Abstract
Infectious pancreatic necrosis virus (IPNV) causes economic losses with a highly variable mortality rate worldwide, especially in rainbow trout. The virus has a double-stranded bi-partite RNA genome designated segment A and B. New complete genome sequences of nine rainbow trout isolates from Turkey were determined and subjected to phylogenetic analysis, identifying all as genotype 5 (serotype Sp). A time-dependent change in the extended pathogenicity motif of VP2 from P217T221A247 (PTA) to PTE P217T221E247 over a period of 10 years was identified. A wider analysis of 99 IPNV sequences from Turkey and Iran revealed the emergence of the motif PTE from 2007 to 2017, inducing significant morbidity in fry by 2013. In fact, displacement of the PTA motif, by the PTE motif in IPNV isolates appeared to be connected to a production peak of rainbow trout in 2013. An additional CAI analysis provided more evidence, indicating that rainbow trout culture in Turkey has an influence on the evolution of IPNV.
1. Introduction
Infectious pancreatic necrosis is a contagious viral disease affecting farmed saltwater and freshwater salmonid fish through vertical and horizontal transmission. Infectious pancreatic necrosis virus (IPNV) belongs to the genus Aquabirnavirus in the family Birnaviridae and has a bi-segmented (segment A and segment B) double-stranded RNA genome with approximately 6000 nucleotides. Segment A has a large open reading frame that encodes a polyprotein. The polyprotein, which is post-translationally cleaved, consists of four viral proteins in the following order: 5′-VP5, VP2, VP4, VP3-3′ [1]. VP5 partially overlaps with the 5′-prime region of the VP2 ORF and is a non-essential non-structural protein of variant size and unclear function and is not found in all isolates [2].
VP2 is the major capsid protein and contains a central variable domain and antigenic site (residues 183–335), as well as two hypervariable regions (residues 239–257 and 271–284) [3]. The central variable region includes a pathogenicity locus at positions 217 and 221 (PT persistent, PA, low virulent, TA virulent). VP3 is involved in activating the RNA-dependent RNA polymerase (RdRp) by binding to both RdRp and the dsRNA genome segments, but it is also involved in interferon alpha induction in salmon cells [4,5]. A VP2-VP3 fusion protein has been shown to induce a humoral response in immunized trout [6]. VP4 is a viral protease which is responsible for the post-translational cleavage of the polyprotein. Segment B has one large open frame (VP1) consisting of 2535 bp encoding the RdRp [3,4].
IPNV has been divided into 11 serotypes (Wb, Sp, Ab, He, Te, C1, C2, C3, Ja, TV-1, and MaBV) and seven genotypes (1–7). TV-1 and MaBV (genotype 7) are classified in Serogroup B. The other serotypes are classified as Serogroup A [7,8]. IPNV is distributed worldwide; common serotypes are Sp (genotype 5) and Ab serotypes (genotype 2) in Europe. The He serotype (genotype 6) and Te serotype (genotype 3) have been described in Germany and the United Kingdom, respectively [9]. The Wb (genotype 1), C1 (genotype 3), C2 (genotype 4), C3 (genotype 4), and Ja serotypes (genotype 5) originated in the USA and Canada [8,9,10,11,12].
In vitro studies indicate that reassortment in Birnaviruses can modulate IPNV virulence [13].
IPNV was first reported in Turkey in 2002 [14], and although it mainly caused asymptomatic infections in medium- and large-sized trout (>10 cm), it can, however, cause fatal outbreaks in fingerlings. IPN was a notifiable disease until 2007, and, initially, the demand to test for IPNV in outbreaks from farms was high, but it has dropped since 2007. Meanwhile, IPNV is considered enzootic in Turkey, and there has been no routine screening since 2017. A licensed vaccine is not in use; however, IPNV continues to be detected in sporadic samples received by the Virology Laboratory of the Faculty of Veterinary Medicine at the Ondokuz Mayıs University.
In our previous study, we described partial VP2 sequences of 62 IPNV isolates of serotype Sp from Turkey [15], confirming the observations reported by others [16,17]. In this study, we determined the whole genome of nine Turkish IPNV isolates by NGS. The overall analysis, including all IPNV sequences published from Turkey, indicates the emergence of IPNV isolates signified by previously described extended pathogenicity locus since 2007.
2. Materials and Methods
2.1. Viruses
Six isolates (Burdur 18, Aydın 21, Denizli 22, Tokat 24, Trabzon 27, and Kayseri 28) which were isolated during routine field screening surveys between 2005 and 2012 were kindly provided by Bornova Veterinary Control Institute/Izmir, Turkey (Gülnur KALAYCI and Buket ÖZKAN). IPNV Sdf-4 was provided by Central Fisheries Research Institute, Trabzon, Turkey (Hakan IŞIDAN). IPNV 1054 and IPNV GRE were isolated from dead fish sent to Ondokuz Mayıs University, Faculty of Veterinary Medicine, Virology Laboratory for virological diagnosis in 2013 and 2016, respectively (Table 1) [15].

Table 1.
IPNV isolates and sequences derived.
2.2. Cell Culture
For cell culture isolation, under 1 g of fish whole body, with the tail and head discarded, were cut with a scalpel and were mixed with 1.5 mL of cold Leibowitz’s (L-15) medium (Gibco, Grand Island, NY, USA, Cat No: 11415-064). The mixture was homogenized for 1 min using a TissueLyser LT (Qiagen, Hilden, Germany). The homogenate was centrifuged at 1500× g for 30 min, and the supernatant was sterile-filtered (0.22 µm) and stored at −20 °C. Epithelioma papulosum cyprini (EPC) cells were used for the virus isolation. Briefly, EPC cells were cultivated at 25 °C in L-15 supplemented with 10% fetal calf serum (FCS), 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA, Cat No: P4333-100ML), and 10 mM HEPES (Sigma -Aldrich, St. Louis, MO, USA, Cat No: H0887-100ML). A 1/100 dilution of the homogenates were inoculated onto 1-day-old EPC cells. The cells were maintained at 15 °C in a cooling incubator and checked daily for cytopathic effects (CPEs).
To check for bacterial infectious agents, the homogenates were also inoculated onto Tryptic Soy Agar (TSA), McConkey Agar (MCA), Cytophaga Agar (CA), and Shotts–Waltman Agar (SWA) and incubated at normal atmospheric conditions at 22 °C for 1–2 days (24–48 h).
2.3. Sequencing
Viral RNA extraction and RT-PCR were performed as described earlier [15]. After that, single-strand cDNA synthesis was generated using SuperScript™ III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA Cat No: 18080044) and ds cDNA was synthesized using the NEBNext® Ultra™ II Directional RNA Second Strand Synthesis Module (New England Biolabs, Ipswich, MA, USA, Cat No: E7550L UK) following the manufacturer’s instructions, and ds cDNAs were cleaned by using RNAClean XP (Beckman Coulter, Brea, CA, USA Cat No: A63987).
