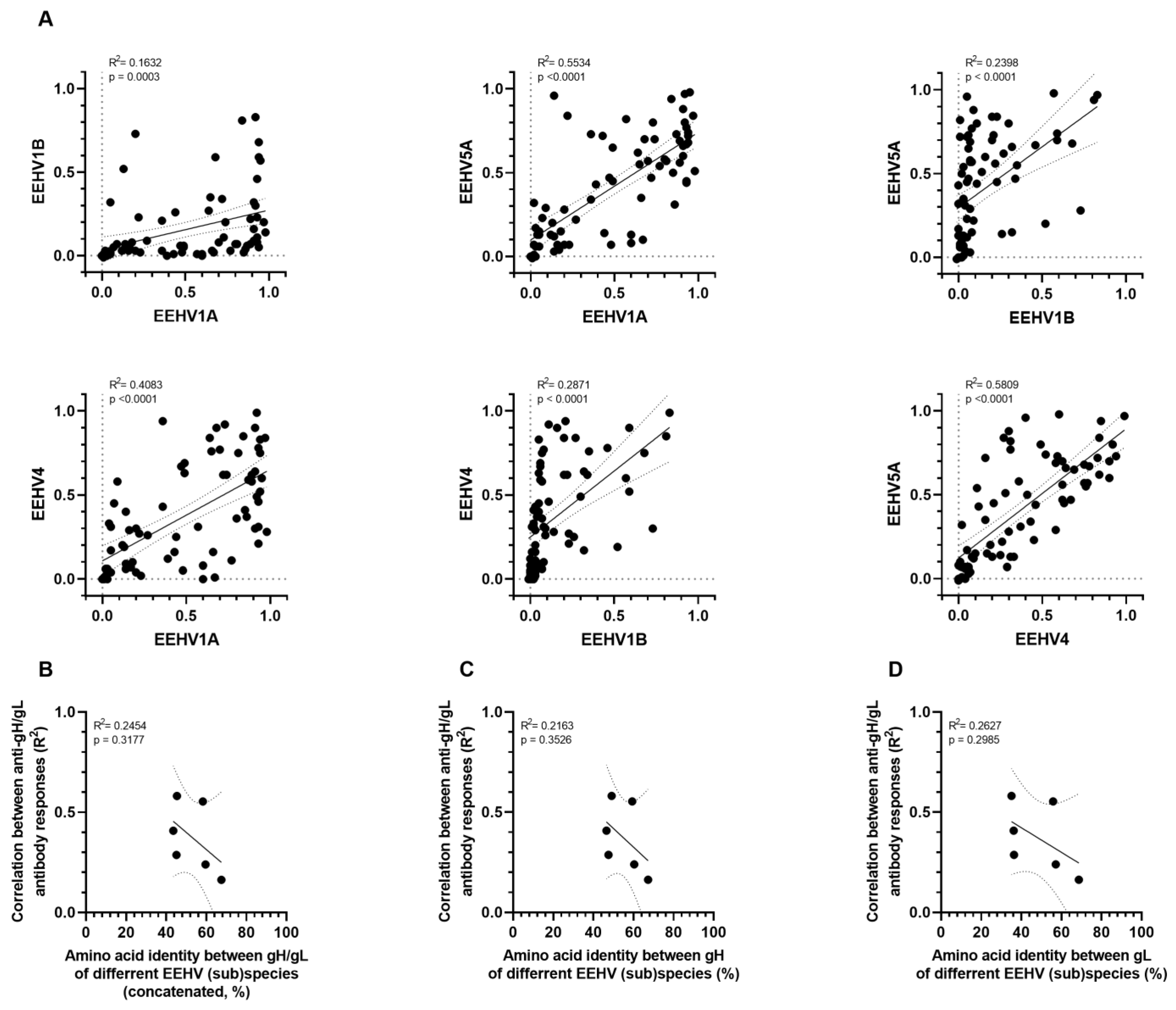
3.3. Correlation amongst gB and gH/gL-Specific Antibody Levels of the Different EEHV (Sub)Species
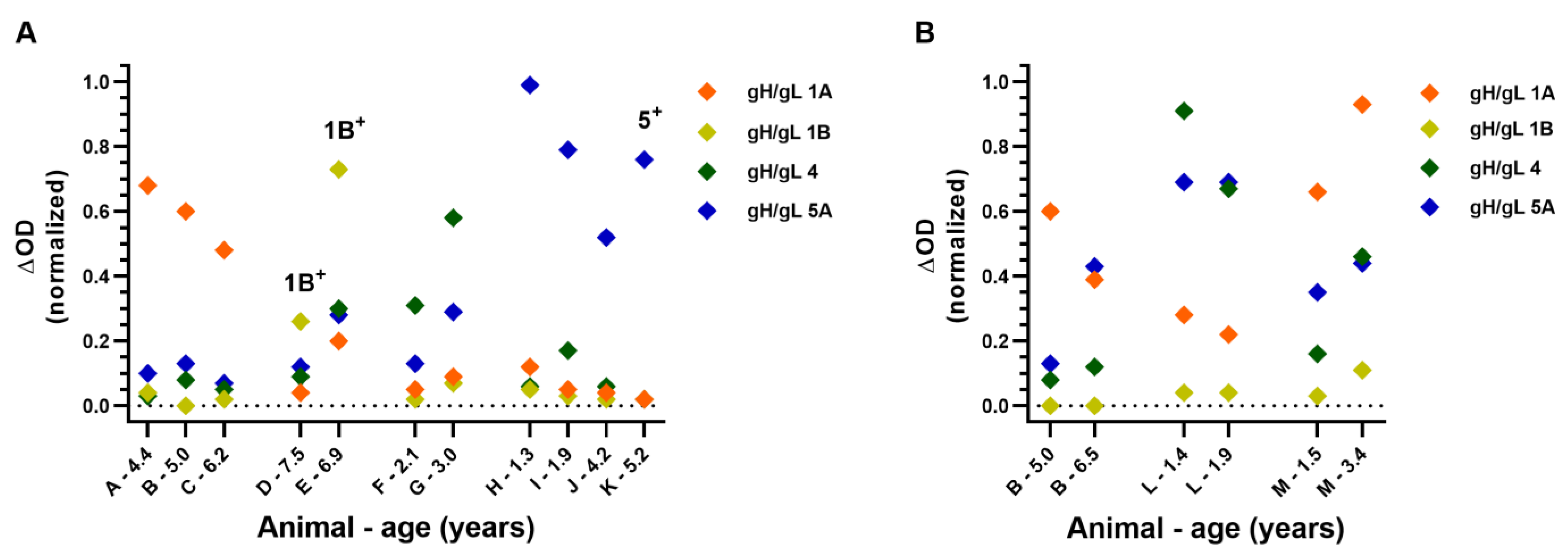
3.4. Antibodies Specific for gH/gL of a Single EEHV (Sub)Species Were Discerned in Several Animals
3.5. EEHV-HD Fatalities Never Have High Antibody Levels to gH/gL of the (Sub)Species They Succumbed to
3.6. Seropositivity to gH/gL of the Different EEHV (Sub)Species Increases with Age
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Long, S.Y.; Latimer, E.M.; Hayward, G.S. Review of Elephant Endotheliotropic Herpesviruses and Acute Hemorrhagic Disease. ILAR J. 2016, 56, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Fuery, A.; Pursell, T.; Tan, J.; Peng, R.; Burbelo, P.D.; Hayward, G.S.; Ling, P.D. Lethal Hemorrhagic Disease and Clinical Illness Associated with Elephant Endotheliotropic Herpesvirus 1 Are Caused by Primary Infection: Implications for the Detection of Diagnostic Proteins. J. Virol. 2020, 94, e01528-19. [Google Scholar] [CrossRef] [PubMed]
- Hoornweg, T.E.; Schaftenaar, W.; Maurer, G.; van den Doel, P.B.; Molenaar, F.M.; Chamouard-Galante, A.; Vercammen, F.; Rutten, V.; de Haan, C.A.M. Elephant Endotheliotropic Herpesvirus Is Omnipresent in Elephants in European Zoos and an Asian Elephant Range Country. Viruses 2021, 13, 283. [Google Scholar] [CrossRef] [PubMed]
- Pursell, T.; Spencer Clinton, J.L.; Tan, J.; Peng, R.; Qin, X.; Doddapaneni, H.; Menon, V.; Momin, Z.; Kottapalli, K.; Howard, L.; et al. Primary Infection May Be an Underlying Factor Contributing to Lethal Hemorrhagic Disease Caused by Elephant Endotheliotropic Herpesvirus 3 in African Elephants (Loxodonta africana). Microbiol. Spectr. 2021, 9, e0098321. [Google Scholar] [CrossRef] [PubMed]
- Hoornweg, T.E.; Perera, V.P.; Karunarathne, R.N.S.; Schaftenaar, W.; Mahakapuge, T.A.N.; Kalupahana, A.W.; Rutten, V.; de Haan, C.A.M. Young elephants in a large herd maintain high levels of elephant endotheliotropic herpesvirus-specific antibodies and do not succumb to fatal haemorrhagic disease. Transbound. Emerg. Dis. 2022, 69, e3379–e3385. [Google Scholar] [CrossRef] [PubMed]
- Stanton, J.J.; Zong, J.C.; Latimer, E.; Tan, J.; Herron, A.; Hayward, G.S.; Ling, P.D. Detection of pathogenic elephant endotheliotropic herpesvirus in routine trunk washes from healthy adult Asian elephants (Elephas maximus) by use of a real-time quantitative polymerase chain reaction assay. Am. J. Vet. Res. 2010, 71, 925–933. [Google Scholar] [CrossRef]
