The Mytilus chilensis Steamer-like Element-1 Retrotransposon Antisense mRNA Harbors an Internal Ribosome Entry Site That Is Modulated by hnRNPK
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cell Culture and Nuclear Extract Preparation
2.3. Mussels RNAs and DNA Samples
2.4. cDNA Synthesis and Quantitative PCR
2.5. In Vitro Transcription
2.6. In Vitro Translation
2.7. DNA, RNA and siRNA Transfection
2.8. Luciferase Assays
2.9. Western Blot
2.10. Statistical Analysis
3. Results
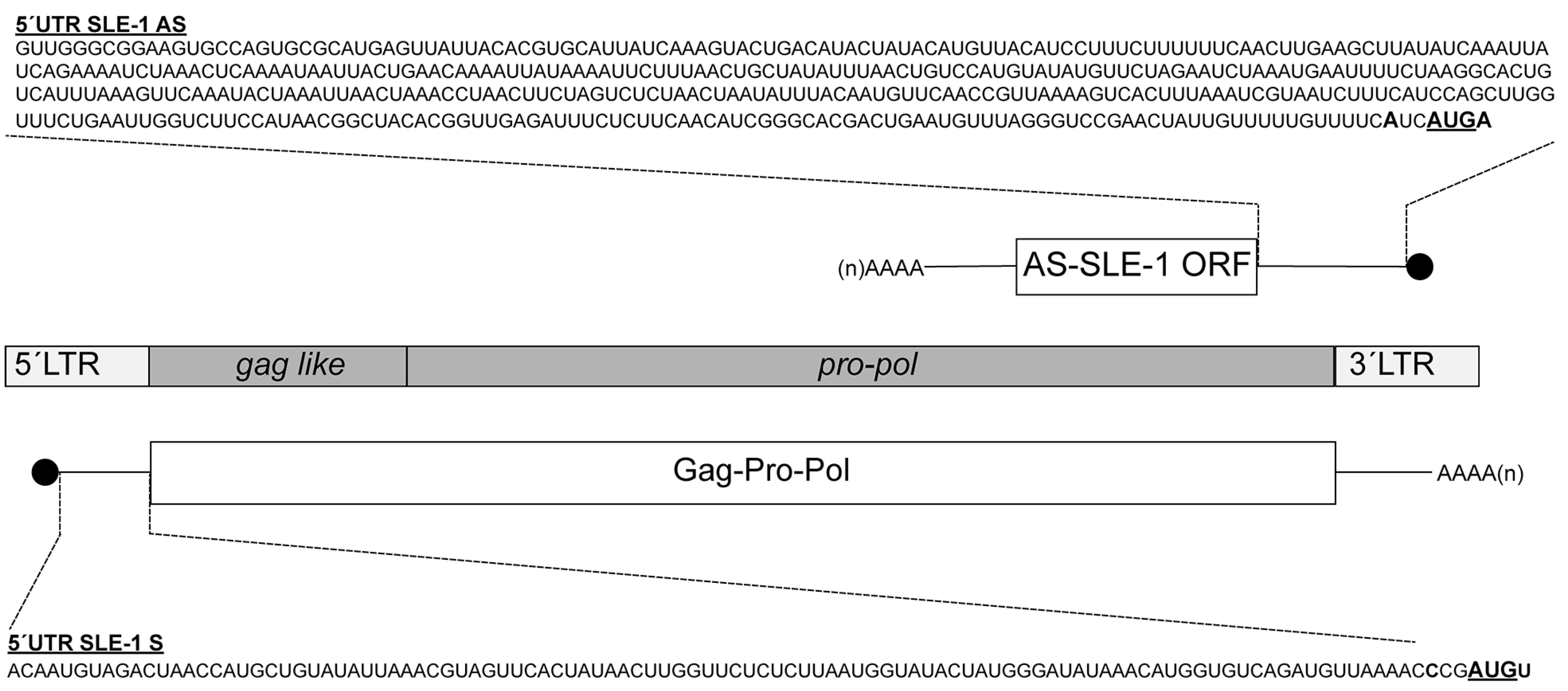
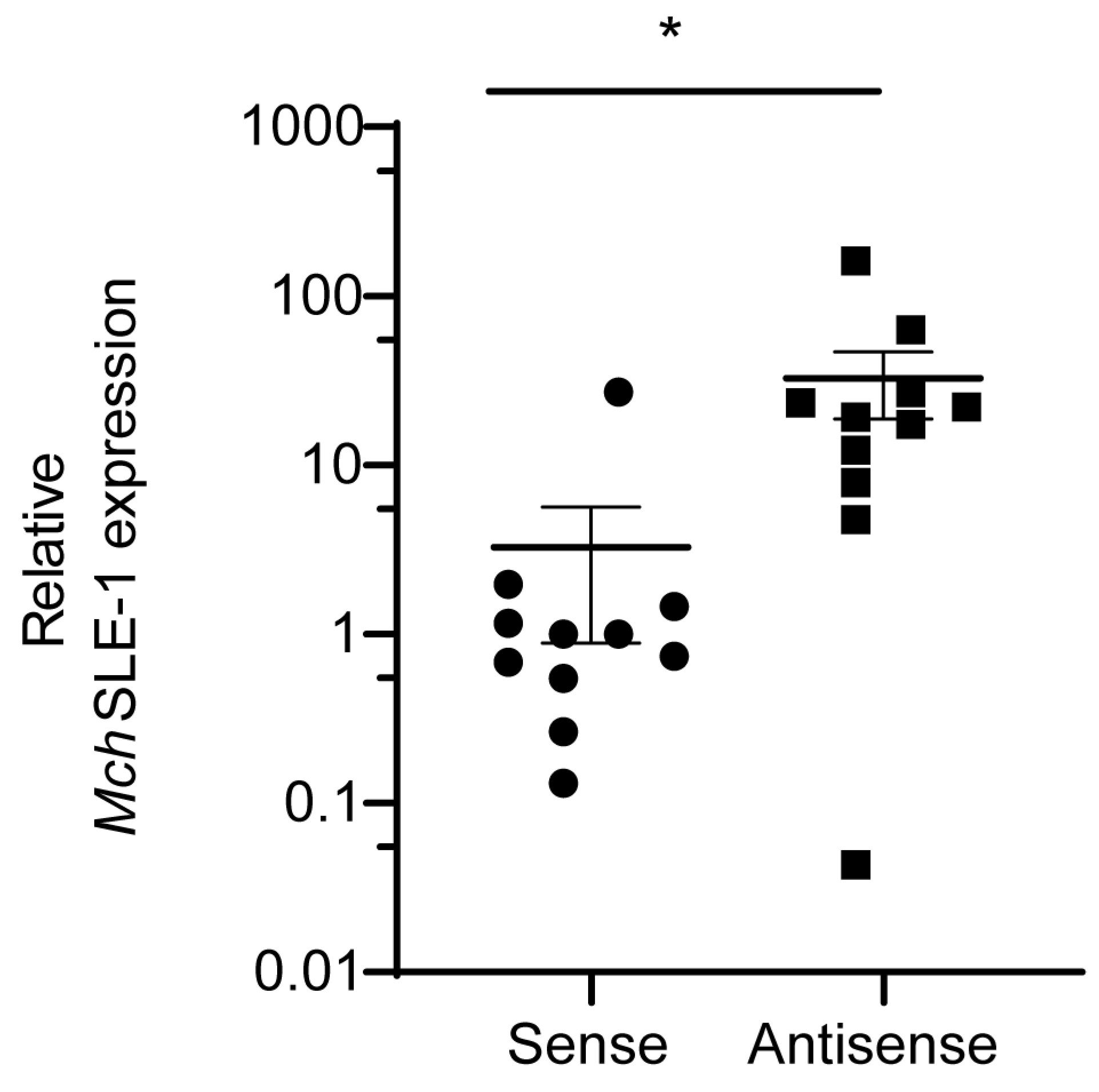
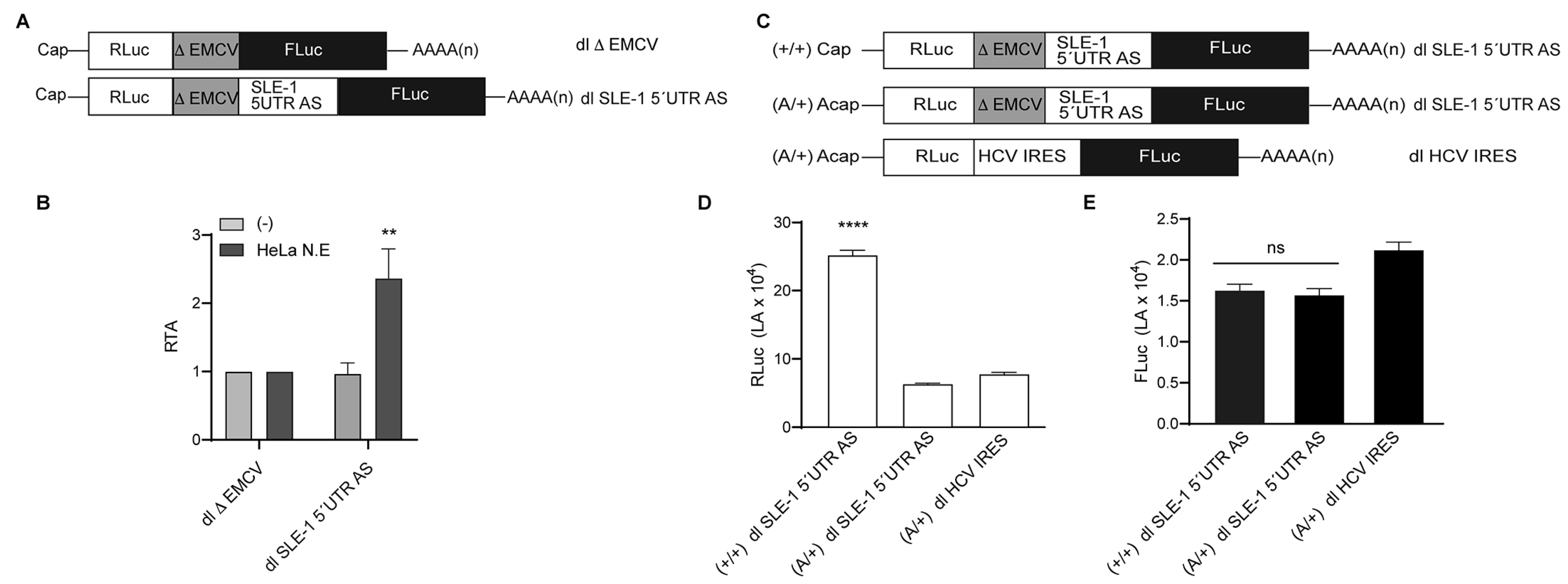
3.1. MchSLE-1 RNA Is Transcribed from Both DNA Strands
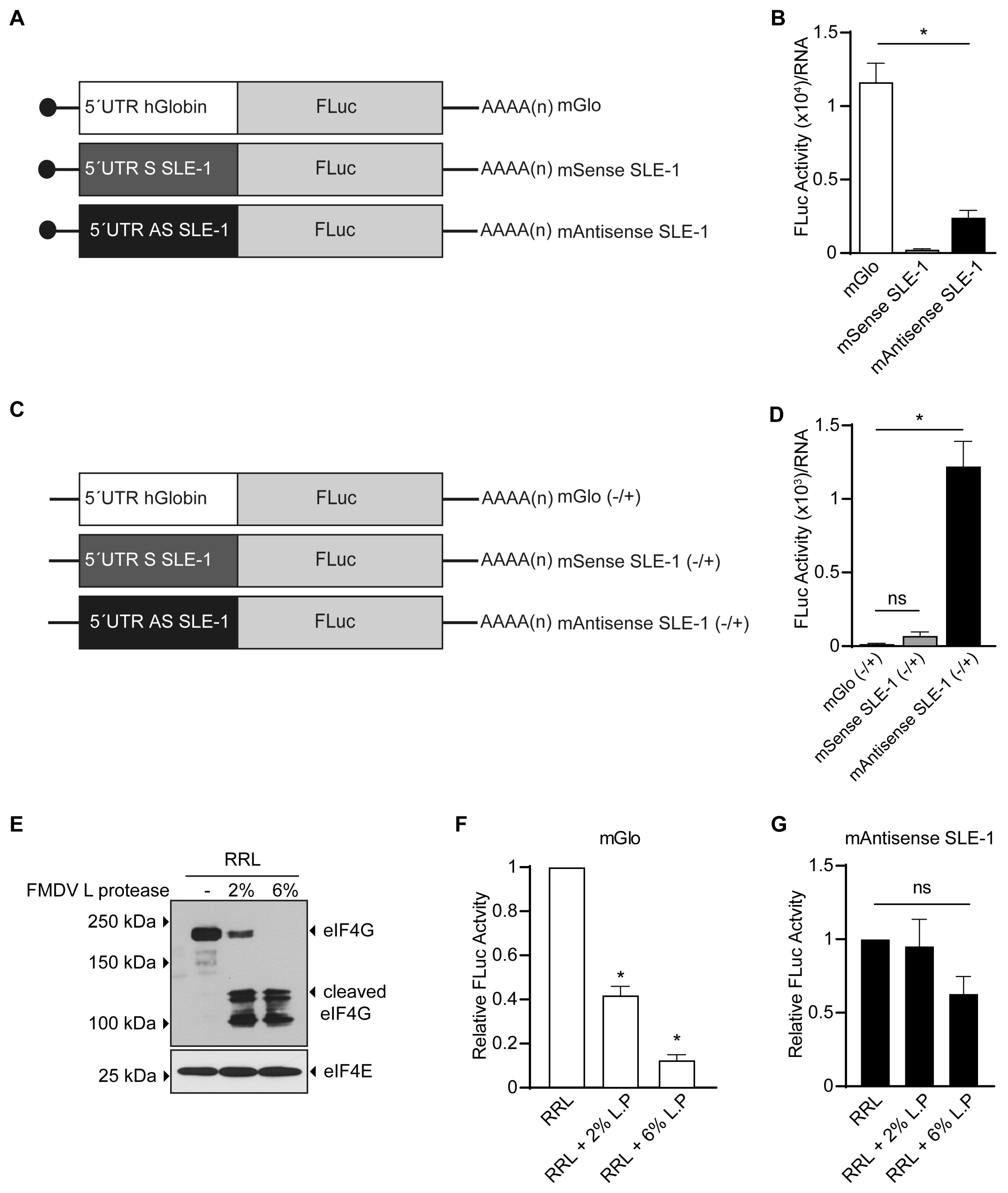
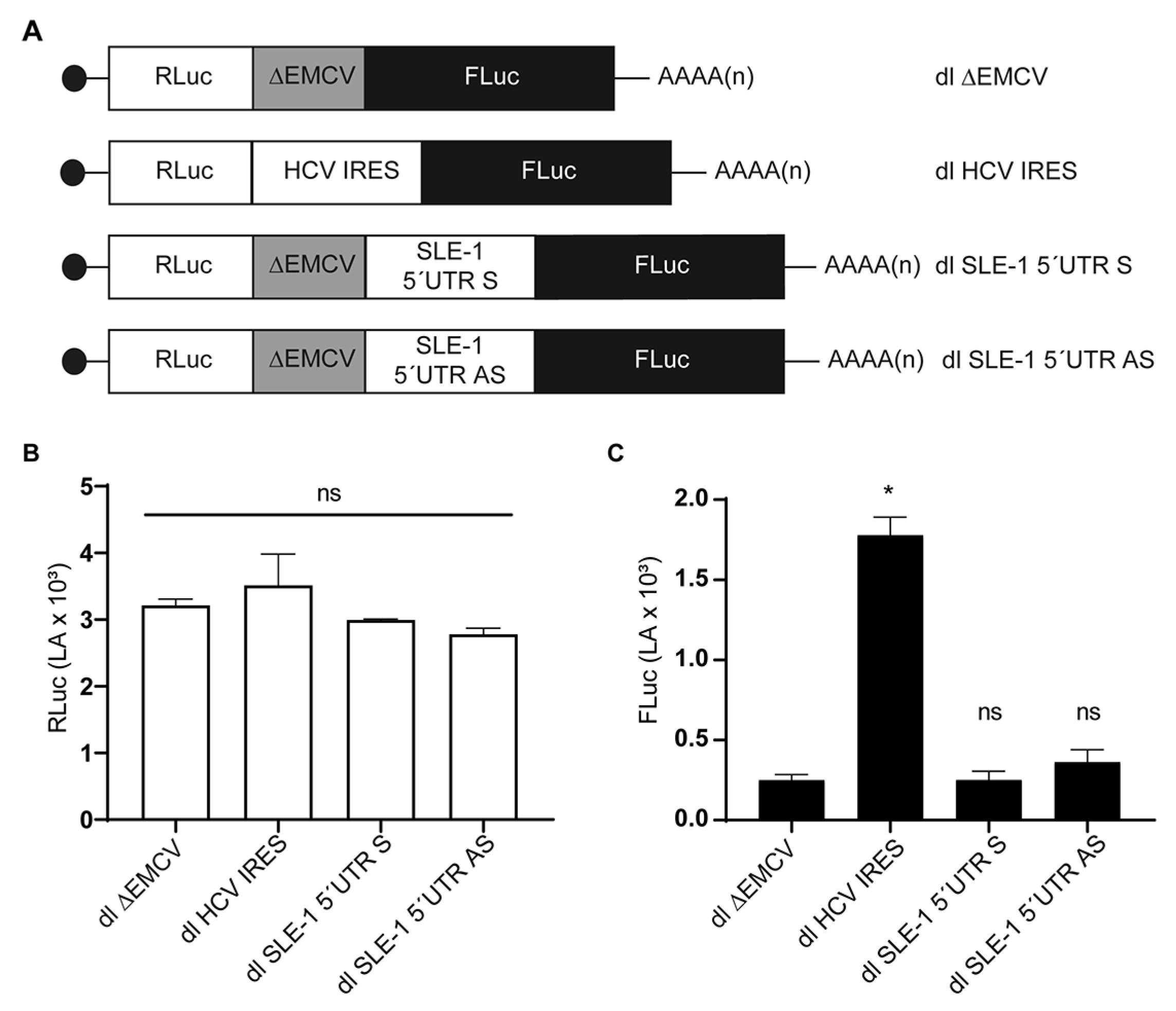
3.2. MchSLE-1 5′ UTR AS Initiate Translation in a Cap-independent Manner In Vitro
3.3. The IRES Activity of the MchSLE-1 5′UTR AS Requires a Nuclear Experience
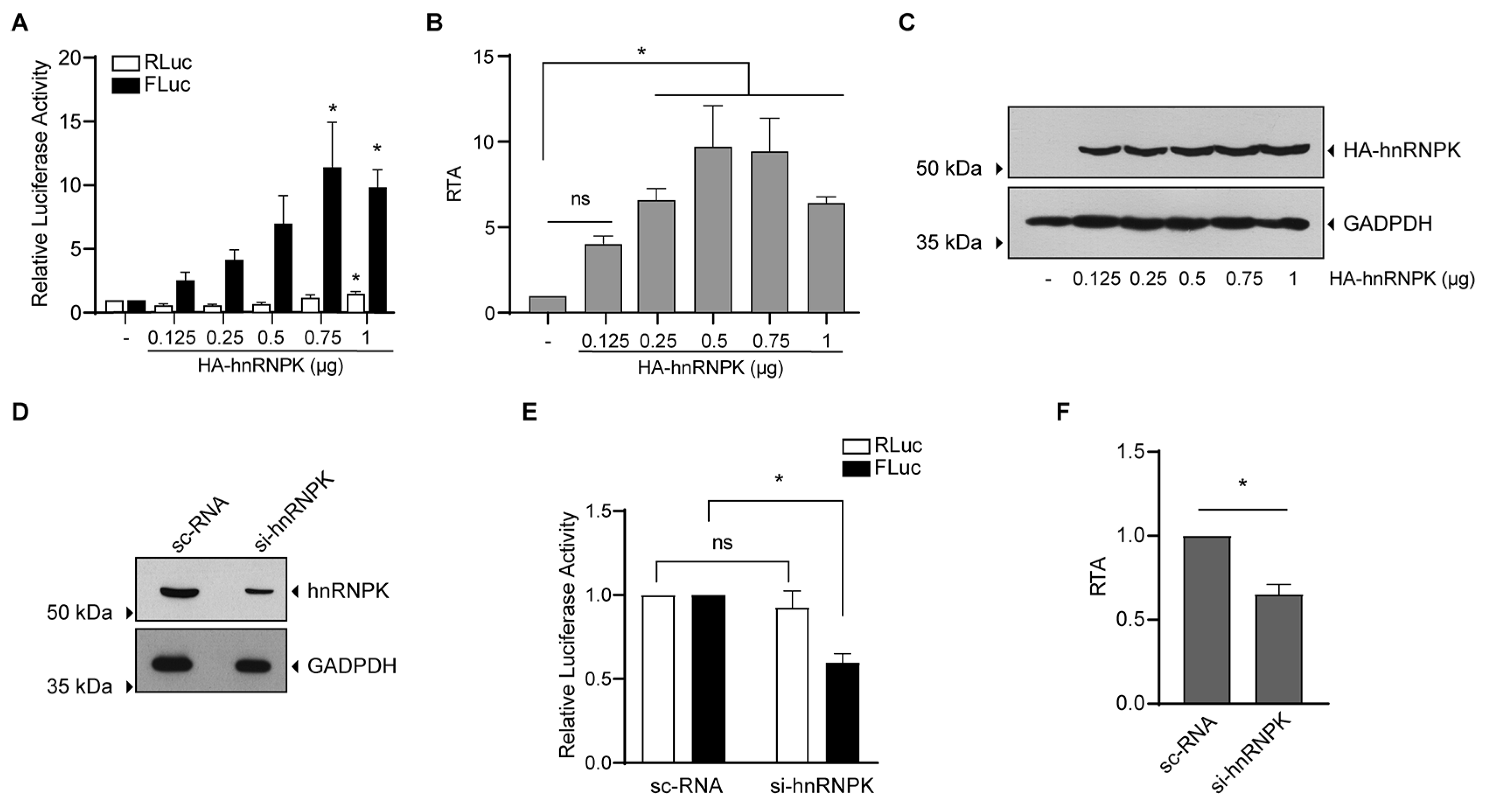
3.4. hnRNPK Is an IRES Trans-Activating Factor of MchSLE-1 IRES
3.5. The AS ORF of MchSLE-1 Can Be Expressed as Protein in Cell Culture
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Platt, R.N., 2nd; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018, 26, 25–43. [Google Scholar] [CrossRef]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71.e11. [Google Scholar] [CrossRef]
- Naville, M.; Warren, I.A.; Haftek-Terreau, Z.; Chalopin, D.; Brunet, F.; Levin, P.; Galiana, D.; Volff, J.N. Not so bad after all: Retroviruses and long terminal repeat retrotransposons as a source of new genes in vertebrates. Clin. Microbiol. Infect. 2016, 22, 312–323. [Google Scholar] [CrossRef]
- Goff, S.P. (Ed.) Retroviridae: The Retroviruses and Their Replication, 4th ed.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2001; pp. 1871–1939. [Google Scholar]
- Arriagada, G.; Metzger, M.J.; Muttray, A.F.; Sherry, J.; Reinisch, C.; Street, C.; Lipkin, W.I.; Goff, S.P. Activation of transcription and retrotransposition of a novel retroelement, Steamer, in neoplastic hemocytes of the mollusk Mya arenaria. Proc. Natl. Acad. Sci. USA 2014, 111, 14175–14180. [Google Scholar] [CrossRef]
- Geis, F.K.; Goff, S.P. Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview. Viruses 2020, 12, 884. [Google Scholar] [CrossRef]