Libraries were generated by using the Illumina Nextera XT DNA library preparation kit, and all the reads were conducted in a MiSeq sequencer (Illumina, Cambridge, UK) following a previously described protocol [17].
2.4. Phylogenetic Analyses
Whole-genome alignment was performed using Clustal W [18], and the bootstrapped phylogenetic tree of the whole-genome sequences was inferred using RAxML [19], both included in the MEGALIGN module of the DNASTAR software package, version 11. The neighbor net and parsimony net analysis were performed in SPLITS TREE 5.0 using a nucleotide sequence block of either the whole-genome segment sequences and from partial nucleotides sequences of segment A [20].
2.5. Codon Adaptation Index Calculation
To investigate if IPNV isolates from Turkey show evidence of codon adaptation to salmonid genomes, we calculated the codon adaptation indices (CAIs) for VP2. To calculate normalized CAIs, we first used the CAIcal program to obtain a “raw” CAI (rCAI). Next, an “expected neutral CAI” (eCAI) value was calculated by generating 1000 random sequences using a similar length, codon composition, and GC content. Normalized CAI values were then compared among different time points and viral lineages using a non-parametric rank test because the central tendencies trend varied throughout time more than each time point variance. To obtain our normalized CAI threshold, rCAI/eCAI values were calculated. Values greater than 1 were taken as evidence for codon adaptation to the reference set of codon preferences [21]. Values lower than 1 were taken as evidence that mutational bias is driving codon selection. The codon usage tables for Oncorhynchus mykiss and Salmon salar were downloaded directly from the publicly available Codon Usage Database (www.kazusa.or.jp/codon, accessed 25 April 2024).
3. Results
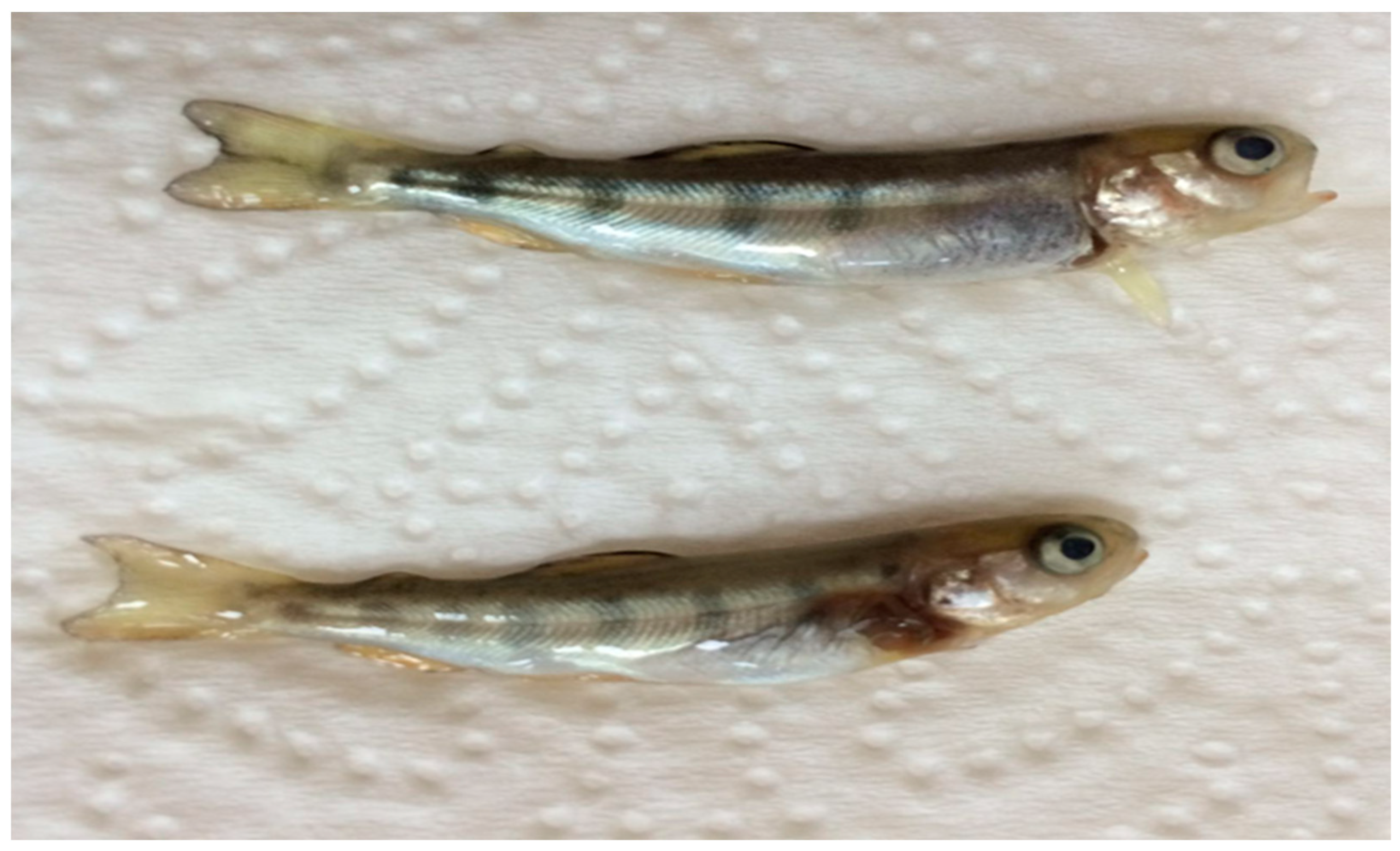
IPNV 1054 and IPNV Gre were isolated from rainbow trout fry of hatchery outbreaks from the Blacksea Region, Turkey, with fry showing abnormal swimming behavior close to the surface of the water. The gross pathology observed included exophthalmia, spinal curvature, skin darkening, and peritonitis with high mortality (Figure 1). All other isolates were isolated during routine screening from 1 g of asymptomatic rainbow trout.
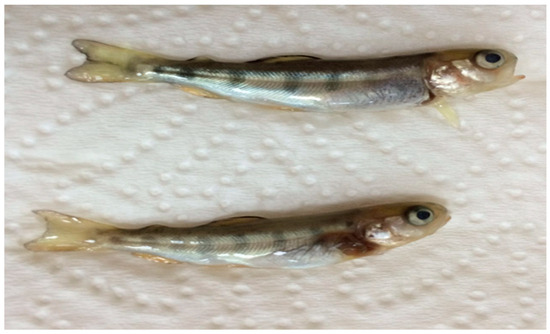
Figure 1.
Gross pathology of rainbow trout fry with exophthalmia and spinal curvature from which IPNV 1054 was isolated. Size 4–4.5 cm; weight 0.7–1 g.
