Respiratory Virus-Induced PARP1 Alters DC Metabolism and Antiviral Immunity Inducing Pulmonary Immunopathology
Abstract
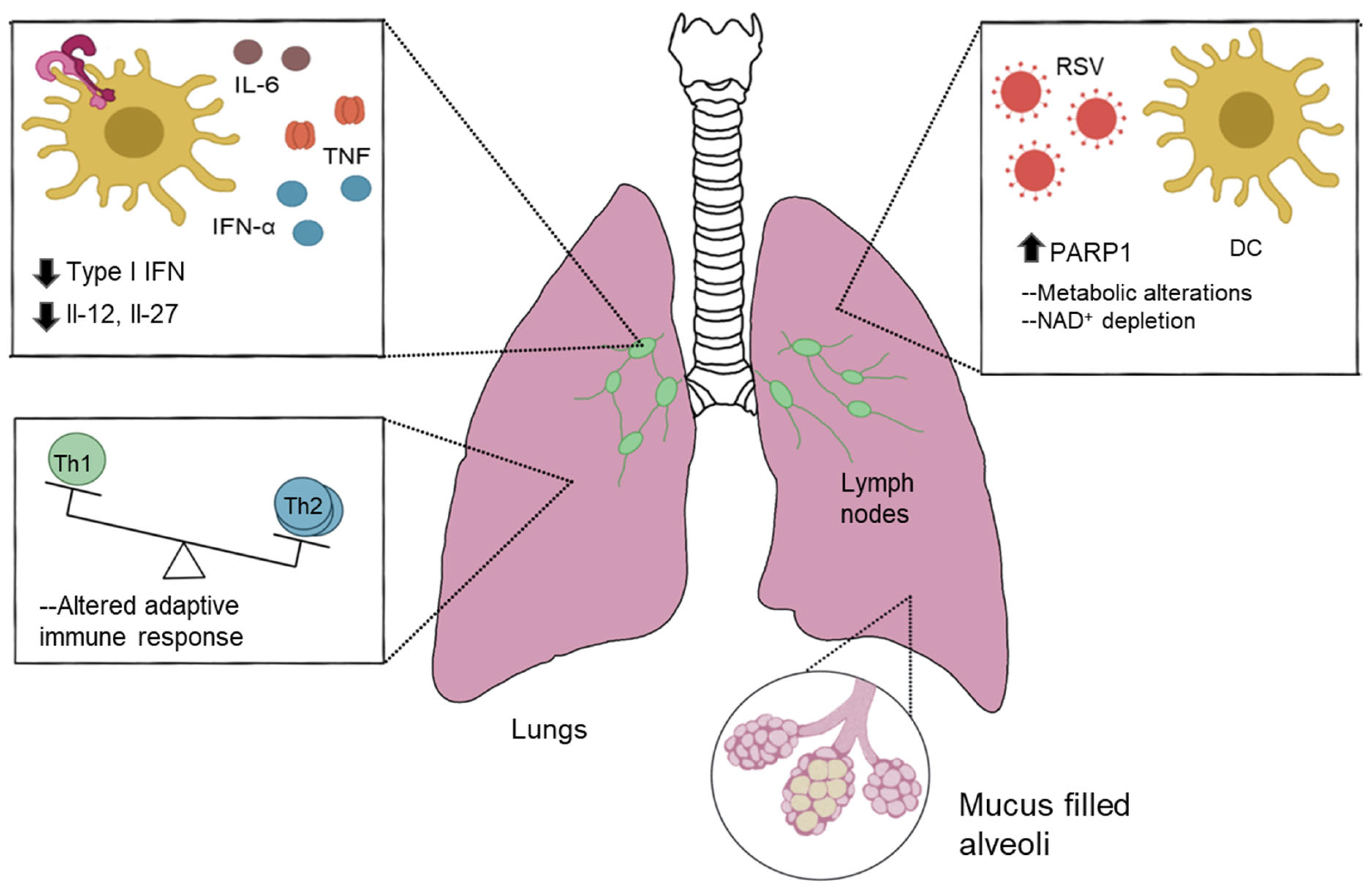
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. PARP1 Inhibitor
2.3. RSV Infection
2.4. Histology
2.5. Lymph Node Restimulation
2.6. Flow Cytometry
2.7. Array-Based Omics Data
2.8. Quantitative rt-PCR
2.9. Real-Time Cell Metabolic Analysis
2.10. Co-Culture Experiments
2.11. Statistical Analysis
3. Results
3.1. RSV Differentially Regulates Key Transcriptomic and Metabolic Targets in Dendritic Cells
3.2. PARP1 Suppresses Key Elements of DC Innate Function during RSV Infection
3.3. PARP1 Modulates Key Aspects of DC Metabolic Processes during RSV Infection
3.4. PARP1-/- Mice Are Protected against RSV-Induced Immunopathology
3.5. Depletion of PARP1 Enzymatic Activity Protected Mice from RSV-Induced Pulmonary Pathology
3.6. Delayed/Therapeutic PARPi Treatment Has an Incomplete Effect
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Smith, D.K.; Seales, S.; Budzik, C. Respiratory Syncytial Virus Bronchiolitis in Children. Am. Fam. Physician 2017, 95, 94–99. [Google Scholar] [PubMed]
- Choi, Y.; Hill-Ricciuti, A.; Branche, A.R.; Sieling, W.D.; Saiman, L.; Walsh, E.E.; Phillips, M.; Falsey, A.R.; Finelli, L. Cost determinants among adults hospitalized with respiratory syncytial virus in the United States, 2017–2019. Influenza Respir. Viruses 2022, 16, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R. Respiratory syncytial virus infection in elderly and high-risk adults. Exp. Lung Res. 2005, 31, 77. [Google Scholar] [CrossRef] [PubMed]
- Openshaw, P.J.M.; Dean, G.S.; Culley, F.J. Links between respiratory syncytial virus bronchiolitis and childhood asthma: Clinical and research approaches. Pediatr. Infect. Dis. J. 2003, 22, S58–S65. [Google Scholar] [CrossRef] [PubMed]
- Nduaguba, S.O.; Tran, P.T.; Choi, Y.; Winterstein, A.G. Respiratory syncytial virus reinfections among infants and young children in the United States, 2011–2019. PLoS ONE 2023, 18, e0281555. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, Y.; Yasui, Y.; Nakayama, T. Development of Acquired Immunity following Repeated Respiratory Syncytial Virus Infections in Cotton Rats. PLoS ONE 2016, 11, e0155777. [Google Scholar] [CrossRef] [PubMed]
- Glezen, W.P.; Taber, L.H.; Frank, A.L.; Kasel, J.A. Risk of Primary Infection and Reinfection With Respiratory Syncytial Virus. Am. J. Dis. Child. 1986, 140, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Lindell, D.M.; Lane, T.E.; Lukacs, N.W. CXCL10/CXCR3-mediated responses promote immunity to respiratory syncytial virus infection by augmenting dendritic cell and CD8+ T cell efficacy. Eur. J. Immunol. 2008, 38, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.; Swanson, M.S.; Lieberman, A.; Reed, M.; Yue, Z.; Lindell, D.M.; Lukacs, N.W. Autophagy-Mediated Dendritic Cell Activation Is Essential for Innate Cytokine Production and APC Function with Respiratory Syncytial Virus Responses. J. Immunol. 2011, 187, 3953–3961. [Google Scholar] [CrossRef]