- Ackermann, M.; Hatt, J.-M.; Schetle, N.; Steinmetz, H. Identification of shedders of elephant endotheliotropic herpesviruses among Asian elephants (Elephas maximus) in Switzerland. PLoS ONE 2017, 12, e0176891. [Google Scholar] [CrossRef] [PubMed]
- Howard, L.; Schaftenaar, W. Elephant Endotheliotropic Herpesvirus. In Fowler’s Zoo and Wild Animal Medicine Current Therapy; Elsevier: Saunders, PA, USA, 2018; Volume 9. [Google Scholar]
- Perrin, K.L.; Saxmose Nielsen, S.; Martinussen, T.; Bertelsen, M.F. Quantification and risk factor analysis of elephant endotheliotropic herpesvirus-haemorrhagic disease fatalities in Asian elephants (Elephas maximus) in Europe (1985–2017). J. Zoo Aquar. Res. 2021, 9, 8–13. [Google Scholar]
- Jesus, S.A.; Doherr, M.G.; Hildebrandt, T.B. Elephant Endotheliotropic Herpesvirus Impact in the European Asian Elephant (Elephas maximus) Population: Are Hereditability and Zoo-Associated Factors Linked with Mortality? Animals 2021, 11, 2816. [Google Scholar] [CrossRef]
- Reid, C.E.; Hildebrandt, T.B.; Marx, N.; Hunt, M.; Thy, N.; Reynes, J.M.; Schaftenaar, W.; Fickel, J. Endotheliotropic elephant herpes virus (EEHV) infection. The first PCR-confirmed fatal case in Asia. Vet. Q. 2006, 28, 61–64. [Google Scholar] [CrossRef]
- Zachariah, A.; Sajesh, P.K.; Santhosh, S.; Bathrachalam, C.; Megha, M.; Pandiyan, J.; Jishnu, M.; Kobragade, R.S.; Long, S.Y.; Zong, J.C.; et al. Extended genotypic evaluation and comparison of twenty-two cases of lethal EEHV1 hemorrhagic disease in wild and captive Asian elephants in India. PLoS ONE 2018, 13, e0202438. [Google Scholar] [CrossRef]
- Oo, Z.M.; Aung, Y.H.; Aung, T.T.; San, N.; Tun, Z.M.; Hayward, G.S.; Zachariah, A. Elephant Endotheliotropic Herpesvirus Hemorrhagic Disease in Asian Elephant Calves in Logging Camps, Myanmar. Emerg. Infect. Dis. 2020, 26, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Fayette, M.A.; Brenner, E.E.; Garner, M.M.; Bowman, M.R.; Latimer, E.; Proudfoot, J.S. Acute hemorrhagic disease due to elephant endotheliotropic herpesvirus 3A infection in five African elephants (Loxodonta africana) at one North American zoological institution. J. Wildl. Wildl. Med. 2021, 52, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Nofs, S.A.; Atmar, R.L.; Keitel, W.A.; Hanlon, C.; Stanton, J.J.; Tan, J.; Flanagan, J.P.; Howard, L.; Ling, P.D. Prenatal passive transfer of maternal immunity in Asian elephants (Elephas maximus). Vet. Immunol. Immunopathol. 