- Balvay, L.; Lopez Lastra, M.; Sargueil, B.; Darlix, J.L.; Ohlmann, T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007, 5, 128–140. [Google Scholar] [CrossRef]
- Kobayashi-Ishihara, M.; Terahara, K.; Martinez, J.P.; Yamagishi, M.; Iwabuchi, R.; Brander, C.; Ato, M.; Watanabe, T.; Meyerhans, A.; Tsunetsugu-Yokota, Y. HIV LTR-Driven Antisense RNA by Itself Has Regulatory Function and May Curtail Virus Reactivation from Latency. Front. Microbiol. 2018, 9, 1066. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, M.H.; Landry, S.; Audet, B.; Arpin-André, C.; Hivin, P.; Paré, M.E.; Thête, J.; Wattel, E.; Marriott, S.J.; Mesnard, J.M.; et al. HTLV-I antisense transcripts initiating in the 3′LTR are alternatively spliced and polyadenylated. Retrovirology 2006, 3, 15. [Google Scholar] [CrossRef]
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 2015, 163, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Nishikawa, Y.; Nishimura, K.; Teng, D.; Takemoto, K.; Ueda, K. Effects of activation of the LINE-1 antisense promoter on the growth of cultured cells. Sci. Rep. 2020, 10, 22136. [Google Scholar] [CrossRef] [PubMed]
- Romerio, F. Origin and functional role of antisense transcription in endogenous and exogenous retroviruses. Retrovirology 2023, 20, 6. [Google Scholar] [CrossRef]
- Miller, R.H.; Zimmer, A.; Moutot, G.; Mesnard, J.M.; Chazal, N. Retroviral Antisense Transcripts and Genes: 33 Years after First Predicted, a Silent Retroviral Revolution? Viruses 2021, 13, 2221. [Google Scholar] [CrossRef]
- Barrera, A.; Olguín, V.; Vera-Otarola, J.; López-Lastra, M. Cap-independent translation initiation of the unspliced RNA of retroviruses. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194583. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G.; Lorsch, J.R. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb. Perspect. Biol. 2012, 4, a011544. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. The scanning model for translation: An update. J. Cell Biol. 1989, 108, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Mailliot, J.; Martin, F. Viral internal ribosomal entry sites: Four classes for one goal. Wiley Interdiscip. Rev. RNA 2018, 9, e1458. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.K.; Kräusslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Sorokin, I.I.; Vassilenko, K.S.; Terenin, I.M.; Kalinina, N.O.; Agol, V.I.; Dmitriev, S.E. Non-Canonical Translation Initiation Mechanisms Employed by Eukaryotic Viral mRNAs. Biochemistry 2021, 86, 1060–1094. [Google Scholar] [CrossRef]