3.1. Virus Isolation and Sequencing
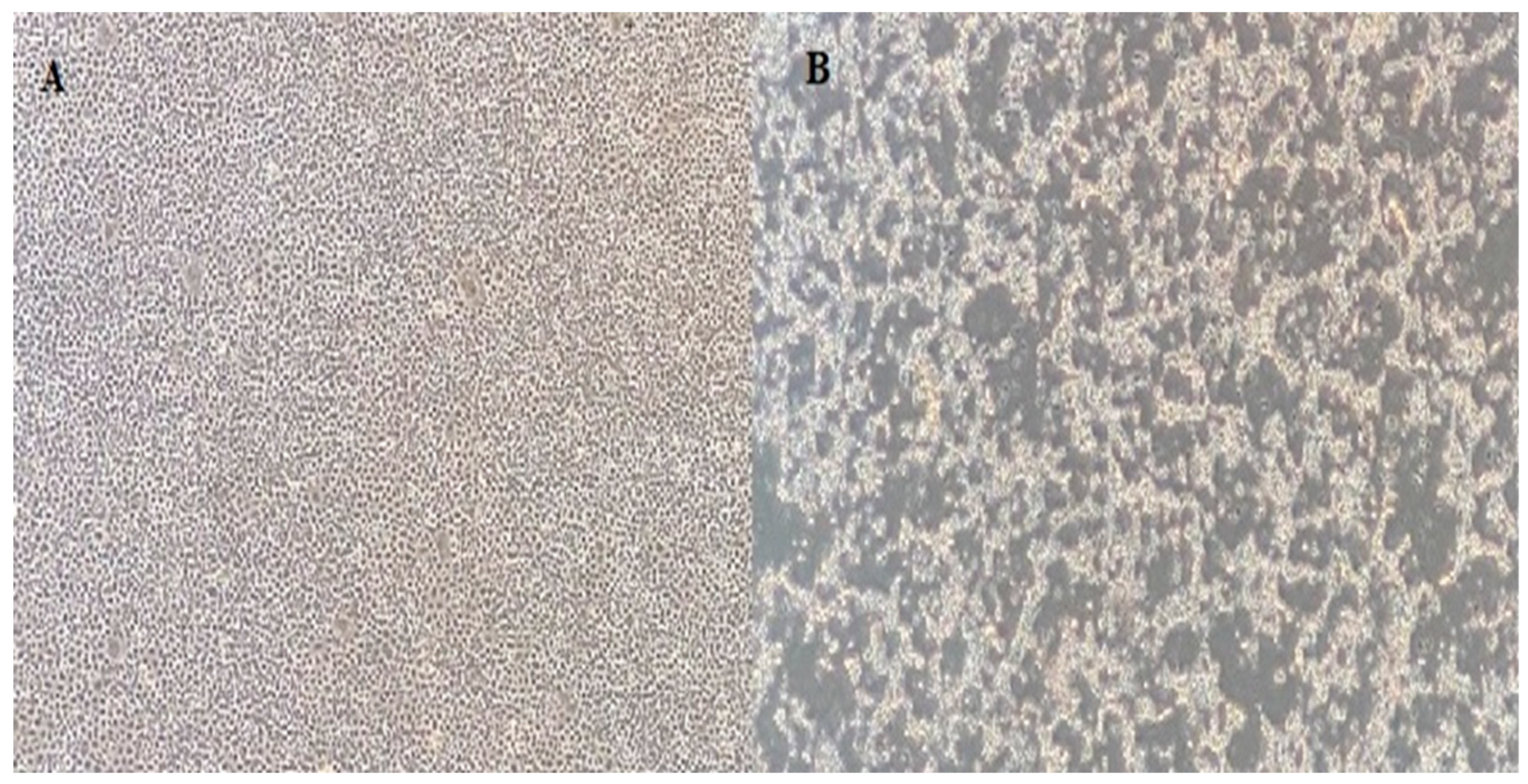
Nine isolates were successfully isolated from EPC cells, and all showed the typical IPNV CPE pattern (Figure 2). The RNA was extracted and subjected to sequencing. The main coverage of the genomes ranged from 20- to 40-fold. Assembled genome sequences were submitted to GenBank, and the accession numbers are listed in Table 1.
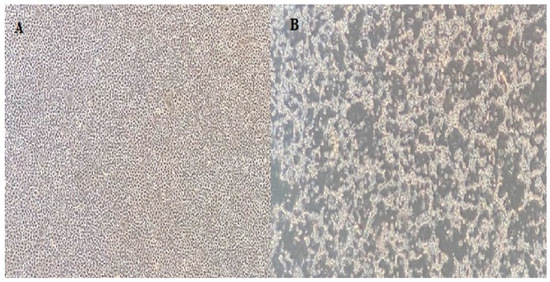
Figure 2.
(A) EPC cell line control; (B) 7th day of inoculation of 1054 IPNV. Cells are visualized at the 40× magnification in an inverted microscope.
3.2. Phylogenetic Analyses
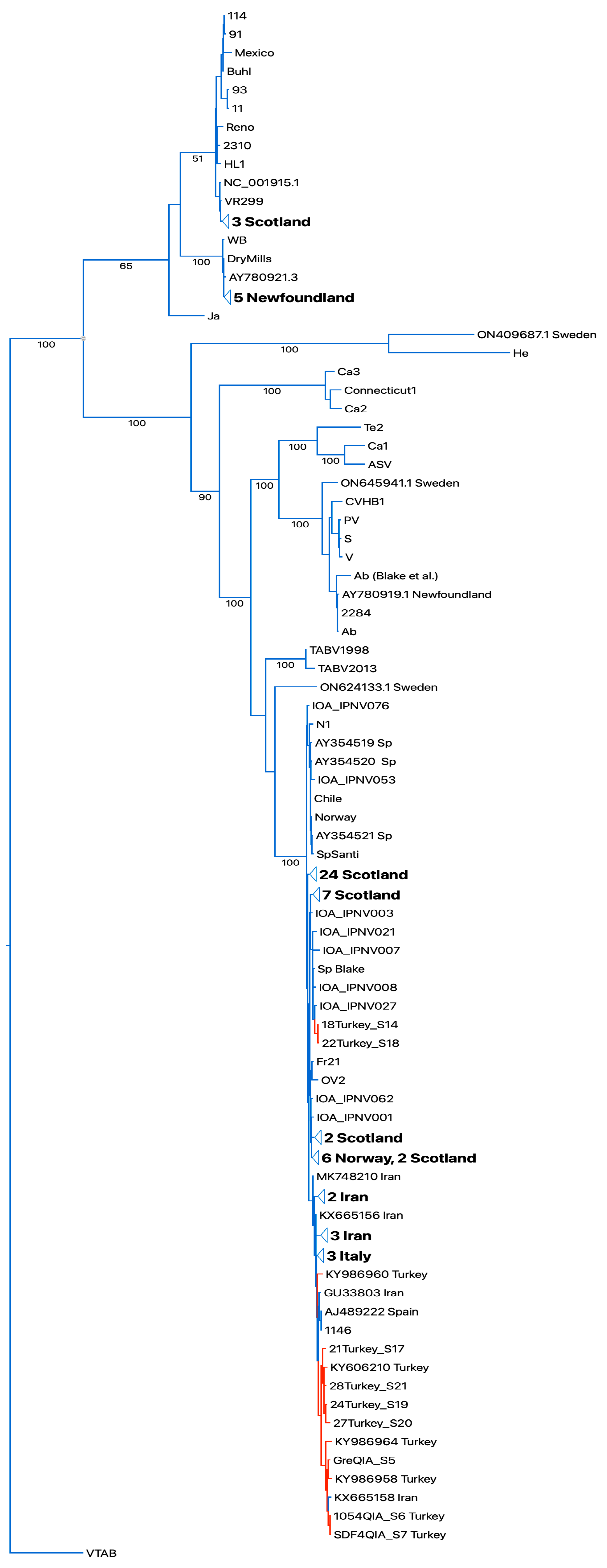
The segment A and segment B sequences were aligned with the respective available full segment IPNV sequences [16,22]. Both the Turkish IPNV isolate segment A and segment B sequences grouped in IPNV genogroup V (Sp serotype). For segment A, a set of two Turkish IPNV isolate sequences grouped with sequences of Scottish isolates in one subclade (segment A and B), and larger number grouped close to Spanish, Italian, and Iranian IPNV isolate sequences in a second independent subclade (segment A, Figure 3). Segment B, which codes for the RNA-dependent RNA polymerase, showed a similar grouping pattern (Figure S1).
