Cross-Reactivity Assessment of Vaccine-Derived SARS-CoV-2 T Cell Responses against BA.2.86 and JN.1
Abstract
1. Introduction
2. Materials and Methods
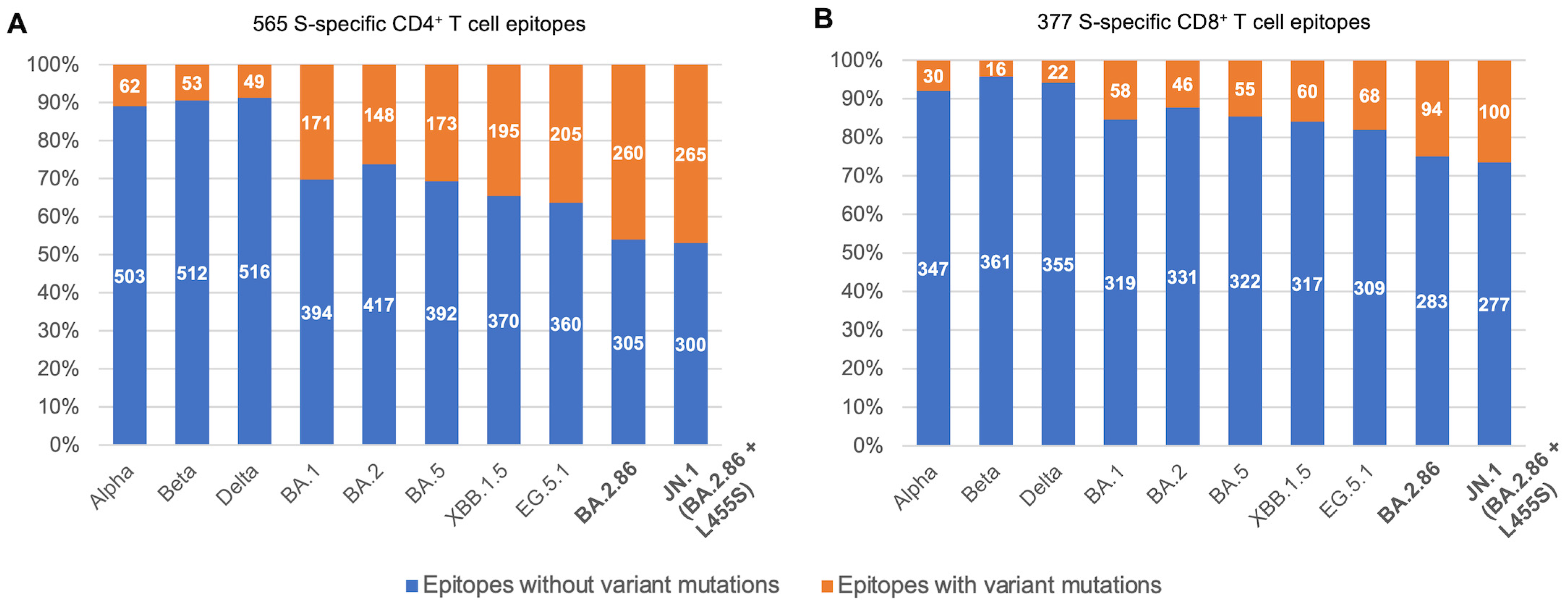
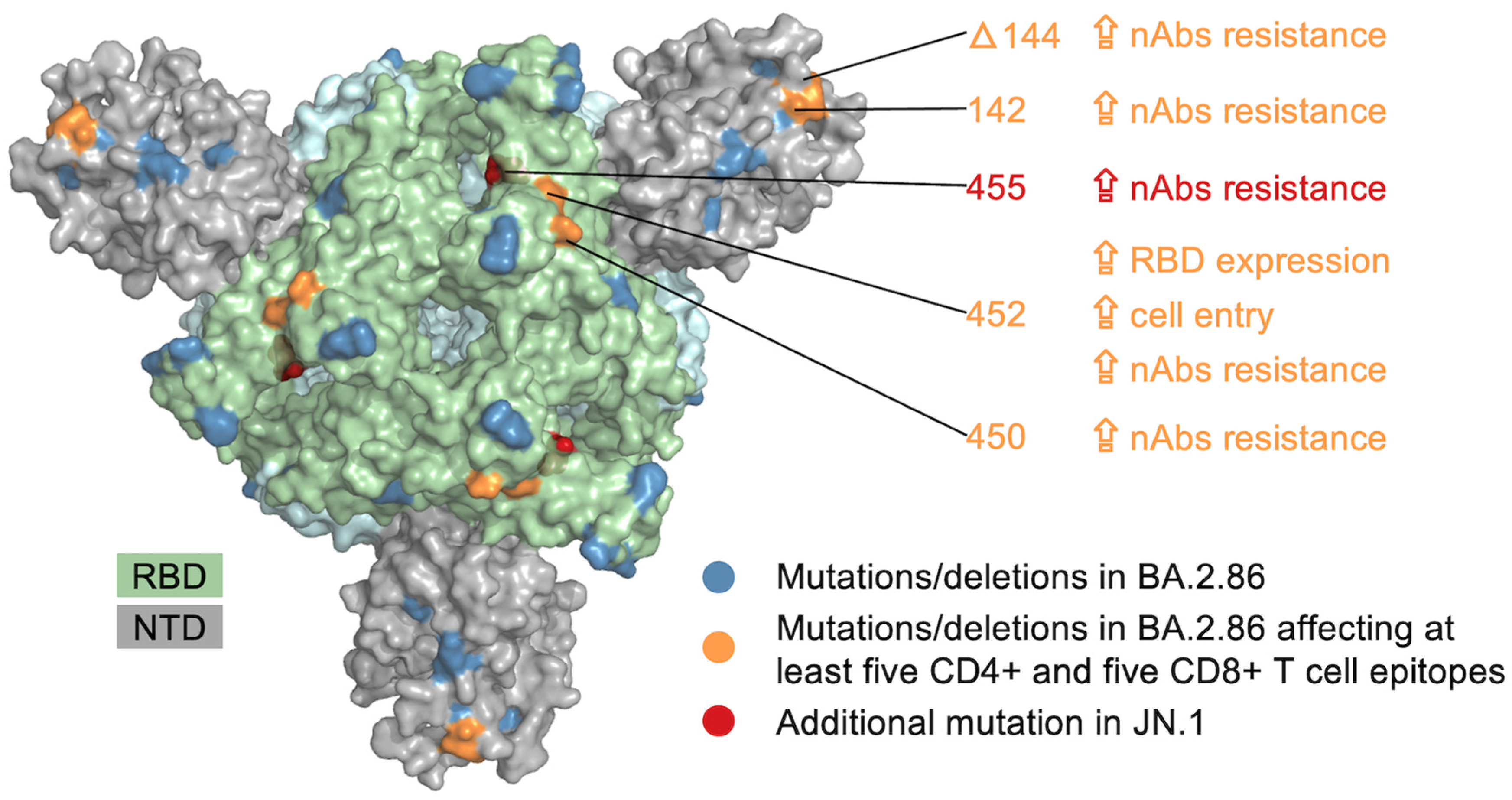
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sheward, D.J.; Yang, Y.; Westerberg, M.; Öling, S.; Muschiol, S.; Sato, K.; Peacock, T.P.; Hedestam, G.B.K.; Albert, J.; Murrell, B. Sensitivity of the SARS-CoV-2 BA.2.86 Variant to Prevailing Neutralising Antibody Responses. Lancet Infect. Dis. 2023, 23, e462–e463. [Google Scholar] [CrossRef]
- Yang, S.; Yu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Chen, X.; Xu, Y.; Wang, P.; Wang, J.; Yu, L.; et al. Antigenicity and Infectivity Characterisation of SARS-CoV-2 BA.2.86. Lancet Infect. Dis. 2023, 23, e457–e459. [Google Scholar] [CrossRef]
- Uriu, K.; Ito, J.; Kosugi, Y.; Tanaka, Y.L.; Mugita, Y.; Guo, Z.; Hinay, A.A.; Putri, O.; Kim, Y.; Shimizu, R.; et al. Transmissibility, Infectivity, and Immune Resistance of the SARS-CoV-2 BA.2.86 Variant. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Liu, L.; Schwanz, L.T.; Li, Z.; Nair, M.S.; Ho, J.; Zhang, R.M.; Iketani, S.; Yu, J.; et al. Antigenicity and Receptor Affinity of SARS-CoV-2 BA.2.86 Spike. Nature 2023, 624, 639–644. [Google Scholar] [CrossRef]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L.; et al. Fast Evolution of SARS-CoV-2 BA.2.86 to JN.1 under Heavy Immune Pressure. Lancet Infect. Dis. 2024, 24, E70–E72. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J.; et al. Virological Characteristics of the SARS-CoV-2 JN.1 Variant. Lancet Infect. Dis. 2024, 24, E82. [Google Scholar] [CrossRef] [PubMed]
- Painter, M.M.; Johnston, T.S.; Lundgreen, K.A.; Santos, J.J.S.; Qin, J.S.; Goel, R.R.; Apostolidis, S.A.; Mathew, D.; Fulmer, B.; Williams, J.C.; et al. Prior Vaccination Promotes Early Activation of Memory T Cells and Enhances Immune Responses during SARS-CoV-2 Breakthrough Infection. Nat. Immunol. 2023, 24, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Upadhyay, R.; Hao, Y.; Samanovic, M.I.; Herati, R.S.; Blair, J.D.; Axelrad, J.; Mulligan, M.J.; Littman, D.R.; Satija, R. Multimodal Single-Cell Datasets Characterize Antigen-Specific CD8+ T Cells across SARS-CoV-2 Vaccination and Infection. Nat. Immunol. 2023, 24, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; Reynaldi, A.; Lee, W.S.; Nguyen, J.; Amarasena, T.; Taiaroa, G.; Kinsella, P.; Liew, K.C.; Tran, T.; Kent, H.E.; et al. SARS-CoV-2 Breakthrough Infection Induces Rapid Memory and de Novo T Cell Responses. Immunity 2023, 56, 879–892.e4. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Samandari, T. Silent Battles: Immune Responses in Asymptomatic SARS-CoV-2 Infection. Cell. Mol. Immunol. 2024, 21, 159–170. [Google Scholar] [CrossRef]
- Ramirez, S.I.; Lopez, P.G.; Faraji, F.; Parikh, U.M.; Heaps, A.; Ritz, J.; Moser, C.; Eron, J.J.; Wohl, D.A.; Currier, J.S.; et al. Early Antiviral CD4 and CD8 T Cell Responses Are Associated with Upper Respiratory Tract Clearance of SARS-CoV-2. bioRxiv 2023. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 Update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Hodcroft, E.B. CoVariants: SARS-CoV-2 Mutations and Variants of Interest. Available online: https://covariants.org/ (accessed on 28 November 2023).