- López-Ulloa, B.; Fuentes, Y.; Pizarro-Ortega, M.S.; López-Lastra, M. RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation. Viruses 2022, 14, 188. [Google Scholar] [CrossRef]
- Francisco-Velilla, R.; Embarc-Buh, A.; Abellan, S.; Martinez-Salas, E. Picornavirus translation strategies. FEBS Open Bio 2022, 12, 1125–1141. [Google Scholar] [CrossRef] [PubMed]
- De Breyne, S.; Ohlmann, T. Focus on Translation Initiation of the HIV-1 mRNAs. Int. J. Mol. Sci. 2018, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Cáceres, C.J.; Angulo, J.; Lowy, F.; Contreras, N.; Walters, B.; Olivares, E.; Allouche, D.; Merviel, A.; Pino, K.; Sargueil, B.; et al. Non-canonical translation initiation of the spliced mRNA encoding the human T-cell leukemia virus type 1 basic leucine zipper protein. Nucleic Acids Res. 2018, 46, 11030–11047. [Google Scholar] [CrossRef] [PubMed]
- Olivares, E.; Landry, D.M.; Caceres, C.J.; Pino, K.; Rossi, F.; Navarrete, C.; Huidobro-Toro, J.P.; Thompson, S.R.; Lopez-Lastra, M. The 5′ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J. Virol. 2014, 88, 5936–5955. [Google Scholar] [CrossRef]
- Li, P.W.; Li, J.; Timmerman, S.L.; Krushel, L.A.; Martin, S.L. The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: Implications for retrotransposition. Nucleic Acids Res. 2006, 34, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Ronfort, C.; De Breyne, S.; Sandrin, V.; Darlix, J.L.; Ohlmann, T. Characterization of two distinct RNA domains that regulate translation of the Drosophila gypsy retroelement. RNA 2004, 10, 504–515. [Google Scholar] [CrossRef]
- López-Lastra, M.; Ulrici, S.; Gabus, C.; Darlix, J.L. Identification of an internal ribosome entry segment in the 5′ region of the mouse VL30 retrotransposon and its use in the development of retroviral vectors. J. Virol. 1999, 73, 8393–8402. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.F.M.; Yonemitsu, M.A.; Giersch, R.M.; Garrett, F.E.S.; Beal, B.F.; Arriagada, G.; Davis, B.W.; Ostrander, E.A.; Goff, S.P.; Metzger, M.J. Centuries of genome instability and evolution in soft-shell clam, Mya arenaria, bivalve transmissible neoplasia. Nat. Cancer 2023, 4, 1561–1574. [Google Scholar] [CrossRef]