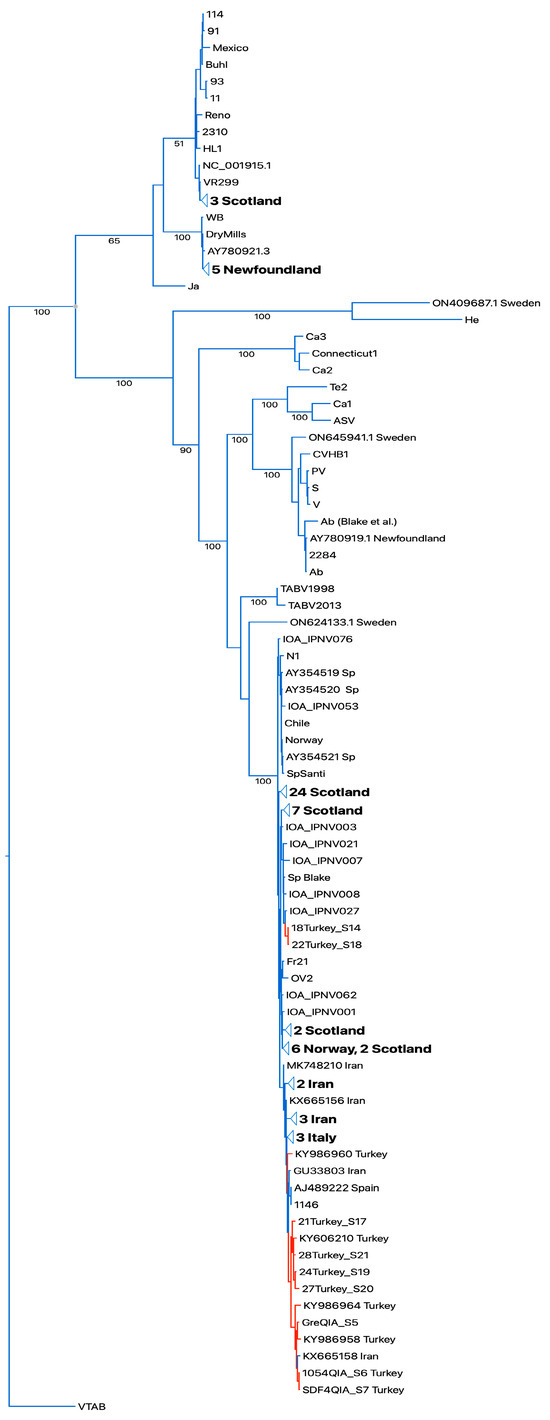
Figure 3.
Bootstrapped RAXML tree of whole-segment A sequences. Sequences of Turkish isolates highlighted in red. Designations of IPNV isolates as in [22]. Bootstrap values on tree branches. Turkish IPNV isolates group in the Sp serogroup clade. TABV: Tasmanian aquabirnaviruses. Outgroup Victorian trout aquabirnavirus (NC_030242.1).
Analysis of the extended pathogenicity locus of VP2 at amino acid positions 217, 221, and 247 [23] revealed that six sequences of isolates from 2005 to 2007 harbored the motif P217T221A247 (PTA), while three from 2010 to 2016 harbored the extended motif PTE P217T221E247 (PTE, Table 2).

Table 2.
Extended pathogenicity locus configurations of sequenced IPNV isolates.
