Whole Genome Sequence-Based Analysis of Bovine Gammaherpesvirus 4 Isolated from Bovine Abortions
Abstract
1. Introduction
2. Material and Methods
2.1. Viruses
2.2. Genomic Sequencing
2.3. Genome Analyses
2.4. ORF Analyses
2.4.1. Sequence Analyses
2.4.2. Phylogenetic Comparisons
2.4.3. Recombination Analyses
3. Results
3.1. Genomic Sequencing
3.2. Genome Analyses
3.3. ORF Analyses
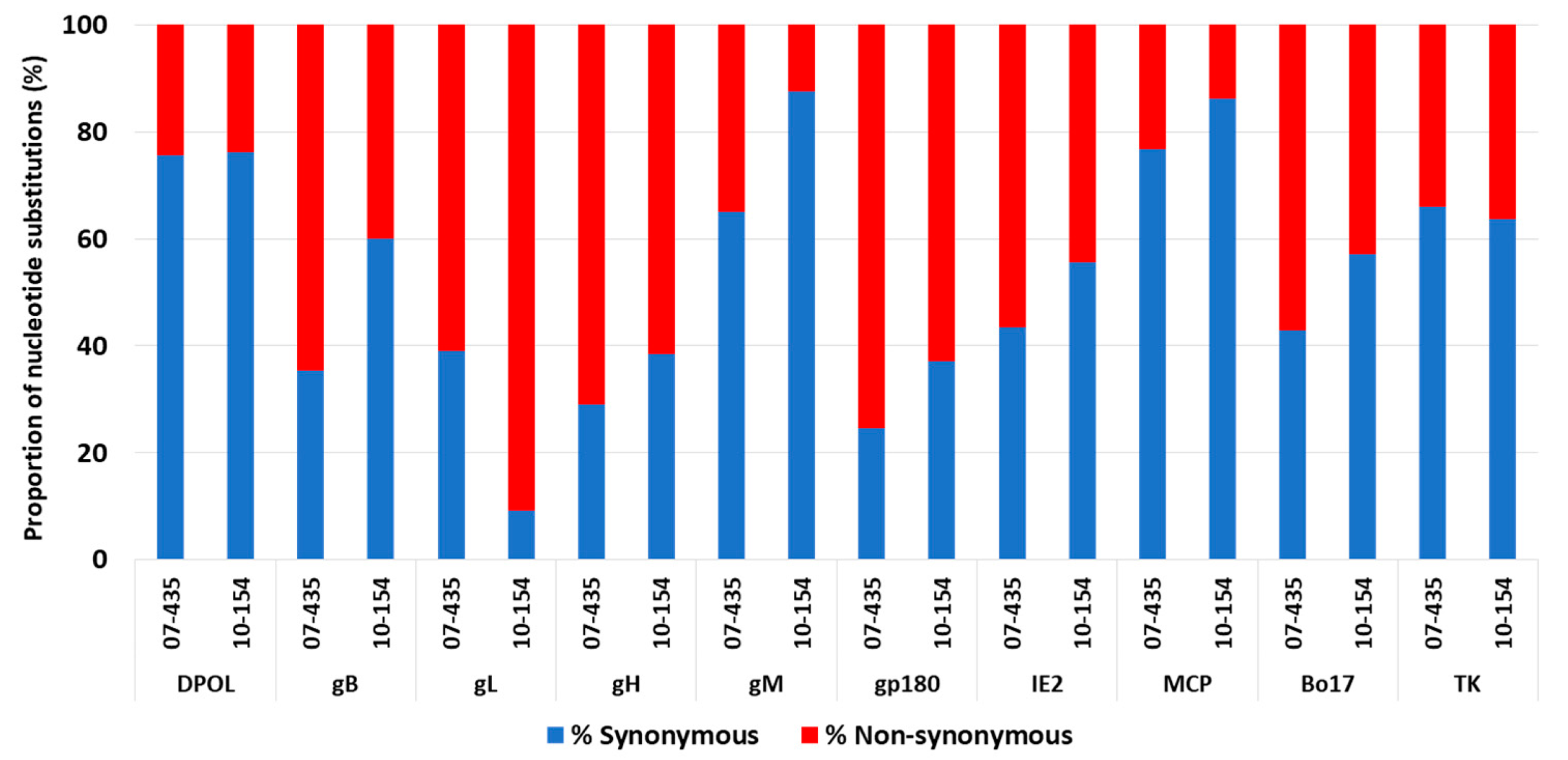
3.3.1. Sequence Analyses
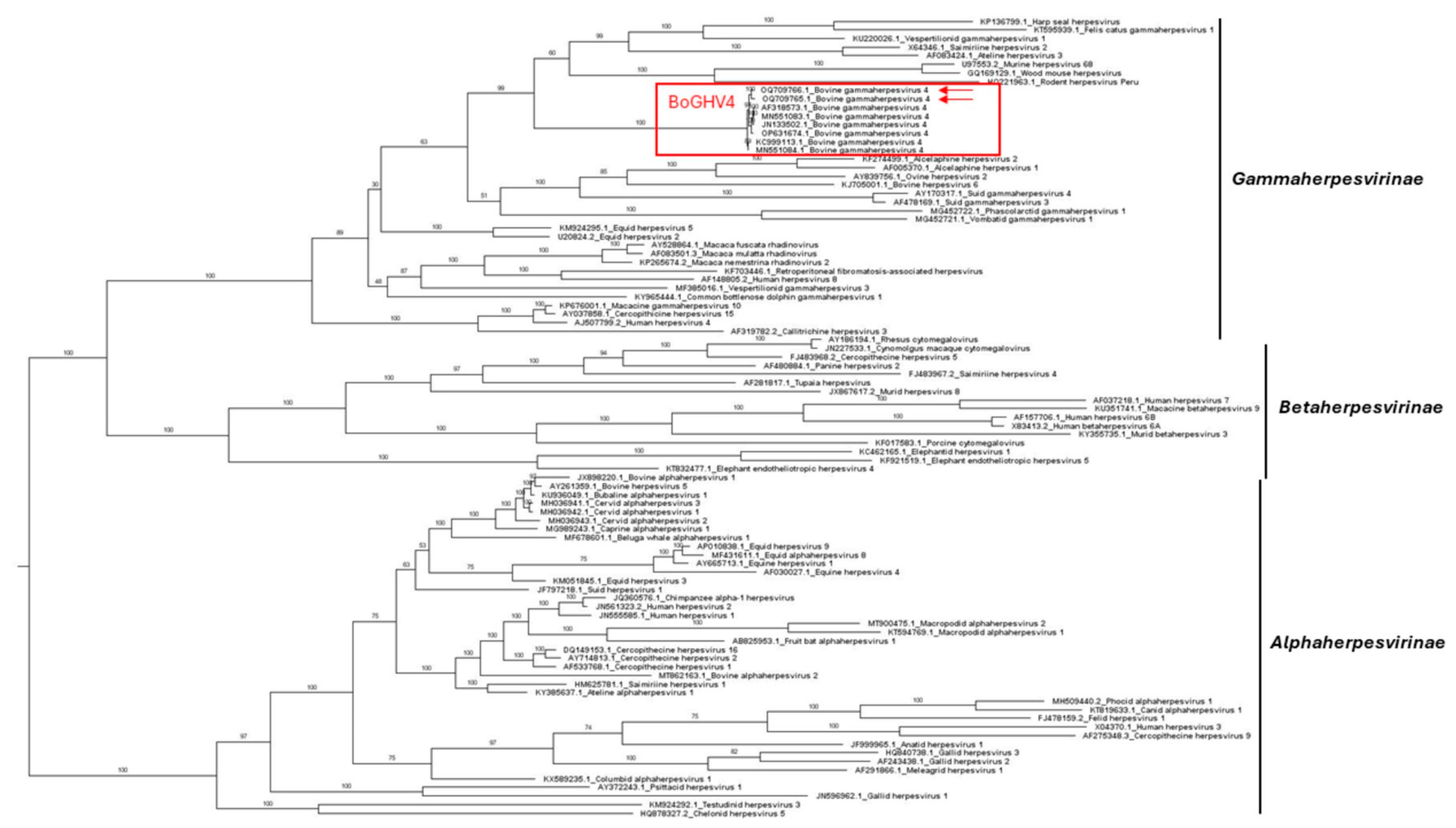
3.3.2. Phylogenetic Comparisons
3.3.3. Recombination Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses (ICTV): 2022 Release. Available online: https://ictv.global/taxonomy (accessed on 17 April 2024).