- Yonemitsu, M.A.; Giersch, R.M.; Polo-Prieto, M.; Hammel, M.; Simon, A.; Cremonte, F.; Avilés, F.T.; Merino-Véliz, N.; Burioli, E.A.; Muttray, A.F.; et al. A single clonal lineage of transmissible cancer identified in two marine mussel species in South America and Europe. Elife 2019, 8, e47788. [Google Scholar] [CrossRef]
- Metzger, M.J.; Paynter, A.N.; Siddall, M.E.; Goff, S.P. Horizontal transfer of retrotransposons between bivalves and other aquatic species of multiple phyla. Proc. Natl. Acad. Sci. USA 2018, 115, E4227–E4235. [Google Scholar] [CrossRef]
- Paynter, A.N.; Metzger, M.J.; Sessa, J.A.; Siddall, M.E. Evidence of horizontal transmission of the cancer-associated Steamer retrotransposon among ecological cohort bivalve species. Dis. Aquat. Organ. 2017, 124, 165–168. [Google Scholar] [CrossRef]
- Gallardo-Escárate, C.; Valenzuela-Muñoz, V.; Nuñez-Acuña, G.; Valenzuela-Miranda, D.; Tapia, F.J.; Yévenes, M.; Gajardo, G.; Toro, J.E.; Oyarzún, P.A.; Arriagada, G.; et al. Chromosome-Level Genome Assembly of the Blue Mussel Mytilus chilensis Reveals Molecular Signatures Facing the Marine Environment. Genes 2023, 14, 876. [Google Scholar] [CrossRef] [PubMed]
- Brasey, A.; Lopez-Lastra, M.; Ohlmann, T.; Beerens, N.; Berkhout, B.; Darlix, J.L.; Sonenberg, N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003, 77, 3939–3949. [Google Scholar] [CrossRef]
- Barría, M.I.; González, A.; Vera-Otarola, J.; León, U.; Vollrath, V.; Marsac, D.; Monasterio, O.; Pérez-Acle, T.; Soza, A.; López-Lastra, M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 2009, 37, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.E.; Powell, M.J.; Hoover, S.E.; Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell Biol. 2000, 20, 4990–4999. [Google Scholar] [CrossRef]