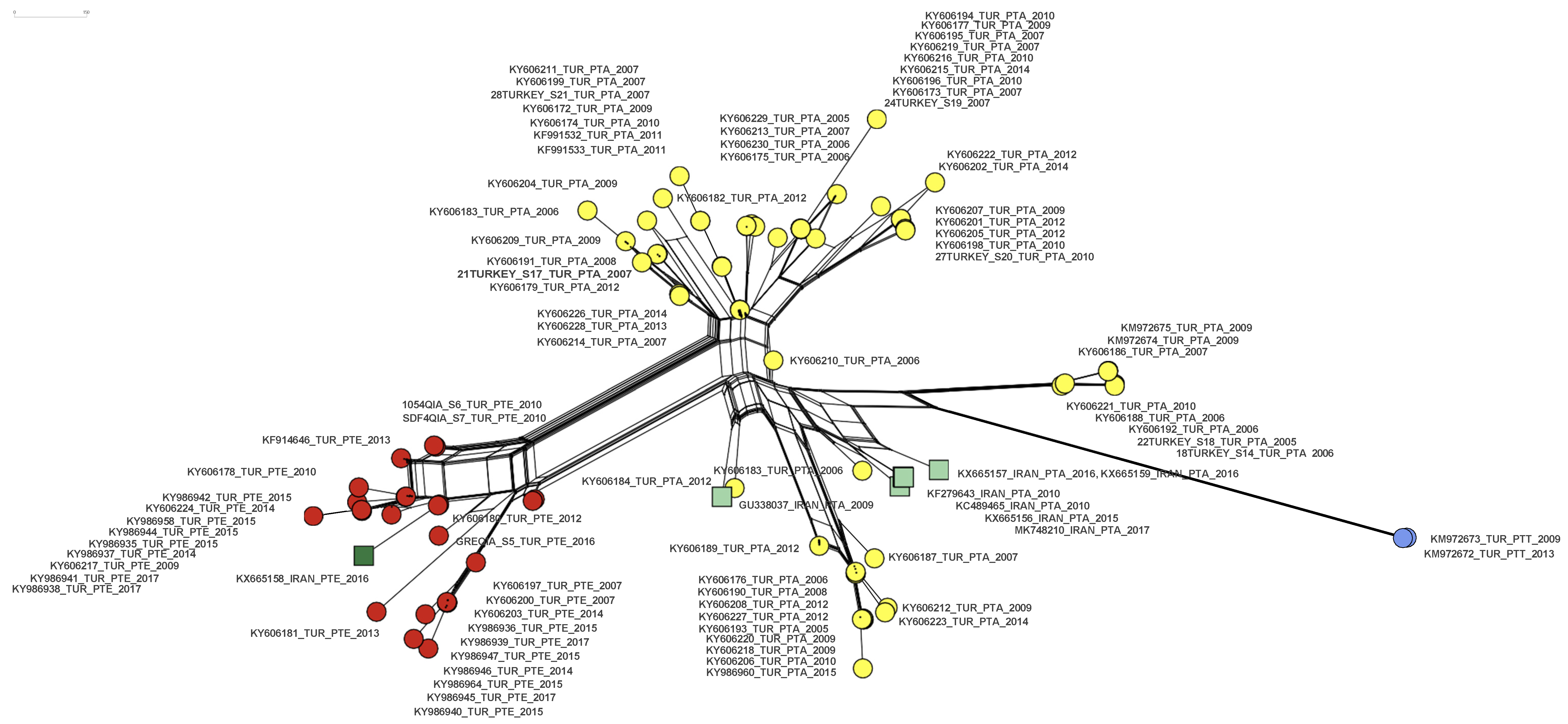
A network analysis of a partial IPNV VP2 gene segment of IPNV trout isolates from Turkey and Iran published so far indicated two distinct emergence events of the PTE motif in Turkey from 2007 to 2017, representing roughly 30% (27/91) of the sequences described. One of the events is connected to the emergence of the PTE motif in Iran. Additionally, a PTT motif emerged in Turkey in 2009 in 2/91 sequences (Figure 4).
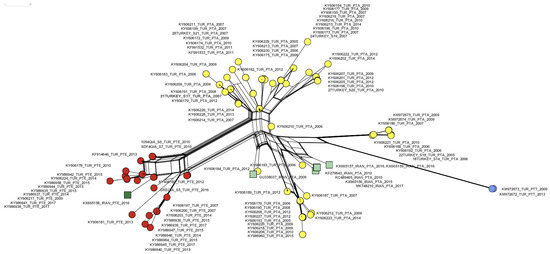
Figure 4.
A neighbor net of 99 partial IPNV VP2 nucleotide set (405 characters from position 620 to 1025 (amino acids 170–303, respectively) of 91 isolates from Turkey ([15,16], this study), and of 7 isolates from Iran [24] (Table S1). Extended motif PTA: yellow circles (Turkey), light green squares (Iran). Extended motif PTE: red circles (Turkey), dark green square (Iran). Extended motif PTT: blue circles (Turkey).
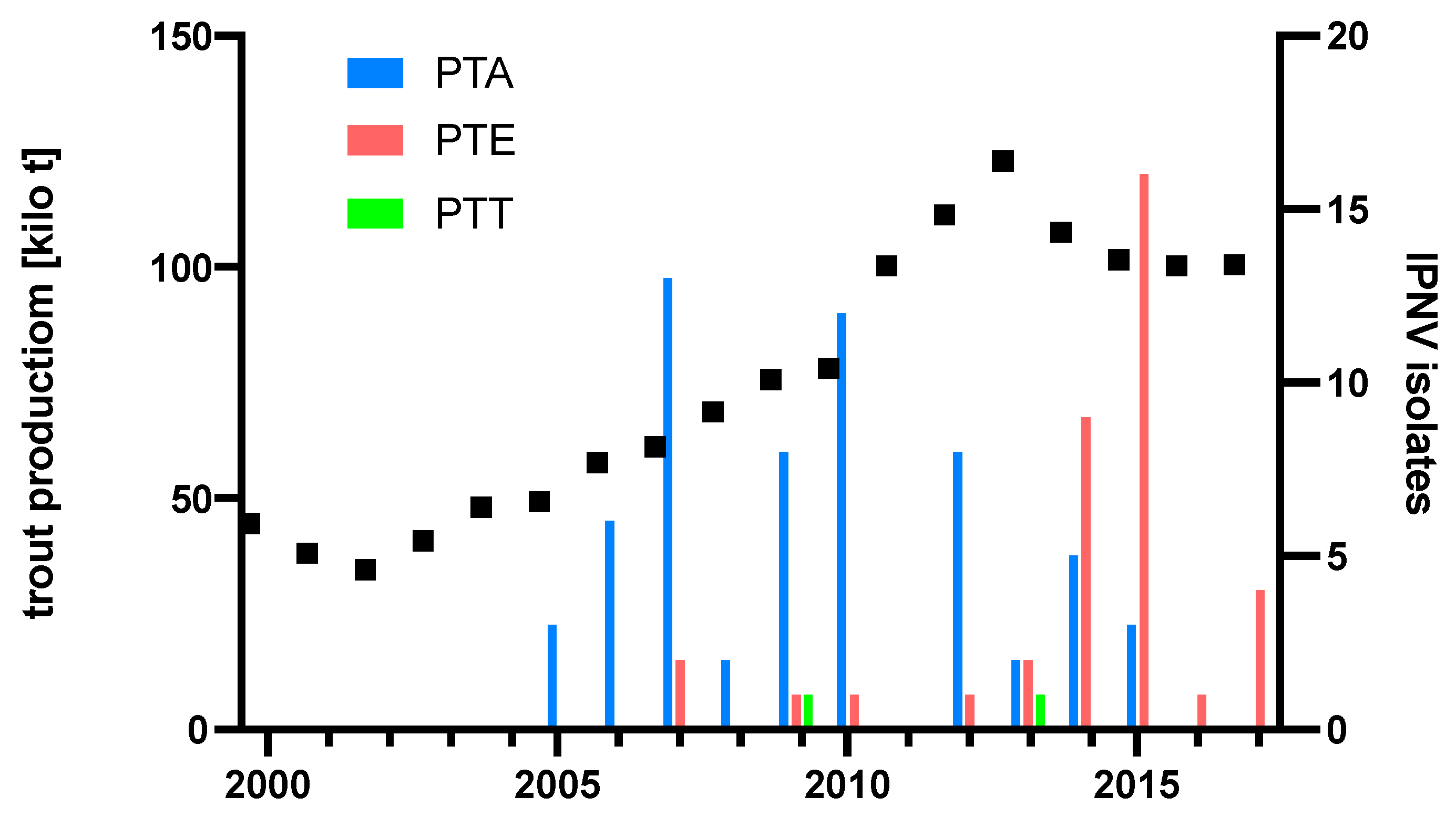
To understand if the emergence of the PTE over time might have been influenced by rainbow trout production levels, diagnostic IPNV isolates and their respective PTA and PTE motives were plotted over time, and rainbow trout production tonnage in Turkey was added. The resulting plot indicates the displacement of PTA isolates by PTE isolates from 2014, preceded by a production peak in 2013 (Figure 5).