- Narayanan, S.; Elesela, S.; Rasky, A.J.; Morris, S.H.; Kumar, S.; Lombard, D.; Lukacs, N.W. ER stress protein PERK promotes inappropriate innate immune responses and pathogenesis during RSV infection. J. Leukoc. Biol. 2022, 111, 379–389. [Google Scholar] [CrossRef]
- Malinczak, C.A.; Fonseca, W.; Mire, M.M.; Parolia, A.; Chinnaiyan, A.; Rasky, A.J.; Morris, S.; Yagi, K.; Bermick, J.R.; Lukacs, N.W. Sex-associated early-life viral innate immune response is transcriptionally associated with chromatin remodeling of type-I IFN-inducible genes. Mucosal Immunol. 2023, 16, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Smit, J.J.; Rudd, B.D.; Lukacs, N.W. Plasmacytoid dendritic cells inhibit pulmonary immunopathology and promote clearance of respiratory syncytial virus. J. Exp. Med. 2006, 203, 1153–1159. [Google Scholar] [CrossRef]
- Morales, J.C.; Li, L.; Fattah, F.J.; Dong, Y.; Bey, E.A.; Patel, M.; Gao, J.; Boothman, D.A. Review of Poly (ADP-ribose) Polymerase (PARP) Mechanisms of Action and Rationale for Targeting in Cancer and Other Diseases. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 15–28. [Google Scholar] [CrossRef]
- Sinha, S.; Molla, S.; Kundu, C.N. PARP1-modulated chromatin remodeling is a new target for cancer treatment. Med. Oncol. 2021, 38, 118. [Google Scholar] [CrossRef]
- Zong, C.; Zhu, T.; He, J.; Huang, R.; Jia, R.; Shen, J. PARP mediated DNA damage response, genomic stability and immune responses. Int. J. Cancer 2022, 150, 1745–1759. [Google Scholar] [CrossRef] [PubMed]
- Realini, C.A.; Althaus, F.R. Histone shuttling by poly(ADP-ribosylation). J. Biol. Chem. 1992, 267, 18858–18865. [Google Scholar] [CrossRef]
- Beneke, S.; Cohausz, O.; Malanga, M.; Boukamp, P.; Althaus, F.; Bürkle, A. Rapid regulation of telomere length is mediated by poly(ADP-ribose) polymerase-1. Nucleic Acids Res. 2008, 36, 6309–6317. [Google Scholar] [CrossRef] [PubMed]
- Dantzer, F.; Amé, J.C.; Schreiber, V.; Nakamura, J.; Ménissier-de Murcia, J.; de Murcia, G. Poly(ADP-ribose) Polymerase-1 Activation During DNA Damage and Repair. Methods Enzymol. 2006, 409, 493–510. [Google Scholar]
- Kamaletdinova, T.; Fanaei-Kahrani, Z.; Wang, Z.Q. The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers. Cells 2019, 8, 1625. [Google Scholar] [CrossRef]
- Amé, J.C.; Spenlehauer, C.; De Murcia, G. The PARP superfamily. Bioessays 2004, 26, 882–893. [Google Scholar] [CrossRef]