2013, 153, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.L.; Molter, C.M.; Flanagan, J.P.; Bauer, K.L.; Bernardy, R.; Hoffman, D.; Parkinson, L.; Brainard, B.M.; Evans, T.S.; Pursell, T.; et al. Novel diagnostic and therapeutic approaches to elephant endotheliotropic herpesvirus 1A hemorrhagic disease in a captive juvenile Asian elephant (Elephas maximus). J. Zoo Wildl. Med. 2022, 53, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232-5. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.A. Immunology and Evolution of Infectious Disease; Princeton University Press: Princeton, NJ, USA, 2002. [Google Scholar]
- Zhong, L.; Zhang, W.; Krummenacher, C.; Chen, Y.; Zheng, Q.; Zhao, Q.; Zeng, M.-S.; Xia, N.; Zeng, Y.-X.; Xu, M.; et al. Targeting herpesvirus entry complex and fusogen glycoproteins with prophylactic and therapeutic agents. Trends Microbiol. 2023, 31, 788–804. [Google Scholar] [CrossRef]
- Chen, W.-H.; Kim, J.; Bu, W.; Board, N.L.; Tsybovsky, Y.; Wang, Y.; Hostal, A.; Andrews, S.F.; Gillespie, R.A.; Choe, M.; et al. Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition. Immunity 2022, 55, 2135–2148.e6. [Google Scholar] [CrossRef]
- Fricke, T.; Großkopf, A.K.; Ensser, A.; Backovic, M.; Hahn, A.S. Antibodies Targeting KSHV gH/gL Reveal Distinct Neutralization Mechanisms. Viruses 2022, 14, 541. [Google Scholar] [CrossRef]
- Zehner, M.; Alt, M.; Ashurov, A.; Goldsmith, J.A.; Spies, R.; Weiler, N.; Lerma, J.; Gieselmann, L.; Stöhr, D.; Gruell, H.; et al. Single-cell analysis of memory B cells from top neutralizers reveals multiple sites of vulnerability within HCMV Trimer and Pentamer. Immunity 2023, 56, 2602–2620.e10. [Google Scholar] [CrossRef]
- Peng, T.; Ponce-de-Leon, M.; Jiang, H.; Dubin, G.; Lubinski, J.M.; Eisenberg, R.J.; Cohen, G.H. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J. Virol. 1998, 72, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Pursell, T.; Spencer Clinton, J.L.; Tan, J.; Peng, R.; Ling, P.D. Modified vaccinia Ankara expressing EEHV1A glycoprotein B elicits humoral and cell-mediated immune responses in mice. PLoS ONE 2022, 17, e0265424. [Google Scholar] [CrossRef] [PubMed]