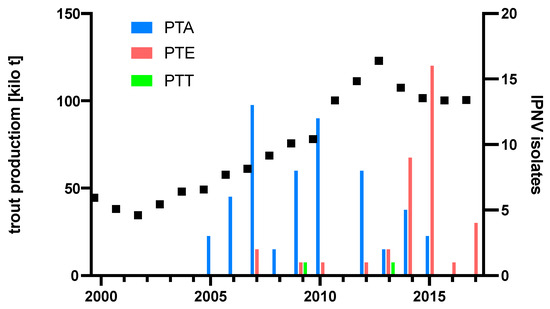
Figure 5.
Production of rainbow trout in Turkey and pathogenicity type of diagnostic IPNV isolates. Production figures [25]. Total 101 isolates: 62 from [15], 30 from [16], and 9 from Table 1.
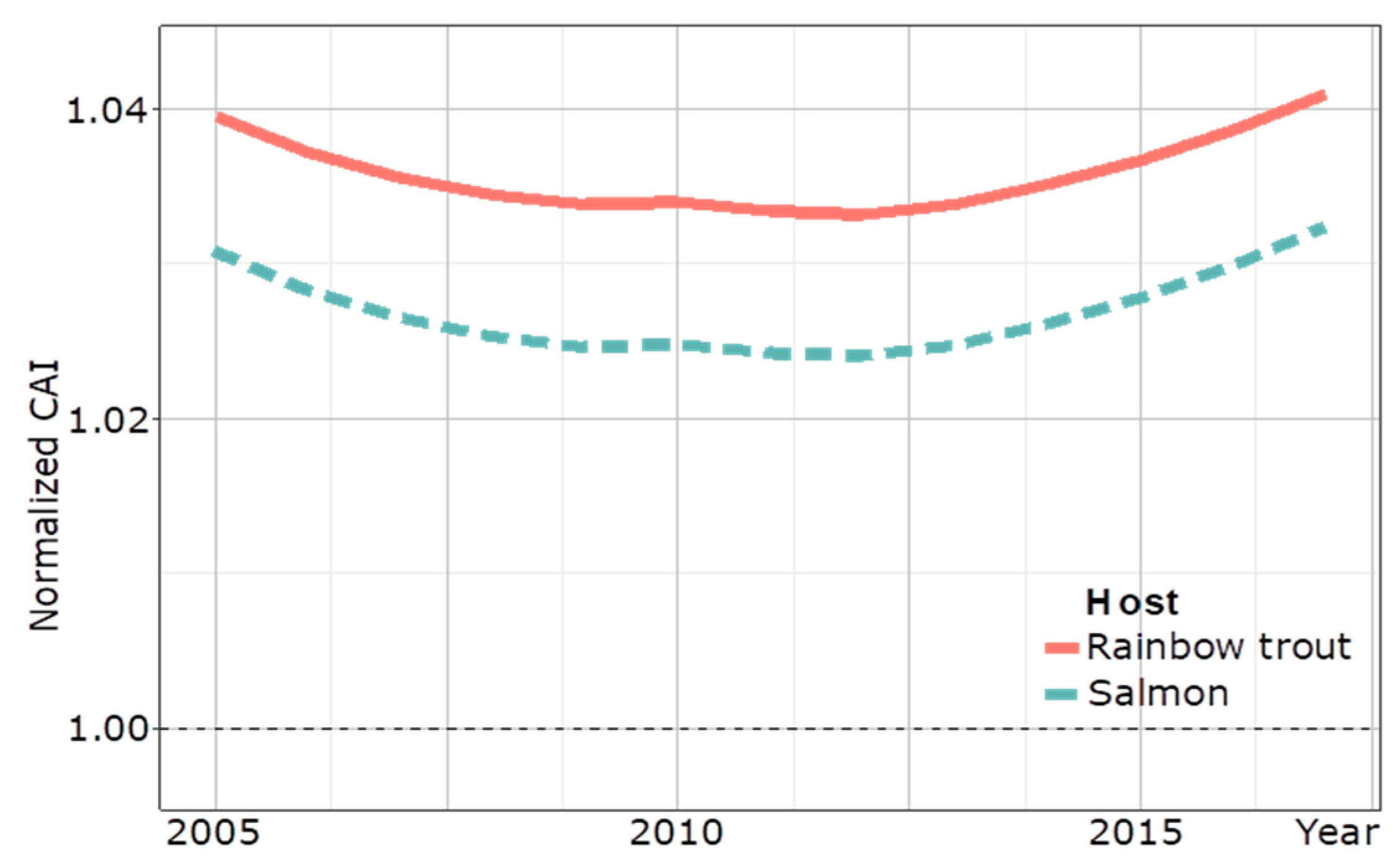
To assess the extent of the adaptation of IPNV to the host over time, a codon adaptation index (CAI) of a partial VP2 amino acid sequence of the Turkish IPNV trout isolates calculated a surge of the CAI towards the trout genome trending to >1.04 between 2013 and 2017, whereas a decreasing trend of adaptation was seen before 2013 (Figure 6).
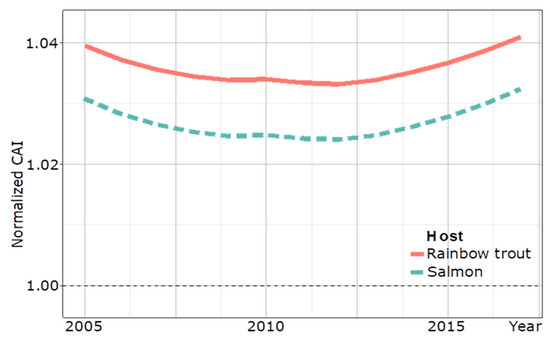
Figure 6.
Temporal CAI analysis of 89 partial VP2 amino acidic sequences (169 amino acids) of IPNV isolates from Turkey to the host genome of rainbow trout from 2005 to 2017. The CAI curve for salmon is a control. A dotted line at 1.00 marks the delineation between evidence for adapted and un-adapted host-specific codon usage.
4. Discussion
Surveillance evidence from farmed and wild fish and cell culture work described the impact of amino acid changes at an identified pathogenicity locus in VP2 within the hypervariable region [26]. By using reverse-engineered infectious IPNV clones in infection experiments, it was eventually shown that the IPNV clones switched from non-virulent T217 T221 to high-virulent T217 A221 when the hosts were subjected to stress [27].