- Tsueng, G.; Mullen, J.L.; Alkuzweny, M.; Cano, M.; Rush, B.; Haag, E.; Lin, J.; Welzel, D.J.; Zhou, X.; Qian, Z.; et al. Outbreak.Info Research Library: A Standardized, Searchable Platform to Discover and Explore COVID-19 Resources. Nat. Methods 2023, 20, 536–540. [Google Scholar] [CrossRef]
- Reynisson, B.; Barra, C.; Kaabinejadian, S.; Hildebrand, W.H.; Peters, B.; Nielsen, M. Improved Prediction of MHC II Antigen Presentation through Integration and Motif Deconvolution of Mass Spectrometry MHC Eluted Ligand Data. J. Proteome Res. 2020, 19, 2304–2315. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved Predictions of MHC Antigen Presentation by Concurrent Motif Deconvolution and Integration of MS MHC Eluted Ligand Data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Keeton, R.; Tincho, M.B.; Ngomti, A.; Baguma, R.; Benede, N.; Suzuki, A.; Khan, K.; Cele, S.; Bernstein, M.; Karim, F.; et al. T Cell Responses to SARS-CoV-2 Spike Cross-Recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Dörnte, C.; Traska, V.; Jansen, N.; Kostyra, J.; Baurmann, H.; Lauer, G.; Huang, Y.-J.; Kramer, S.; Brauns, O.; Winkels, H.; et al. Vaccines against the Original Strain of SARS-CoV-2 Provide T Cell Memory to the B.1.1.529 Variant. Commun. Med. 2022, 2, 140. [Google Scholar] [CrossRef]
- Sang, Y.; Zhang, Z.; Liu, F.; Lu, H.; Yu, C.; Sun, H.; Long, J.; Cao, Y.; Mai, J.; Wang, X.; et al. Monkeypox Virus Quadrivalent mRNA Vaccine Induces Antibody Responses and Cellular Immunity and Protects Mice against Vaccinia Virus. bioRxiv 2022. [Google Scholar] [CrossRef]
- Agerer, B.; Koblischke, M.; Gudipati, V.; Montaño-Gutierrez, L.F.; Smyth, M.; Popa, A.; Genger, J.-W.; Endler, L.; Florian, D.M.; Mühlgrabner, V.; et al. SARS-CoV-2 Mutations in MHC-I-Restricted Epitopes Evade CD8+ T Cell Responses. Sci. Immunol. 2021, 6, eabg6461. [Google Scholar] [CrossRef]
- Dolton, G.; Rius, C.; Hasan, M.S.; Wall, A.; Szomolay, B.; Behiry, E.; Whalley, T.; Southgate, J.; Fuller, A.; Morin, T.; et al. Emergence of Immune Escape at Dominant SARS-CoV-2 Killer T Cell Epitope. Cell 2022, 185, 2936–2951.e19. [Google Scholar] [CrossRef]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive Analysis of T Cell Immunodominance and Immunoprevalence of SARS-CoV-2 Epitopes in COVID-19 Cases. Cell Rep. Med. 2021, 2, 100204. [Google Scholar] [CrossRef]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 Escape Antibodies Elicited by Omicron Infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 Humoral Immunity Induces Convergent Omicron RBD Evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef] [PubMed]
- Lusvarghi, S.; Wang, W.; Herrup, R.; Neerukonda, S.N.; Vassell, R.; Bentley, L.; Eakin, A.E.; Erlandson, K.J.; Weiss, C.D. Key Substitutions in the Spike Protein of SARS-CoV-2 Variants Can Predict Resistance to Monoclonal Antibodies, but Other Substitutions Can Modify the Effects. J. Virol. 2022, 96, e01110-21. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hannon, W.W.; Loes, A.N.; Hauser, K.; Dillen, J.R.; Ferri, E.; Farrell, A.G.; Dadonaite, B.; McCallum, M.; et al. Shifting Mutational Constraints in the SARS-CoV-2 Receptor-Binding Domain during Viral Evolution. Science 2022, 377, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Quadeer, A.A.; Ahmed, S.F.; McKay, M.R. Landscape of Epitopes Targeted by T Cells in 852 Individuals Recovered from COVID-19: Meta-Analysis, Immunoprevalence, and Web Platform. Cell Rep. Med. 2021, 2, 100312. [Google Scholar] [CrossRef] [PubMed]