- d’Amours, D.; Desnoyers, S.; d’Silva, I.; Poirier, G.G. Poly (ADP-ribosyl) ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.M.; Kong, X.; Moncada, E.; Chen, Y.; Imamura, H.; Wang, P.; Berns, M.W.; Yokomori, K.; Digman, M.A. NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. Mol. Biol. Cell 2019, 30, 2584–2597. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Bagès, S.; Knobloch, G.; Ladurner, A.G.; Buschbeck, M. The taming of PARP1 and its impact on NAD+ metabolism. Mol. Metab. 2020, 38, 100950. [Google Scholar] [CrossRef] [PubMed]
- Namgaladze, D.; Brüne, B. Rapid glycolytic activation accompanying innate immune responses: Mechanisms and function. Front. Immunol. 2023, 14, 1180488. [Google Scholar] [CrossRef] [PubMed]
- Herlocher, M.L.; Ewasyshyn, M.; Sambhara, S.; Gharaee-Kermani, M.; Cho, D.; Lai, J.; Klein, M.; Maassab, H.F. Immunological properties of plaque purified strains of live attenuated respiratory syncytial virus (RSV) for human vaccine. Vaccine 1999, 17, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.L.; Chi, M.H.; Luongo, C.; Lukacs, N.W.; Polosukhin, V.V.; Huckabee, M.M.; Newcomb, D.C.; Buchholz, U.J.; Crowe, J.E.; Goleniewska, K.; et al. A chimeric A2 strain of respiratory syncytial virus (RSV) with the fusion protein of RSV strain line 19 exhibits enhanced viral load, mucus, and airway dysfunction. J. Virol. 2009, 83, 4185–4194. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Smit, J.; Kallal, L.E.; Lukacs, N.W. Respiratory syncytial virus infection modifies and accelerates pulmonary disease via DC activation and migration. J. Leukoc. Biol. 2013, 94, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Elesela, S.; Morris, S.B.; Narayanan, S.; Kumar, S.; Lombard, D.B.; Lukacs, N.W. Sirtuin 1 regulates mitochondrial function and immune homeostasis in respiratory syncytial virus infected dendritic cells. PLoS Pathog. 2020, 16, e1008319. [Google Scholar] [CrossRef] [PubMed]
- Malinczak, C.-A.; Parolia, A.; Fonseca, W.; Morris, S.; Rasky, A.J.; Bawa, P.; Zhang, Y.; Mire, M.M.; Ziegler, S.F.; Ptaschinski, C.; et al. TSLP-Driven Chromatin Remodeling and Trained Systemic Immunity after Neonatal Respiratory Viral Infection. J. Immunol. 2021, 206, 1315–1328. [Google Scholar] [CrossRef]
- Smyth, G.K. limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Miller, A.L.; Strieter, R.M.; Gruber, A.D.; Ho, S.B.; Lukacs, N.W. CXCR2 Regulates Respiratory Syncytial Virus-Induced Airway Hyperreactivity and Mucus Overproduction. J. Immunol. 2003, 170, 3348–3356. [Google Scholar] [CrossRef] [PubMed]
- Todosenko, N.; Khaziakhmatova, O.; Malashchenko, V.; Yurova, K.; Bograya, M.; Beletskaya, M.; Vulf, M.; Gazatova, N.; Litvinova, L. Mitochondrial Dysfunction Associated with mtDNA in Metabolic Syndrome and Obesity. Int. J. Mol. Sci. 2023, 24, 12012. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Flynn, C.; Niepel, M.; Hafner, M.; Muhlich, J.L.; Fernandez, N.F.; Rouillard, A.D.; Tan, C.M.; Chen, E.Y.; Golub, T.R.; et al. LINCS Canvas Browser: Interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 2014, 42, W449–W460. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Sanin, D.E.; Everts, B.; Chen, Q.; Qiu, J.; Buck, M.D.; Patterson, A.; Smith, A.M.; Chang, C.H.; Liu, Z.; et al. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity 2016, 44, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Montoya, M.; Schiavoni, G.; Mattel, F.; Gresser, I.; Belardelli, F.; Borrow, P.; Tough, D.F. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood 2002, 99, 3263–3271. [Google Scholar] [CrossRef] [PubMed]
- Pantel, A.; Teixeira, A.; Haddad, E.; Wood, E.G.; Steinman, R.M.; Longhi, M.P. Direct Type I IFN but Not MDA5/TLR3 Activation of Dendritic Cells Is Required for Maturation and Metabolic Shift to Glycolysis after Poly IC Stimulation. PLoS Biol. 2014, 12, e1001759. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, J.; Hulpiau, P.; Saelens, X. Mx Proteins: Antiviral Gatekeepers That Restrain the Uninvited. Microbiol. Mol. Biol. Rev. 2013, 77, 551. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, D.F.; Zlotnik, A.; Vieira, P.; Mosmann, T.R.; Howard, M.; Moore, K.W.; O’garra, A. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J. Immunol. 1991, 146, 3444–3451. [Google Scholar] [CrossRef] [PubMed]
- Amjad, S.; Nisar, S.; Bhat, A.A.; Shah, A.R.; Frenneaux, M.P.; Fakhro, K.; Haris, M.; Reddy, R.; Patay, Z.; Baur, J.; et al. Role of NAD+ in regulating cellular and metabolic signaling pathways. Mol. Metab. 2021, 49, 101195. [Google Scholar] [CrossRef] [PubMed]
- Guak, H.; al Habyan, S.; Ma, E.H.; Aldossary, H.; Al-Masri, M.; Won, S.Y.; Ying, T.; Fixman, E.D.; Jones, R.G.; McCaffrey, L.M.; et al. Glycolytic metabolism is essential for CCR7 oligomerization and dendritic cell migration. Nat. Commun. 2018, 9, 2463. [Google Scholar] [CrossRef]
- Krawczyk, C.M.; Holowka, T.; Sun, J.; Blagih, J.; Amiel, E.; DeBerardinis, R.J.; Cross, J.R.; Jung, E.; Thompson, C.B.; Jones, R.G.; et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010, 115, 4742–4749. [Google Scholar] [CrossRef] [PubMed]
- Everts, B.; Amiel, E.; van der Windt GJ, W.; Freitas, T.C.; Chott, R.; Yarasheski, K.E.; Pearce, E.L.; Pearce, E.J. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood 2012, 120, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Everts, B.; Amiel, E.; Huang, S.C.; Smith, A.M.; Chang, C.H.; Lam, W.Y.; Redmann, V.; Freitas, T.C.; Blagih, J.; Van Der Windt, G.J.; et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKε supports the anabolic demands of dendritic cell activation. Nat. Immunol. 2014, 15, 323–332. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.; Ramanathan, R.K.; Mina, L.; Chugh, R.; Glaspy, J.; Rafii, S.; Kaye, S.; Sachdev, J.; Heymach, J.; Smith, D.C.; et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. 2017, 7, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Hijano, D.R.; Siefker, D.T.; Shrestha, B.; Jaligama, S.; Vu, L.D.; Tillman, H.; Finkelstein, D.; Saravia, J.; You, D.; Cormier, S.A. Type I Interferon Potentiates IgA Immunity to Respiratory Syncytial Virus Infection During Infancy. Sci. Rep. 2018, 8, 11034. [Google Scholar] [CrossRef] [PubMed]