The idea that the pathogenicity locus P217 and A221 linked to virulence might be extended by A247 (PTA) was reported in field studies from various salmonids in northern Europe, which linked PTA variously to subclinical infection, or to low and high virulence in salmon [9].
Turkey and Iran reported the PTA motif from IPNV isolates of farmed rainbow trout. Given the fact of the vertical transmission of IPNV via fertilized trout eggs [28], an inadvertent importation of IPNV carrying the PTA motif is obvious, and IPNV carrying this motif has spread widely in Turkey and Iran, as suggested by the interleaved clades of the phylogenetic trees of IPNV isolate sequences from Europe, Turkey, and Iran [15,16,24] (Figure 3 and Figure S1). IPNV isolates with the PTA motif from Turkey have been experimentally shown to induce moderate virulence in rainbow trout [29].
This study focused on nine additional IPNV isolates from Turkey and complemented the existing IPNV sequence dataset now allowing to describe the gradual emergence of the PTE motif in IPNV isolates from 2007 to 2017. This is evident from the network analysis which suggests two independent transmission chains inside Turkey after the PTE motif evolved in 2007 (Figure 4, red circles). It additionally corroborates that IPNV carrying the PTE motif was most likely introduced to Iran from Turkey (Figure 4), as suggested earlier [24].
A switch from PTA to PTE occurred just after a peak in trout production in 2013 as the PTE motif dominated the diagnostic IPNV isolates from 2014 onward (Figure 5). This is reminiscent of the switch of IPNV serotype Ab to Sp, which occurred during the massive expansion phase of the Scottish salmon industry in the 1990s, concomitant with a significant increase in IPNV VP2 CAI to >1.05 [20]. A CAI analysis of a partial amino acid VP2 sequence of IPNV isolates from Turkey indicated that VP2 codon adaptation to rainbow trout in Turkey may only have started to surge after this production peak in 2013 to a CAI >1.04 by 2017, and it is to be expected that the analysis of future isolates would confirm this trend for the full-length VP2 sequence. Although the nCAI of IPNV isolates from Turkey suggests host-specific codon adaption since 2005, the initial decrease in nCAI likely resulted from other evolutionary factors, such as modulating gene expression to evade immune surveillance [30]. Notably, it is of concern that only the most recent PTE-carrying IPNV isolates of this study induced gross pathology; however, experimental evidence for the pathogenicity of the PTE motif carrying IPNV isolates has not yet been reported. Further characterization of IPNV–host dynamics is needed to help elucidate relationships between genotypes and phenotypes.
A recent analysis of IPNV isolated from diseased, vaccinated, and resistant freshwater salmon in Chile described eight configurations at VP2 positions 217, 221, and 247, which, however, do not include the PTE motif. All of these isolates showed evidence of pathogenicity in gross pathology, histopathology, and cell culture [31]. Our analysis additionally identified two IPNV isolates from a turbot and a rainbow trout carrying yet another motif (PTT), which apparently is rare but points to the fact that IPNV infecting seawater and freshwater salmonids can adapt the hypervariable region in the VP2 subject to influences in various natural and farmed environments. Evidently, IPNV is continually evolving in the rainbow trout culture setting in Turkey.
Turkey’s fish product exports and imports are worth USD 1.3 billion and USD 200,000 annually. In 2022, for example, 144,347 tons of freshwater rainbow trout and 45,454 tons of seawater rainbow trout were produced in Turkey. Thanks to the increasing demand, rainbow trout are exported to Europe and Russia [32]. Since 2007, IPN has no longer been a notifiable disease in Turkey, and, unfortunately, the requests for IPNV isolation and analysis have subsequently dropped significantly, and rainbow trout breeders now only request tests for fish diseases listed by the WOAH for fish exportation (such as VHSV, IHNV, and ISAV).
In light of the evidence presented here, the expansion of the rainbow trout industry since the described time window (until 2017) may have continued to foster conditions for the evolution of the VP2 pathogenicity locus; however, the absence of adequate surveillance will most likely impact on safeguarding fish health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v16060994/s1, Figure S1: Bootstrapped RAXML tree of whole-segment B IPNV sequences. Sequences of Turkish isolates highlighted in red. Designations of IPNV isolates as in [24]. TABV: Tasmanian aquabirnaviruses. Outgroup Victorian trout aquabirnavirus (NC_030244.1). Table S1: Sequences used for network analysis.
Author Contributions
Conceptualization, M.W. and H.A.; methodology, M.W.; software, N.D.P. and M.W.; validation, M.W.; formal analysis, N.D.P. and M.W.; investigation, C.T., K.U., H.N.K. and H.A.; writing—original draft preparation, C.T., H.A. and M.W.; writing—review and editing, C.T., H.A. and M.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was conducted under the protocols and procedures following the national legislative rules and ethical standards under validation order by the Samsun Veterinary Control Institute Scientific Ethics Committee, Ministry of Agriculture and Forestry, Republic of Turkey. Code:1653. Date: 21 October 2013.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequence data or data used to construct the maps are available from the corresponding authors upon reasonable request. Representative sequences have been submitted to the NCBI GenBank database and will be released on 14 November 2024.
Acknowledgments
We would like to thank Gülnur Kalaycı, Buket Özkan (18 Turkey S14, 21 Turkey S17, 22 Turkey S18, 24 turkey S19, 27 Turkey S20, 28 Turkey S21), and Hakan Işıdan (Sdf-4) who provided seven isolates. We would like to thank Stefanie Wehner and Michael Bekaert who compiled the sequencing data.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- ICTV. International Committee on Taxonomy of Viruses. Available online: https://ictv.global/taxonomy (accessed on 3 March 2024).