- Gillet, L.; Dewals, B.; Farnir, F.; De Leval, L.; Vanderplasschen, A. Bovine herpesvirus 4 induces apoptosis of human carcinoma cell lines in vitro and in vivo. Cancer Res. 2005, 65, 9463–9472. [Google Scholar] [CrossRef] [PubMed]
- Egyed, L. Bovine herpesvirus type 4: A special herpesvirus (review article). Acta Vet. Hung. 2000, 48, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Donofrio, G.; Cavirani, S.; Taddei, S.; Flammini, C.F. Activation of bovine herpesvirus 4 lytic replication in a non-permissive cell line by overexpression of BoHV-4 immediate early (IE) 2 gene. J. Virol. Methods. 2004, 15, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Jiang, M.; Eng, H.; Shi, G.; Lai, L.; Huang, B.; Huang, K.; Wu, H. Experimental infection with bovine herpesvirus-4 enhances atherosclerotic process in rabbits. Lab. Investig. 2000, 80, 3–11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thiry, E.; Meurens, F.; Muylkens, B.; McVoy, M.; Gogev, S.; Thiry, J.; Vanderplasschen, A.; Epstein, A.; Keil, G.; Schynts, F. Recombination in alphaherpesviruses. Rev. Med. Virol. 2005, 15, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, W.; Broll, H.; Ehlers, B.; Buhk, H.J.; Rosenthal, A.; Goltz, M. Genome sequence of bovine herpesvirus 4, a bovine Rhadinovirus, and identification of an origin of DNA replication. J. Virol. 2001, 75, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Roizman, B.; Baines, J. The diversity and unity of Herpesviridae. Comp. Immunol. Microbiol. Infect. Dis. 1991, 14, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Lété, C.; Palmeira, L.; Leroy, B.; Mast, J.; Machiels, B.; Wattiez, R.; Vanderplasschen, A.; Gillet, L. Proteomic characterization of bovine herpesvirus 4 extracellular virions. J. Virol. 2012, 86, 11567–11580. [Google Scholar] [CrossRef] [PubMed]
- Machiels, B.; Lété, C.; Guillaume, A.; Mast, J.; Stevenson, P.G.; Vanderplasschen, A.; Gillet, L. Antibody Evasion by a Gammaherpesvirus O-Glycan Shield. PLoS Pathog. 2011, 7, e1002387. [Google Scholar] [CrossRef]
- Vanderplasschen, A.; Bublot, M.; Dubuisson, J.; Pastoret, P.P.; Thiry, E. Attachment of the gammaherpesvirus bovine herpesvirus 4 is mediated by the interaction of gp8 glycoprotein with heparinlike moieties on the cell surface. Virology 1993, 196, 232–240. [Google Scholar] [CrossRef]
- Vanderplasschen, A.; Markine-Goriaynoff, N.; Lomonte, P.; Suzuki, M.; Hiraoka, N.; Yeh, J.C.; Bureau, F.; Willems, L.; Thiry, E.; Fukuda, M.; et al. A multipotential beta-1,6-N-acetylglucosaminyltransferase is encoded by bovine herpesvirus type 4. Proc. Natl. Acad. Sci. USA 2000, 97, 5756–5761. [Google Scholar] [CrossRef] [PubMed]
- Markine-Goriaynoff, N.; Georgin, J.P.; Goltz, M.; Zimmermann, W.; Broll, H.; Wamwayi, H.M.; Pastoret, P.P.; Sharp, P.M.; Vanderplasschen, A. The core 2 beta-1,6-N-acetylglucosaminyltransferase-mucin encoded by bovine herpesvirus 4 was acquired from an ancestor of the African buffalo. J. Virol. 2003, 77, 1784–1792. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bublot, M.; Van Bressem, M.F.; Thiry, E.; Dubuisson, J.; Pastoret, P.P. Bovine herpesvirus 4 genome: Cloning, mapping and strain variation analysis. J. Gen. Virol. 1990, 71, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Palmeira, L.; Machiels, B.; Lété, C.; Vanderplasschen, A.; Gillet, L. Sequencing of bovine herpesvirus 4 v.test strain reveals important genome features. Virol. J. 2011, 8, 406. [Google Scholar] [CrossRef] [PubMed]