- Goritzka, M.; Durant, L.R.; Pereira, C.; Salek-Ardakani, S.; Openshaw, P.J.M.; Johansson, C. Alpha/Beta Interferon Receptor Signaling Amplifies Early Proinflammatory Cytokine Production in the Lung during Respiratory Syncytial Virus Infection. J. Virol. 2014, 88, 6128. [Google Scholar] [CrossRef] [PubMed]
- Rezinciuc, S.; Bezavada, L.; Bahadoran, A.; Duan, S.; Wang, R.; Lopez-Ferrer, D.; Finkelstein, D.; McGargill, M.A.; Green, D.R.; Pasa-Tolic, L.; et al. Dynamic metabolic reprogramming in dendritic cells: An early response to influenza infection that is essential for effector function. PLoS Pathog. 2020, 16, e1008957. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.J.; Filla, M.B.; Fultz, M.J.; Vogel, S.N.; Russell, S.W.; Murphy, W.J. Autocrine/Paracrine IFN-αβ Mediates the Lipopolysaccharide-Induced Activation of Transcription Factor Stat1α in Mouse Macrophages: Pivotal Role of Stat1α in Induction of the Inducible Nitric Oxide Synthase Gene. J. Immunol. 1998, 161, 4803–4810. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Chen, K.; Zhu, H.; Li, Z.; Cao, X. CCR7 Chemokine Receptor-Inducible lnc-Dpf3 Restrains Dendritic Cell Migration by Inhibiting HIF-1α-Mediated Glycolysis. Immunity 2019, 50, 600–615. [Google Scholar]
- Inamdar, S.; Suresh, A.P.; Mangal, J.L.; Ng, N.D.; Sundem, A.; Wu, C.; Lintecum, K.; Thumsi, A.; Khodaei, T.; Halim, M.; et al. Rescue of dendritic cells from glycolysis inhibition improves cancer immunotherapy in mice. Nat. Commun. 2023, 14, 5333. [Google Scholar] [CrossRef] [PubMed]
- Zevini, A.; Palermo, E.; di Carlo, D.; Alexandridi, M.; Rinaldo, S.; Paone, A.; Cutruzzola, F.; Etna, M.P.; Coccia, E.M.; Olagnier, D.; et al. Inhibition of Glycolysis Impairs Retinoic Acid-Inducible Gene I–Mediated Antiviral Responses in Primary Human Dendritic Cells. Front. Cell. Infect. Microbiol. 2022, 12, 910864. [Google Scholar] [CrossRef]
- Ren, K.; Lv, Y.; Zhuo, Y.; Chen, C.; Shi, H.; Guo, L.; Yang, G.; Hou, Y.; Tan, R.X.; Li, E. Suppression of IRG-1 reduces inflammatory cell infiltration and lung injury in respiratory syncytial virus infection by reducing production of reactive oxygen species. J. Virol. 2016, 90, 7313–7322. [Google Scholar] [CrossRef] [PubMed]
- Muraro, S.P.; De Souza, G.F.; Gallo, S.W.; Da Silva, B.K.; De Oliveira, S.D.; Vinolo, M.A.; Saraiva, E.M.; Porto, B.N. Respiratory Syncytial Virus induces the classical ROS-dependent NETosis through PAD-4 and necroptosis pathways activation. Sci. Rep. 2018, 8, 14166. [Google Scholar] [CrossRef] [PubMed]