- Augusto, D.G.; Murdolo, L.D.; Chatzileontiadou, D.S.M.; Sabatino, J.J.; Yusufali, T.; Peyser, N.D.; Butcher, X.; Kizer, K.; Guthrie, K.; Murray, V.W.; et al. A Common Allele of HLA Is Associated with Asymptomatic SARS-CoV-2 Infection. Nature 2023, 620, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Artiles, A.; Calvo-Calle, J.M.; Co, M.D.; Nanaware, P.P.; Cruz, J.; Weaver, G.C.; Lu, L.; Forconi, C.; Finberg, R.W.; Moormann, A.M.; et al. Broadly Recognized, Cross-Reactive SARS-CoV-2 CD4 T Cell Epitopes Are Highly Conserved across Human Coronaviruses and Presented by Common HLA Alleles. Cell Rep. 2022, 39, 110952. [Google Scholar] [CrossRef]
- Kedzierska, K.; Thomas, P.G. Count on Us: T Cells in SARS-CoV-2 Infection and Vaccination. Cell Rep. Med. 2022, 3, 100562. [Google Scholar] [CrossRef]
- Mudd, P.A.; Minervina, A.A.; Pogorelyy, M.V.; Turner, J.S.; Kim, W.; Kalaidina, E.; Petersen, J.; Schmitz, A.J.; Lei, T.; Haile, A.; et al. SARS-CoV-2 mRNA Vaccination Elicits a Robust and Persistent T Follicular Helper Cell Response in Humans. Cell 2022, 185, 603–613.e15. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Sohail, M.S.; Quadeer, A.A.; McKay, M.R. Identification of Potential SARS-CoV-2 CD8+ T Cell Escape Mutants. Vaccines 2022, 10, 542. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control Updated COVID-19 Vaccine Recommendations Now Available|CDC. Available online: https://www.cdc.gov/respiratory-viruses/whats-new/covid-vaccine-recommendations-9-12-2023.html (accessed on 9 November 2023).
- Chalkias, S.; McGhee, N.; Whatley, J.L.; Essink, B.; Brosz, A.; Tomassini, J.E.; Girard, B.; Wu, K.; Edwards, D.K.; Nasir, A.; et al. Safety and Immunogenicity of XBB.1.5-Containing mRNA Vaccines. MedRxiv 2023. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The Nucleocapsid Protein of SARS–CoV-2: A Target for Vaccine Development. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. SARS-CoV-2 T Cell Responses Elicited by COVID-19 Vaccines or Infection Are Expected to Remain Robust against Omicron. Viruses 2022, 14, 79. [Google Scholar] [CrossRef]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. SARS-CoV-2 Vaccination Induces Immunological T Cell Memory Able to Cross-Recognize Variants from Alpha to Omicron. Cell 2022, 185, 847–859.e11. [Google Scholar] [CrossRef] [PubMed]
- Naranbhai, V.; Nathan, A.; Kaseke, C.; Berrios, C.; Khatri, A.; Choi, S.; Getz, M.A.; Tano-Menka, R.; Ofoman, O.; Gayton, A.; et al. T Cell Reactivity to the SARS-CoV-2 Omicron Variant Is Preserved in Most but Not All Individuals. Cell 2022, 185, 1041–1051.e6. [Google Scholar] [CrossRef] [PubMed]