- Bublot, M.; Dubuisson, J.; Van Bressem, M.F.; Danyi, S.; Pastoret, P.P.; Thiry, E. Antigenic and genomic identity between simian herpesvirus aotus type 2 and bovine herpesvirus type 4. J. Gen. Virol. 1991, 72, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Donofrio, G.; Flammini, C.F.; Scatozza, F.; Cavirani, S. Detection of bovine herpesvirus 4 (BoHV-4) DNA in the cell fraction of milk of dairy cattle with history of BoHV-4 infection. J. Clin. Microbiol. 2000, 38, 4668–4671. [Google Scholar] [CrossRef] [PubMed]
- Verna, A.E.; Manrique, J.M.; Pérez, S.E.; Leunda, M.R.; Pereyra, S.B.; Jones, L.R.; Odeón, A.C. Genomic analysis of bovine herpesvirus type 4 (BoHV-4) from Argentina: High genetic variability and novel phylogenetic groups. Vet. Microbiol. 2012, 160, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pérez, S.E.; Verna, A.E.; Leunda, M.R.; Favier, P.A.; Ceriani, C.; Morán, P.A.; Odeón, A.C.; Esteban, E.N. High prevalence of bovine herpesvirus type 4 (BoHV-4) DNA in Argentinean Holstein cattle from Santiago del Estero, Argentina. Braz. J. Vet. Res. Anim. Sci. 2011, 48, 454–463. [Google Scholar] [CrossRef]
- Morán, P.E.; Favier, P.A.; Lomónaco, M.; Catena, M.C.; Chiapparrone, M.L.; Odeón, A.C.; Verna, A.E.; Pérez, S.E. Search for the genome of bovine herpesvirus types 1, 4 and 5 in bovine semen. Open Vet. J. 2013, 3, 126–130. [Google Scholar]
- González Altamiranda, E.; Manrique, J.M.; Pérez, S.E.; Ríos, G.L.; Odeón, A.C.; Leunda, M.R.; Jones, L.R.; Verna, A. Molecular characterization of the first bovine herpesvirus 4 (BoHV-4) strains isolated from in vitro bovine embryos production in Argentina. PLoS ONE 2015, 15, e0132212. [Google Scholar] [CrossRef]
- Romeo, F.; Manrique, J.; Pérez, S.; Lounge Uriarte, E.; Marin, M.; Cantón, G.; Leunda, M.R.; González Altamiranda, E.; Pereyra, S.; Spetter, M.; et al. Characterization of the first bovine gammaherpesvirus 4 strain isolated from an aborted bovine fetus in Argentina. Arch. Virol. 2020, 165, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Romeo, F.; Spetter, M.J.; Moran, P.; Pereyra, S.; Odeón, A.; Perez, S.E.; Verna, A.E. Analysis of the transcripts encoding for antigenic proteins of bovine gammaherpesvirus 4. J. Vet. Sci. 2020, 21, e5. [Google Scholar] [CrossRef] [PubMed]
- Romeo, F.; Louge-Uriarte, E.; Gonzalez-Altamiranda, E.; Delgado, S.; Pereyra, S.; Morán, P.; Odeón, A.; Pérez, S.; Verna, A.E. Gene expression and in vitro replication of bovine gammaherpesvirus type 4. Arch. Virol. 2021, 166, 535–544. [Google Scholar] [CrossRef]
- Verna, A.E.; Morán, P.E.; Gonzalez Altamiranda, E.A.; Marin, M.S.; Leunda, M.R.; Spetter Lucas, M.J.; Pereyra, S.; Odeón, A.; Perez, S. Experimental infection of calves with Argentinean strains of bovine herpesvirus type 4 belonging to different genotypes. J. Vet. Res. Ani. 2020, 3, 101–106. [Google Scholar]
- Dubuisson, J.; Guillaume, J.; Boulanger, D.; Thiry, E.; Bublot, M.; Pastoret, P.P. Neutralization of bovine herpesvirus type 4 by pairs of monoclonal antibodies raised against two glycoproteins and identification of antigenic determinants involved in neutralization. J. Gen. Virol. 1990, 71, 647–653. [Google Scholar] [CrossRef]