- Komaravelli, N.; Tian, B.; Ivanciuc, T.; Mautemps, N.; Brasier, A.R.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus infection down-regulates antioxidant enzyme expression by triggering deacetylation-proteasomal degradation of Nrf2. Free Radic. Biol. Med. 2015, 88, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Hosakote, Y.M.; Jantzi, P.D.; Esham, D.L.; Spratt, H.; Kurosky, A.; Casola, A.; Garofalo, R.P. Viral-mediated inhibition of antioxidant enzymes contributes to the pathogenesis of severe respiratory syncytial virus bronchiolitis. Am. J. Respir. Crit. Care Med. 2011, 183, 1550–1560. [Google Scholar] [CrossRef] [PubMed]
- Ohol, Y.M.; Wang, Z.; Kemble, G.; Duke, G. Direct Inhibition of Cellular Fatty Acid Synthase Impairs Replication of Respiratory Syncytial Virus and Other Respiratory Viruses. PLoS ONE 2015, 10, e0144648. [Google Scholar] [CrossRef] [PubMed]
- Alemasova, E.E.; Lavrik, O.I. Poly(ADP-ribosyl)ation by PARP1: Reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019, 47, 3811–3827. [Google Scholar] [CrossRef] [PubMed]
- Fouquerel, E.; Goellner, E.M.; Yu, Z.; Gagne, J.P.; de Moura, M.B.; Feinstein, T.; Wheeler, D.; Redpath, P.; Li, J.; Romero, G.; et al. ARTD1/PARP1 Negatively Regulates Glycolysis by Inhibiting Hexokinase 1 Independent of NAD+ Depletion. Cell Rep. 2014, 8, 1819–1831. [Google Scholar] [CrossRef]
- Stein, L.R.; Imai, S.I. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol. Metab. 2012, 23, 420–428. [Google Scholar] [CrossRef]
- Bai, P.; Cantó, C.; Oudart, H.; Brunyánszki, A.; Cen, Y.; Thomas, C.; Yamamoto, H.; Huber, A.; Kiss, B.; Houtkooper, R.H.; et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011, 13, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Scheibye-Knudsen, M.; Brace, L.E.; Kassahun, H.; Sengupta, T.; Nilsen, H.; Mitchell, J.R.; Croteau, D.L.; Bohr, V.A. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell 2014, 157, 882–896. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Garnier, P.; Swanson, R.A. NAD+ repletion prevents PARP-1-induced glycolytic blockade and cell death in cultured mouse astrocytes. Biochem. Biophys. Res. Commun. 2003, 308, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Devalaraja-Narashimha, K.; Padanilam, B.J. PARP-1 Inhibits Glycolysis in Ischemic Kidneys. J. Am. Soc. Nephrol. 2009, 20, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Du, M.; Tan, X.; Yang, L.; Li, X.; Jiang, Y.; Wang, C.; Zhang, F.; Zhu, F.; Cheng, M.; et al. PARP1-mediated PPARα poly(ADP-ribosyl)ation suppresses fatty acid oxidation in non-alcoholic fatty liver disease. J. Hepatol. 2017, 66, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Prolla, T.A.; Denu, J.M. NAD+ deficiency in age-related mitochondrial dysfunction. Cell Metab. 2014, 19, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Z.; Tan, Y.; Li, Q.; Yang, H.; Wang, P.; Lu, J.; Liu, P. PARP1 interacts with STAT3 and retains active phosphorylated-STAT3 in nucleus during pathological myocardial hypertrophy. Mol. Cell. Endocrinol. 2018, 474, 137–150. [Google Scholar] [CrossRef]