- de Prost, N.; Audureau, E.; Guillon, A.; Handala, L.; Préau, S.; Guigon, A.; Uhel, F.; Hingrat, Q.L.; Delamaire, F.; Grolhier, C.; et al. Clinical Phenotypes and Outcomes Associated with SARS-CoV-2 Omicron Variant JN.1 in Critically Ill COVID-19 Patients: A Prospective, Multicenter Cohort Study. MedRxiv 2024. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; Barton, J.P.; McKay, M.R. Cross-Serotypically Conserved Epitope Recommendations for a Universal T Cell-Based Dengue Vaccine. PLoS Negl. Trop. Dis. 2020, 14, e0008676. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. COVIDep: A Web-Based Platform for Real-Time Reporting of Vaccine Target Recommendations for SARS-CoV-2. Nat. Protoc. 2020, 15, 2141–2142. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Sohail, M.S.; Quadeer, A.A.; McKay, M.R. Vaccinia-Virus-Based Vaccines Are Expected to Elicit Highly Cross-Reactive Immunity to the 2022 Monkeypox Virus. Viruses 2022, 14, 1960. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.S.; Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. In Silico T Cell Epitope Identification for SARS-CoV-2: Progress and Perspectives. Adv. Drug Deliv. Rev. 2021, 171, 29–47. [Google Scholar] [CrossRef] [PubMed]
- Nesamari, R.; Omondi, M.A.; Baguma, R.; Höft, M.A.; Ngomti, A.; Nkayi, A.A.; Besethi, A.S.; Magugu, S.F.J.; Mosala, P.; Walters, A.; et al. Post-Pandemic Memory T Cell Response to SARS-CoV-2 Is Durable, Broadly Targeted, and Cross-Reactive to the Hypermutated BA.2.86 Variant. Cell Host Microbe 2024, 32, 162–169e3. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.R.; Gao, Y.; Wu, J.; Ribeiro, O.; Chen, P.; Bergman, P.; Blennow, O.; Hansson, L.; Mielke, S.; Nowak, P.; et al. Memory T Cells Effectively Recognize the SARS-CoV-2 Hypermutated BA.2.86 Variant. Cell Host Microbe 2024, 32, 156–161.e3. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.; Nielsen, M.; Sette, A. T Cell Epitope Predictions. Annu. Rev. Immunol. 2020, 38, 123–145. [Google Scholar] [CrossRef]
- Mazor, R.; Vassall, A.N.; Eberle, J.A.; Beers, R.; Weldon, J.E.; Venzon, D.J.; Tsang, K.Y.; Benhar, I.; Pastan, I. Identification and Elimination of an Immunodominant T-Cell Epitope in Recombinant Immunotoxins Based on Pseudomonas Exotoxin A. Proc. Natl. Acad. Sci. USA 2012, 109, E3597–E3603. [Google Scholar] [CrossRef]
- Haj, A.K.; Breitbach, M.E.; Baker, D.A.; Mohns, M.S.; Moreno, G.K.; Wilson, N.A.; Lyamichev, V.; Patel, J.; Weisgrau, K.L.; Dudley, D.M.; et al. High-Throughput Identification of MHC Class I Binding Peptides Using an Ultradense Peptide Array. J. Immunol. 2020, 204, 1689–1696. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohail, M.S.; Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Cross-Reactivity Assessment of Vaccine-Derived SARS-CoV-2 T Cell Responses against BA.2.86 and JN.1. Viruses 2024, 16, 473. https://doi.org/10.3390/v16030473
Sohail MS, Ahmed SF, Quadeer AA, McKay MR. Cross-Reactivity Assessment of Vaccine-Derived SARS-CoV-2 T Cell Responses against BA.2.86 and JN.1. Viruses. 2024; 16(3):473. https://doi.org/10.3390/v16030473
Chicago/Turabian StyleSohail, Muhammad Saqib, Syed Faraz Ahmed, Ahmed Abdul Quadeer, and Matthew R. McKay. 2024. "Cross-Reactivity Assessment of Vaccine-Derived SARS-CoV-2 T Cell Responses against BA.2.86 and JN.1" Viruses 16, no. 3: 473. https://doi.org/10.3390/v16030473
APA StyleSohail, M. S., Ahmed, S. F., Quadeer, A. A., & McKay, M. R. (2024). Cross-Reactivity Assessment of Vaccine-Derived SARS-CoV-2 T Cell Responses against BA.2.86 and JN.1. Viruses, 16(3), 473. https://doi.org/10.3390/v16030473