- Gagnon, C.A.; Traesel, C.K.; Music, N.; Laroche, J.; Tison, N.; Auger, J.P.; Music, S.; Provost, C.; Bellehumer, C.; Abrahamyan, L.; et al. Whole genome sequencing of a Canadian bovine gammaherpesvirus 4 strain and the possible link between the viral infection and respiratory and reproductive clinical manifestations in dairy cattle. Front. Vet. Sci. 2017, 4, 92. [Google Scholar] [CrossRef]
- Bauermann, F.V.; Falkenberg, S.M.; Martins, M.; Dassanayake, R.P.; Neill, J.D.; Ridpath, J.F.; Silveira, S.; Palmer, M.; Buysse, A.; Mohr, A.; et al. Genome sequence and experimental infection of calves with bovine gammaherpesvirus 4 (BoGHV-4). Arch. Virol. 2022, 167, 1659–1668. [Google Scholar] [CrossRef]
- Guo, W.; Sun, T.; Liu, Y.; Duan, X.; Tian, C.; Zhou, Z.; Jung, Y.-S.; Liu, J.; Chen, H. Characterization and phylogenetic analysis of bovine gammaherpesvirus 4 isolated in China, 2022. Virus Genes 2023, 59, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Pérez, S.; Manrique, J.; Morán, P.; Romeo, F.; Angelini, H.; Leunda, M.R.; Pereyra, S.; Spetter, M.; González Altamiranda, E.; Odeón, A.; et al. Genetic characterization of bovine herpesvirus 4 (BoHV-4) isolates from Argentine cattle suggests a complex evolutionary scenario. Mol. Biol. Rep. 2020, 47, 4905–4909. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11, molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.4.4 Software. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. 2018. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 18 April 2024).
- Verna, A.E.; Perez, S.E.; Manrique, J.M.; Leunda, M.R.; Odeon, A.C.; Jones, L.R. Comparative study on the in vitro replication and genomic variability of Argentinean field isolates of bovine herpesvirus type 4 (BoHV-4). Virus Genes 2016, 52, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Romeo, F.; Delgado, S.; Uriarte, E.L.; Storani, L.; Cuesta, L.M.; Morán, P.; González Altamiranda, E.; Odeón, A.; Perez, S.E.; Verna, A. Study of the dynamics of in vitro infection with bovine gammaherpesvirus type 4 and apoptosis markers in susceptible cells. Microb. Pathog. 2022, 169, 105645. [Google Scholar] [CrossRef] [PubMed]
- Dağalp, S.B.; Dogan, F.; Babaoglu, A.R.; Farzani, T.A.; Alkan, F. Genetic variability of bovine herpesvirus type 4 (BoHV-4) field strains from Turkish cattle herds. Vet. Ital. 2021, 57, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Chen, R.H.; Yang, M.J.; Zhu, Y.M.; Xue, F. Isolation and molecular characterization of bovine herpesvirus 4 from cattle in mainland China. Arch. Virol. 2021, 166, 619–626. [Google Scholar] [CrossRef] [PubMed]
- McGeoch, D.J.; Cook, S.; Dolan, A.; Jamieson, F.E.; Telford, E.A. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 1995, 247, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Fábián, K.; Egyed, L. Detection of bovine gammaherpesviruses by a nested duplex PCR. J. Virol. Methods 2004, 115, 93–98. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Machiels, B.; Lété, C.; de Fays, K.; Mast, J.; Dewals, B.; Stevenson, P.G.; Vanderplasschen, A.; Gillet, L. The bovine herpesvirus 4 Bo10 gene encodes a nonessential viral envelope protein that regulates viral tropism through both positive and negative effects. J. Virol. 2011, 85, 1011–1024. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 310–322. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.B.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef]