- Fernández-García, L.; Angulo, J.; Ramos, H.; Barrera, A.; Pino, K.; Vera-Otarola, J.; López-Lastra, M. The internal ribosome entry site of the Dengue virus mRNA is active when cap-dependent translation initiation is inhibited. J. Virol. 2021, 95, e01998-20. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, Y.; Olguín, V.; López-Ulloa, B.; Mendonça, D.; Ramos, H.; Abdalla, A.L.; Guajardo-Contreras, G.; Niu, M.; Rojas-Araya, B.; Mouland, A.J.; et al. Heterogeneous nuclear ribonucleoprotein K promotes cap-independent translation initiation of retroviral mRNAs. Nucleic Acids Res. 2024, gkad1221. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N., Jr. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef]
- Loeb, D.D.; Hutchison, C.A., 3rd; Edgell, M.H.; Farmerie, W.G.; Swanstrom, R. Mutational analysis of human immunodeficiency virus type 1 protease suggests functional homology with aspartic proteinases. J. Virol. 1989, 63, 111–121. [Google Scholar] [CrossRef]
- Yuki, S.; Ishimaru, S.; Inouye, S.; Saigo, K. Identification of genes for reverse transcriptase-like enzymes in two Drosophila retrotransposons, 412 and gypsy; a rapid detection method of reverse transcriptase genes using YXDD box probes. Nucleic Acids Res. 1986, 14, 3017–3030. [Google Scholar] [CrossRef] [PubMed]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and mechanism. DNA Repair 2019, 84, 102672. [Google Scholar] [CrossRef] [PubMed]
- Kulkosky, J.; Jones, K.S.; Katz, R.A.; Mack, J.P.; Skalka, A.M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell Biol. 1992, 12, 2331–2338. [Google Scholar] [CrossRef] [PubMed]
- Ohlmann, T.; Rau, M.; Pain, V.M.; Morley, S.J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996, 15, 1371–1382. [Google Scholar] [CrossRef] [PubMed]
- Lamphear, B.J.; Kirchweger, R.; Skern, T.; Rhoads, R.E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995, 270, 21975–21983. [Google Scholar] [CrossRef] [PubMed]
- Gradi, A.; Foeger, N.; Strong, R.; Svitkin, Y.V.; Sonenberg, N.; Skern, T.; Belsham, G.J. Cleavage of eukaryotic translation initiation factor 4GII within foot-and-mouth disease virus-infected cells: Identification of the L-protease cleavage site in vitro. J. Virol. 2004, 78, 3271–3278. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.R. So you want to know if your message has an IRES? Wiley Interdiscip. Rev. RNA 2012, 3, 697–705. [Google Scholar] [CrossRef]
- Angulo, J. Polypyrimidine-Tract-Binding Protein Isoforms Differentially Regulate the Hepatitis C Virus Internal Ribosome Entry Site. Viruses 2022, 15, 10008. [Google Scholar] [CrossRef]