- Chrisikos, T.T.; Zhou, Y.; Kahn, L.M.; Patel, B.; Denne, N.L.; Brooks, A.; Shen, L.; Wang, J.; Watowich, S.S. STAT3 Inhibits Autocrine IFN Signaling in Type I Conventional Dendritic Cells. J. Immunol. 2022, 209, 1286–1299. [Google Scholar] [CrossRef]
- Melillo, J.A.; Song, L.; Bhagat, G.; Blazquez, A.B.; Plumlee, C.R.; Lee, C.; Berin, C.; Reizis, B.; Schindler, C. Dendritic Cell (DC)-Specific Targeting Reveals Stat3 as a Negative Regulator of DC Function. J. Immunol. 2010, 184, 2638–2645. [Google Scholar] [CrossRef] [PubMed]
- Melillo, J.A.; Song, L.; Bhagat, G.; Blazquez, A.B.; Plumlee, C.R.; Lee, C.; Berin, C.; Reizis, B.; Schindler, C. Activation of Dendritic Cells via Inhibition of Jak2/STAT3 Signaling. J. Immunol. 2005, 175, 4338–4346. [Google Scholar]
- Vasir, B.; Rajabi, H.; Mills, H.; Rosenblatt, J.; Stroopinsky, D.; Luptakova, K.; Coll, M.D.; Kristen, P.; Kufe, D.; Avigan, D. STAT3 Inhibition Promotes Potent Th1 Responses By Down Regulating Pdl-1 Expression On Tumor Cells. Blood 2013, 122, 3217. [Google Scholar] [CrossRef]
- Zhang, C.; Yue, C.; Herrmann, A.; Song, J.; Egelston, C.; Wang, T.; Zhang, Z.; Li, W.; Lee, H.; Aftabizadeh, M.; et al. STAT3 Activation-Induced Fatty Acid Oxidation in CD8+ T Effector Cells Is Critical for Obesity-Promoted Breast Tumor Growth. Cell Metab. 2020, 31, 148–161.e5. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.B.; Nemkov, T.; Stefanoni, D.; Benavides, G.A.; Bassal, M.A.; Crown, B.L.; Matkins, V.R.; Camacho, V.; Kuznetsova, V.; Hoang, A.T.; et al. Metabolic alterations mediated by STAT3 promotes drug persistence in CML. Leukemia 2021, 35, 3371–3382. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mire, M.M.; Elesela, S.; Morris, S.; Corfas, G.; Rasky, A.; Lukacs, N.W. Respiratory Virus-Induced PARP1 Alters DC Metabolism and Antiviral Immunity Inducing Pulmonary Immunopathology. Viruses 2024, 16, 910. https://doi.org/10.3390/v16060910
Mire MM, Elesela S, Morris S, Corfas G, Rasky A, Lukacs NW. Respiratory Virus-Induced PARP1 Alters DC Metabolism and Antiviral Immunity Inducing Pulmonary Immunopathology. Viruses. 2024; 16(6):910. https://doi.org/10.3390/v16060910
Chicago/Turabian StyleMire, Mohamed M., Srikanth Elesela, Susan Morris, Gabriel Corfas, Andrew Rasky, and Nicholas W. Lukacs. 2024. "Respiratory Virus-Induced PARP1 Alters DC Metabolism and Antiviral Immunity Inducing Pulmonary Immunopathology" Viruses 16, no. 6: 910. https://doi.org/10.3390/v16060910
APA StyleMire, M. M., Elesela, S., Morris, S., Corfas, G., Rasky, A., & Lukacs, N. W. (2024). Respiratory Virus-Induced PARP1 Alters DC Metabolism and Antiviral Immunity Inducing Pulmonary Immunopathology. Viruses, 16(6), 910. https://doi.org/10.3390/v16060910