- Owen, M.; Provan, J.S. A fast algorithm for computing geodesic distances in tree space. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Goluch, T.; Bogdanowicz, D.; Giaro, K. Visual TreeCmp: Comprehensive comparison of phylogenetic trees on the web. Methods Ecol. Evol. 2020, 11, 494–499. [Google Scholar] [CrossRef]
- Robinson, O.; Dylus, D.; Dessimoz, C. Phylo.io: Interactive viewing and comparison of large phylogenetic trees on the web. Mol. Biol. Evol. 2016, 33, 2163–2166. [Google Scholar] [CrossRef] [PubMed]
- Dewals, B.; Thirion, M.; Markine-Goriaynoff, N.; Gillet, L.; de Fays, K.; Minner, F.; Daix, V.; Sharp, P.M.; Vanderplasschen, A. Evolution of Bovine herpesvirus 4: Recombination and transmission between African buffalo and cattle. J. Gen. Virol. 2006, 87, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrel, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Ehlers, B.; Dural, G.; Yasmum, N.; Lembo, T.; de Thoisy, B.; Ryser-Degiorgis, M.P.; Ulrich, R.G.; McGeoch, D.J. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: Cospeciation and interspecies transfer. J. Virol. 2008, 82, 3509–3516. [Google Scholar] [CrossRef]
- Black, W.; Troyer, R.M.; Coutu, J.; Wong, K.; Wolff, P.; Gilbert, M.; Yuan, J.; Wise, A.G.; Wang, S.; Xu, D.; et al. Identification of gammaherpesvirus infection in free-ranging black bears (Ursus americanus). Virus Res. 2019, 259, 46–53. [Google Scholar] [CrossRef]
- Maness, H.T.; Nollens, H.H.; Jensen, E.D.; Goldstein, T.; LaMere, S.; Childress, A.; Sykes, J.; St Leger, J.; Lacave, G.; Latson, F.E.; et al. Phylogenetic analysis of marine mammal herpesviruses. Vet. Microbiol. 2011, 21, 23–29. [Google Scholar] [CrossRef]
- Spear, P.G.; Longnecker, R. Herpesvirus entry: An update. J. Virol. 2003, 77, 10179–10185. [Google Scholar] [CrossRef]
- Heldwein, E.E.; Krummenacher, C. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 2008, 65, 1653–1668. [Google Scholar] [CrossRef]
- Thiry, E.; Dubuisson, J.; Bublot, M.; Van Bressem, M.F.; Pastoret, P.P. The biology of bovine herpesvirus-4 infection of cattle. Dtsch. Tierarztl. Wochenschr. 1990, 97, 72–77. [Google Scholar]
- Frazier, K.S.; Baldwin, C.A.; Pence, M.; West, J.; Bernard, J.; Liggett, A.; Miller, D.; Hines, M.E., II. Seroprevalence and comparison of isolates of endometriotropic bovine herpesvirus-4. J. Vet. Diagn. Investig. 2002, 14, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Asano, A.; Inoshima, Y.; Murakami, K.; Iketani, Y.; Yamamoto, Y.; Sentsui, H. Latency and persistence of bovine herpesvirus type 4, strain B11-41, in bovine nervous tissues. J. Vet. Med. Sci. 2003, 65, 87–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McGeoch, D.J.; Dolan, A.; Ralph, A.C. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 2000, 74, 10401–10406. [Google Scholar] [CrossRef] [PubMed]
- Albà, M.M.; Das, R.; Orengo, C.A.; Kellam, P. Genomewide function conservation and phylogeny in the Herpesviridae. Genome Res. 2001, 11, 43–54. [Google Scholar] [CrossRef]
- Franceschi, V.; Capocefalo, A.; Ravanetti, L.; Vanderplasschen, A.; Gillet, L.; Cavirani, S.; van Santen, V.L.; Donofrio, G. Bovine herpesvirus 4 immediate early 2 (Rta) gene is an essential gene and is duplicated in bovine herpesvirus 4 isolate U. Vet. Microbiol. 2011, 148, 219–231. [Google Scholar] [CrossRef]
- Rokas, A.; Williams, B.L.; King, N.; Carroll, S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 2003, 23, 798–804. [Google Scholar] [CrossRef]