- Angulo, J.; Ulryck, N.; Deforges, J.; Chamond, N.; Lopez-Lastra, M.; Masquida, B.; Sargueil, B. LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 2016, 44, 1309–1325. [Google Scholar] [CrossRef]
- Barría, M.I.; Vera-Otarola, J.; León, U.; Vollrath, V.; Marsac, D.; Riquelme, A.; López-Lastra, M.; Soza, A. Influence of extrahepatic viral infection on the natural history of hepatitis C. Ann. Hepatol. 2008, 7, 136–143. [Google Scholar] [CrossRef]
- Cáceres, C.J.; Angulo, J.; Contreras, N.; Pino, K.; Vera-Otarola, J.; López-Lastra, M. Targeting deoxyhypusine hydroxylase activity impairs cap-independent translation initiation driven by the 5′untranslated region of the HIV-1, HTLV-1, and MMTV mRNAs. Antivir. Res. 2016, 134, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, M.; Carvajal, F.; Pino, K.; Navarrete, C.; Ferres, M.; Huidobro-Toro, J.P.; Sargueil, B.; López-Lastra, M. Functional and structural analysis of the internal ribosome entry site present in the mRNA of natural variants of the HIV-1. PLoS ONE 2012, 7, e35031. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, M.; Deforges, J.; Plank, T.D.; Letelier, A.; Ramdohr, P.; Abraham, C.G.; Valiente-Echeverría, F.; Kieft, J.S.; Sargueil, B.; López-Lastra, M. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 2011, 39, 6186–6200. [Google Scholar] [CrossRef] [PubMed]
- Shatsky, I.N.; Terenin, I.M.; Smirnova, V.V.; Andreev, D.E. Cap-Independent Translation: What’s in a Name? Trends Biochem. Sci. 2018, 43, 882–895. [Google Scholar] [CrossRef]
- Merrick, W.C.; Barth-Baus, D. Use of reticulocyte lysates for mechanistic studies of eukaryotic translation initiation. Methods Enzym. 2007, 429, 1–21. [Google Scholar] [CrossRef]
- Bergamini, G.; Preiss, T.; Hentze, M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 2000, 6, 1781–1790. [Google Scholar] [CrossRef]
- King, H.A.; Cobbold, L.C.; Willis, A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010, 38, 1581–1586. [Google Scholar] [CrossRef]
- Spriggs, K.A.; Bushell, M.; Willis, A.E. Translational regulation of gene expression during conditions of cell stress. Mol. Cell 2010, 40, 228–237. [Google Scholar] [CrossRef]