- Spencer Clinton, J.L.; Hoornweg, T.E.; Tan, J.; Peng, R.; Schaftenaar, W.; Rutten, V.; de Haan, C.A.M.; Ling, P.D. EEHV1A glycoprotein B subunit vaccine elicits humoral and cell-mediated immune responses in mice. Vaccine 2022, 40, 5131–5140. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.S.; Herold, B.C.; Permar, S.R. A new era in cytomegalovirus vaccinology: Considerations for rational design of next-generation vaccines to prevent congenital cytomegalovirus infection. Npj Vaccines 2018, 3, 38. [Google Scholar] [CrossRef]


| Protein | EEHV Subspecies | Strain | GenBank Accession Number | Amino Acid Residues Included | Induced Amino Acid Substitutions | Reference |
|---|---|---|---|---|---|---|
| gB | 1A | Case 8 | AAN03667 | 43-685 | F126H, Y128T, W209A, R432K, R433K, R434K, R436K | [3] |
| gB | 1B | Emelia | AGE09921 | 30-671 | F113H, Y115T, W196A, R418K, R419K, R421K | - |
| gB | 4 | Baylor | YP_009179298 | 55-700 | Y145H, Y147T, W227A, R447K, R450K | - |
| gB | 5A | Vijay | YP_009052019 | 29-671 | V112H, Y114T, W195A, R417K, R420K | - |
| gH | 1A | Kimba | AGG16086 | 30-706 | - | [3] |
| gH | 1B | Emelia | AGE09937 | 26-704 | - | - |
| gH | 4 | Baylor | YP_009179315 | 43-723 | - | - |
| gH | 5A | Vijay | YP_009052002 | 28-712 | - | - |
| gL | 1A | Kimba | AGG16117 | 57-304 | - | [3] |
| gL | 1B | Emelia | AGE09964 | 19-264 | - | - |
| gL | 4 | Baylor | YP_009179345 | 21-275 | - | - |
| gL | 5A | Vijay | YP_009051970 | 11-266 | - | - |
| EEHV (Sub)Species | 1A | 6 | 1B | 5A | 5B | 2 | 3A | 3B | 4 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Natural host | EM | Lox | EM | EM | EM | Lox | Lox | Lox | EM | ||
| Genbank accession number | AGG16070 | AEM72556 | AGE09921 | YP_009052019 | UVZ35238 | ADK70917 | QOE74422 | UVZ34364 | YP_009179298 | ||
| 1A | EM | AGG16070 | 89.6 | 87.4 | 79.7 | 80.6 | 81.1 | 64.5 | 65.3 | 64.1 | |
| 6 | Lox | AEM72556 | 89.6 | 87.9 | 81.2 | 82.3 | 82.1 | 64.8 | 64.9 | 64.5 | |
| 1B | EM | AGE09921 | 87.4 | 87.9 | 81.0 | 81.8 | 82.0 | 65.0 | 65.6 | 64.3 | |
| 5A | EM | YP_009052019 | 79.7 | 81.2 | 81.0 | 95.8 | 92.6 | 67.0 | 67.4 | 65.4 | |
| 5B | EM | UVZ35238 | 80.6 | 82.3 | 81.8 | 95.8 | 93.5 | 67.7 | 68.6 | 66.8 | |
| 2 | Lox | ADK70917 | 81.1 | 82.1 | 82.0 | 92.6 | 93.5 | 67.2 | 67.8 | 66.4 | |
| 3A | Lox | QOE74422 | 64.5 | 64.8 | 65.0 | 67.0 | 67.7 | 67.2 | 94.3 | 89.9 | |