- Areda, D.; Chigerwe, M.; Crossley, B. Bovine herpes virus type-4 infection among postpartum dairy cows in California: Risk factors and phylogenetic analysis. Epidemiol. Infect. 2018, 146, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Renner, D.W.; Szpara, M.L. Impacts of genome-wide analyses on our understanding of human herpesvirus diversity and evolution. J. Virol. 2017, 92, e00908-17. [Google Scholar] [CrossRef] [PubMed]
- Kakoola, D.N.; Sheldon, J.; Byabazaire, N.; Bowden, R.J.; Katongole-Mbidde, E.; Schulz, T.F.; Davison, A.J. Recombination in human herpesvirus-8 strains from Uganda and evolution of the K15 gene. J. Gen. Virol. 2001, 82, 2393–2404. [Google Scholar] [CrossRef] [PubMed]
- Schynts, F.; Meurens, F.; Detry, B.; Vanderplasschen, A.; Thiry, E. Rise and survival of bovine herpesvirus 1 recombinants after primary infection and reactivation from latency. J. Virol. 2003, 77, 12535–12542. [Google Scholar] [CrossRef] [PubMed]
- Bowden, R.; Sakaoka, H.; Donnelly, P.; Ward, R. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect. Genet. Evol. 2004, 4, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Kwok, H.; Wu, C.W.; Palser, A.L.; Kellam, P.; Sham, P.C.; Kwong, D.L.W.; Chiang, A.K.S. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J. Virol. 2014, 88, 10662–10672. [Google Scholar] [CrossRef]
- Santpere, G.; Darre, F.; Blanco, S.; Alcami, A.; Villoslada, P.; Mar Albà, M.; Navarro, A. Genome-wide analysis of wild-type Epstein-Barr virus genomes derived from healthy individuals of the 1000 Genomes Project. Genome Biol. Evol. 2014, 6, 846–860. [Google Scholar] [CrossRef]



| 66-p-347 | SD16-38 | HB-ZJK | FMV09 | SD16-49 | V.test | 10-154 | 07-435 | |
|---|---|---|---|---|---|---|---|---|
| 66-p-347 | - | |||||||
| SD16-38 | 0.03 | - | ||||||
| HB-ZJK | 1.03 | 1.03 | - | |||||
| FMV09 | 1.14 | 1.14 | 1.07 | - | ||||
| SD16-49 | 1.13 | 1.12 | 1.04 | 0.06 | - | |||
| V.test | 0.82 | 0.82 | 0.87 | 1.05 | 1.04 | - | ||
| 10-154 | 1.32 | 1.33 | 1.43 | 1.39 | 1.39 | 1.29 | - | |
| 07-435 | 3.27 | 3.29 | 3.21 | 3.02 | 3.01 | 3.33 | 3.34 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romeo, F.; Spetter, M.J.; Pereyra, S.B.; Morán, P.E.; González Altamiranda, E.A.; Louge Uriarte, E.L.; Odeón, A.C.; Pérez, S.E.; Verna, A.E. Whole Genome Sequence-Based Analysis of Bovine Gammaherpesvirus 4 Isolated from Bovine Abortions. Viruses 2024, 16, 739. https://doi.org/10.3390/v16050739
Romeo F, Spetter MJ, Pereyra SB, Morán PE, González Altamiranda EA, Louge Uriarte EL, Odeón AC, Pérez SE, Verna AE. Whole Genome Sequence-Based Analysis of Bovine Gammaherpesvirus 4 Isolated from Bovine Abortions. Viruses. 2024; 16(5):739. https://doi.org/10.3390/v16050739
Chicago/Turabian StyleRomeo, Florencia, Maximiliano Joaquín Spetter, Susana Beatriz Pereyra, Pedro Edgardo Morán, Erika Analía González Altamiranda, Enrique Leopoldo Louge Uriarte, Anselmo Carlos Odeón, Sandra Elizabeth Pérez, and Andrea Elizabeth Verna. 2024. "Whole Genome Sequence-Based Analysis of Bovine Gammaherpesvirus 4 Isolated from Bovine Abortions" Viruses 16, no. 5: 739. https://doi.org/10.3390/v16050739
APA StyleRomeo, F., Spetter, M. J., Pereyra, S. B., Morán, P. E., González Altamiranda, E. A., Louge Uriarte, E. L., Odeón, A. C., Pérez, S. E., & Verna, A. E. (2024). Whole Genome Sequence-Based Analysis of Bovine Gammaherpesvirus 4 Isolated from Bovine Abortions. Viruses, 16(5), 739. https://doi.org/10.3390/v16050739