- Poenisch, M.; Metz, P.; Blankenburg, H.; Ruggieri, A.; Lee, J.Y.; Rupp, D.; Rebhan, I.; Diederich, K.; Kaderali, L.; Domingues, F.S.; et al. Identification of HNRNPK as regulator of hepatitis C virus particle production. PLoS Pathog. 2015, 11, e1004573. [Google Scholar] [CrossRef]
- Meyuhas, O.; Kahan, T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 2015, 1849, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Ogami, K.; Oishi, Y.; Sakamoto, K.; Okumura, M.; Yamagishi, R.; Inoue, T.; Hibino, M.; Nogimori, T.; Yamaguchi, N.; Furutachi, K.; et al. mTOR- and LARP1-dependent regulation of TOP mRNA poly(A) tail and ribosome loading. Cell Rep. 2022, 41, 111548. [Google Scholar] [CrossRef] [PubMed]
- Gonatopoulos-Pournatzis, T.; Cowling, V.H. Cap-binding complex (CBC). Biochem. J. 2014, 457, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Toro-Ascuy, D.; Rojas-Araya, B.; García-de-Gracia, F.; Rojas-Fuentes, C.; Pereira-Montecinos, C.; Gaete-Argel, A.; Valiente-Echeverría, F.; Ohlmann, T.; Soto-Rifo, R. A Rev-CBP80-eIF4AI complex drives Gag synthesis from the HIV-1 unspliced mRNA. Nucleic Acids Res. 2018, 46, 11539–11552. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, C.F.; Mortreux, F.; Auboeuf, D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Witten, J.T.; Ule, J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011, 27, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002, 16, 2906–2922. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, M.; Ramdohr, P.; Valiente-Echeverria, F.; Tapia, K.; Rodriguez, F.E.; Lowy, F.; Huidobro-Toro, J.P.; Dangerfield, J.A.; Lopez-Lastra, M. The 5′-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Res. 2010, 38, 618–632. [Google Scholar] [CrossRef]
- Barrera, A.; Ramos, H.; Vera-Otarola, J.; Fernández-García, L.; Angulo, J.; Olguín, V.; Pino, K.; Mouland, A.J.; López-Lastra, M. Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation. Nucleic Acids Res. 2020, 48, 10479–10499. [Google Scholar] [CrossRef]
- Peddigari, S.; Li, P.W.; Rabe, J.L.; Martin, S.L. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013, 41, 575–585. [Google Scholar] [CrossRef]