| 3B | Lox | UVZ34364 | 65.3 | 64.9 | 65.6 | 67.4 | 68.6 | 67.8 | 94.3 | 90.9 | |
| 4 | EM | YP_009179298 | 64.1 | 64.5 | 64.3 | 65.4 | 66.8 | 66.4 | 89.9 | 90.9 | |
| EEHV (Sub)Species | 1A | 6 | 1B | 5B | 2 | 5A | 4 | 3A | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Natural host | EM | Lox | EM | EM | Lox | EM | EM | Lox | ||
| Genbank accession number | AGG16086/AGG16117 | AEW50143/AGZ17149 | AGE09937/AGE09964 | UVZ35255/UVZ35286 | ADK70927/AGL61575 | YP_009052002/YP_009051970 | YP_009179315/YP_009179345 | QOE74439/QOE74469 | ||
| 1A | EM | AGG16086/AGG16117 | 82.0 | 67.5 | 60.8 | 58.9 | 58.2 | 43.8 | 42.0 | |
| 6 | Lox | AEW50143/AGZ17149 | 82.0 | 67.3 | 60.4 | 58.9 | 59.1 | 44.5 | 42.9 | |
| 1B | EM | AGE09937/AGE09964 | 67.5 | 67.3 | 62.1 | 60.3 | 59.5 | 44.7 | 44.7 | |
| 5B | EM | UVZ35255/UVZ35286 | 60.8 | 60.4 | 62.1 | 64.2 | 64.0 | 44.6 | 43.9 | |
| 2 | Lox | ADK70927/AGL61575 | 58.9 | 58.9 | 60.3 | 64.2 | 87.0 | 46.5 | 43.8 | |
| 5A | EM | YP_009052002/YP_009051970 | 58.2 | 59.1 | 59.5 | 64.0 | 87.0 | 45.9 | 43.0 | |
| 4 | EM | YP_009179315/YP_009179345 | 43.8 | 44.5 | 44.7 | 44.6 | 46.5 | 45.9 | 67.7 | |
| 3A | Lox | QOE74439/QOE74469 | 42.0 | 42.9 | 44.7 | 43.9 | 43.8 | 43.0 | 67.7 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoornweg, T.E.; Schaftenaar, W.; Rutten, V.P.M.G.; de Haan, C.A.M. Low gH/gL (Sub)Species-Specific Antibody Levels Indicate Elephants at Risk of Fatal Elephant Endotheliotropic Herpesvirus Hemorrhagic Disease. Viruses 2024, 16, 268. https://doi.org/10.3390/v16020268
Hoornweg TE, Schaftenaar W, Rutten VPMG, de Haan CAM. Low gH/gL (Sub)Species-Specific Antibody Levels Indicate Elephants at Risk of Fatal Elephant Endotheliotropic Herpesvirus Hemorrhagic Disease. Viruses. 2024; 16(2):268. https://doi.org/10.3390/v16020268
Chicago/Turabian StyleHoornweg, Tabitha E., Willem Schaftenaar, Victor P. M. G. Rutten, and Cornelis A. M. de Haan. 2024. "Low gH/gL (Sub)Species-Specific Antibody Levels Indicate Elephants at Risk of Fatal Elephant Endotheliotropic Herpesvirus Hemorrhagic Disease" Viruses 16, no. 2: 268. https://doi.org/10.3390/v16020268
APA StyleHoornweg, T. E., Schaftenaar, W., Rutten, V. P. M. G., & de Haan, C. A. M. (2024). Low gH/gL (Sub)Species-Specific Antibody Levels Indicate Elephants at Risk of Fatal Elephant Endotheliotropic Herpesvirus Hemorrhagic Disease. Viruses, 16(2), 268. https://doi.org/10.3390/v16020268