- Lin, J.Y.; Li, M.L.; Huang, P.N.; Chien, K.Y.; Horng, J.T.; Shih, S.R. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 2008, 89, 2540–2549. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yang, D.; Sun, C.; Wang, H.; Zhao, B.; Zhou, G.; Yu, L. hnRNP K Is a Novel Internal Ribosomal Entry Site-Transacting Factor That Negatively Regulates Foot-and-Mouth Disease Virus Translation and Replication and Is Antagonized by Viral 3C Protease. J. Virol. 2020, 94, e00803-20. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Lu, K.Y.; Reymond Sutandy, F.X.; Chen, Y.W.; Konan, K.; Zhu, H.; Kao, C.C.; Chen, C.S. A human proteome microarray identifies that the heterogeneous nuclear ribonucleoprotein K (hnRNP K) recognizes the 5′ terminal sequence of the hepatitis C virus RNA. Mol. Cell Proteom. 2014, 13, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Anderson, P. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 2008, 20, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Holcik, M.; Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Astuya, A.; Carrera, C.; Ulloa, V.; Aballay, A.; Nunez-Acuna, G.; Hegaret, H.; Gallardo-Escarate, C. Saxitoxin Modulates Immunological Parameters and Gene Transcription in Mytilus chilensis Hemocytes. Int. J. Mol. Sci. 2015, 16, 15235–15250. [Google Scholar] [CrossRef] [PubMed]
- Monette, A.; Ajamian, L.; López-Lastra, M.; Mouland, A.J. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: Implications for HIV-1 gene expression. J. Biol. Chem. 2009, 284, 31350–31362. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-García, L.; Ahumada-Marchant, C.; Lobos-Ávila, P.; Brauer, B.; Bustos, F.J.; Arriagada, G. The Mytilus chilensis Steamer-like Element-1 Retrotransposon Antisense mRNA Harbors an Internal Ribosome Entry Site That Is Modulated by hnRNPK. Viruses 2024, 16, 403. https://doi.org/10.3390/v16030403
Fernández-García L, Ahumada-Marchant C, Lobos-Ávila P, Brauer B, Bustos FJ, Arriagada G. The Mytilus chilensis Steamer-like Element-1 Retrotransposon Antisense mRNA Harbors an Internal Ribosome Entry Site That Is Modulated by hnRNPK. Viruses. 2024; 16(3):403. https://doi.org/10.3390/v16030403
Chicago/Turabian StyleFernández-García, Leandro, Constanza Ahumada-Marchant, Pablo Lobos-Ávila, Bastián Brauer, Fernando J. Bustos, and Gloria Arriagada. 2024. "The Mytilus chilensis Steamer-like Element-1 Retrotransposon Antisense mRNA Harbors an Internal Ribosome Entry Site That Is Modulated by hnRNPK" Viruses 16, no. 3: 403. https://doi.org/10.3390/v16030403