- Santi, N.; Song, H.; Vakharia, V.N.; Evensen, Ø. Infectious pancreatic necrosis virus VP5 is dispensable for virulence and persistence. J. Virol. 2005, 79, 9206–9216. [Google Scholar] [CrossRef] [PubMed]
- Blake, S.; Ma, J.Y.; Caporale, D.A.; Jairath, S.; Nicholson, B.L. Phylogenetic relationships of aquatic birnaviruses based on deduced amino acid sequences of genome segment A cDNA. Dis. Aquat. Organ. 2001, 45, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Lauksund, S.; Greiner-Tollersrud, L.; Chang, C.J.; Robertsen, B. Infectious pancreatic necrosis virus proteins VP2, VP3, VP4 and VP5 antagonize IFNa1 promoter activation while VP1 induces IFNa1. Virus Res. 2015, 196, 113–121. [Google Scholar] [CrossRef][Green Version]
- Pedersen, T.; Skjesol, A.; Jørgensen, J.B. VP3, a structural protein of infectious pancreatic necrosis virus, interacts with RNA-dependent RNA polymerase VP1 and with double-stranded RNA. J. Virol. 2007, 81, 6652–6663. [Google Scholar] [CrossRef] [PubMed]
- Dadar, M.; Memari, H.R.; Vakharia, V.N.; Peyghan, R.; Shapouri, M.S.; Mohammadian, T.; Hasanzadeh, R.; Ghasemi, M. Protective and immunogenic effects of Escherichia coli-expressed infectious pancreatic necrosis virus (IPNV) VP2-VP3 fusion protein in rainbow trout. Fish Shellfish Immunol. 2015, 47, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Hill, B.J.; Way, K. Serological classification of infectious pancreatic necrosis (IPN) virus and other aquatic birnaviruses. Annu. Rev. Fish Dis. 1995, 5, 55–77. [Google Scholar] [CrossRef]
- Mutoloki, S.; Evensen, Ø. Sequence similarities of the capsid gene of Chilean and European isolates of infectious pancreatic necrosis virus point towards a common origin. J. Gen. Virol. 2011, 92, 1721–1726. [Google Scholar] [CrossRef]
- Dopazo, C.P. The Infectious Pancreatic Necrosis Virus (IPNV) and its Virulence Determinants: What is Known and What Should be Known. Pathogens 2020, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Benkaroun, J.; Muir, K.F.; Allshire, R.; Tamer, C.; Weidmann, M. Isolation of a New Infectious Pancreatic Necrosis Virus (IPNV) Variant from a Fish Farm in Scotland. Viruses 2021, 13, 385. [Google Scholar] [CrossRef]
- Dadar, M.; Peyghan, R.; Memari, H.R.; Shapouri, M.R.; Hasanzadeh, R.; Goudarzi, L.M.; Vakharia, V.N. Sequence analysis of infectious pancreatic necrosis virus isolated from Iranian reared rainbow trout (Oncorhynchus mykiss) in 2012. Virus Genes 2013, 47, 574–578. [Google Scholar] [CrossRef]
- Glenney, G.W.; Barbash, P.A.; Coll, J.A.; Quartz, W.M. Isolation and molecular characterization of a novel infectious pancreatic necrosis virus strain in returning Atlantic salmon Salmo salar from the Connecticut River, USA. J. Aquat. Anim. 2012, 24, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Lago, M.; Bandín, I.; Olveira, J.G.; Dopazo, C.P. In vitro reassortment between Infectious Pancreatic Necrosis Virus (IPNV) strains: The mechanisms involved and its effect on virulence. Virology 2017, 501, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Candan, A. First report on the diagnosis of infectious pancreatic necrosis (IPN) based on reverse transcription polymerase chain reaction (RT-PCR) in Turkey. Bull. Eur. Assoc. Fish Pathol. 2002, 22, 45–47. [Google Scholar]
- Tamer, C.; Isıdan, H.; Kalaycı, G.; Ozan, E.; Ozkan, B.; Albayrak, H. Determination of VP2 sequence-based virulence motifs and phylogenetic analysis of domestic Turkish IPNV ısolates. J. Fish Dis. 2022, 45, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Büyükekiz, A.G.; Altun, S.; Hansen, E.F.; Satıcıoğlu, I.B.; Duman, M.; Markussen, T.; Rimstad, E. Infectious pancreatic necrosis virus (IPNV) serotype Sp is prevalent in Turkish rainbow trout farms. J. Fish Dis. 2018, 41, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Durmaz, Y.; Albayrak, H. Whole genome molecular characterization of Infectious Pancreatic Necrosis Viruses isolated in Turkey. Ank. Üniv. Vet. Fakültesi Derg. 2019, 67, 1–10. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Puigbo, P.; Bravo, I.G.; Garcia-Vallvé, S. E-CAI: A novel server to estimate an expected value of Codon Adaptation Index (eCAI). BMC Bioinform. 2008, 9, 65. [Google Scholar] [CrossRef]
- Ulrich, K.; Wehner, S.; Bekaert, M.; Di Paola, N.; Dilcher, M.; Muir, K.F.; Taggart, J.B.; Matejusova, I.; Weidmann, M. Molecular epidemiological study on Infectious Pancreatic Necrosis Virus isolates from aquafarms in Scotland over three decades. J. Gen. Virol. 2018, 99, 1567–1581. [Google Scholar] [CrossRef] [PubMed]
- Mutoloki, S.; Jøssund, T.B.; Ritchie, G.; Munang’andu, H.M.; Evensen, Ø. Infectious Pancreatic Necrosis Virus Causing Clinical and Subclinical Infections in Atlantic Salmon Have Different Genetic Fingerprints. Front. Microbiol. 2016, 7, 1393. [Google Scholar] [CrossRef] [PubMed]
- Ahmadivand, S.; Weidmann, M.; El-Matbouli, M.; Rahmati-Holasoo, H. Low Pathogenic Strain of Infectious Pancreatic Necrosis Virus (IPNV) Associated with Recent Outbreaks in Iranian Trout Farms. Pathogens 2020, 9, 782. [Google Scholar] [CrossRef]
- Feap, Federation of European Aquaculture Producers. Available online: https://feap.info/ (accessed on 5 May 2024).
- Santi, N.; Vakharia, V.N.; Evensen, Ø. Identification of putative motifs involved in the virulence of infectious pancreatic necrosis virus. Virology 2004, 322, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Gadan, K.; Sandtrø, A.; Marjara, I.S.; Santi, N.; Munang’andu, H.M.; Evensen, Ø. Stress-induced reversion to virulence of infectious pancreatic necrosis virus in naïve fry of Atlantic salmon (Salmo salar L.). PLoS ONE 2013, 8, e54656. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K. Fish Viruses and Fish Viral Diseases; Cornell University Press: Ithaca, NY, USA, 1988; p. 476. [Google Scholar]
- Tamer, C.; Durmaz, Y.; Ozan, E.; Kadi, H.; Cavunt, A.; Muftuoglu, B.; Albayrak, H. Pathogenicity trials regarding Turkish isolates of infectious pancreatic necrosis virus and viral haemorrhagic septicaemia virus in rainbow trout. Aquac. Res. 2021, 52, 1395–1400. [Google Scholar] [CrossRef]
- Shin, Y.C.; Bischof, G.F.; Lauer, W.A.; Desrosiers, R.C. Importance of codon usage for the temporal regulation of viral gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 14030–14035. [Google Scholar] [CrossRef]
- Godoy, M.; Kibenge, M.J.T.; Montes de Oca, M.; Pontigo, J.P.; Coca, Y.; Caro, D.; Kusch, K.; Suarez, R.; Burbulis, I.; Kibenge, F.S.B. Isolation of a New Infectious Pancreatic Necrosis Virus (IPNV) Variant from Genetically Resistant Farmed Atlantic Salmon (Salmo salar) during 2021–2022. Pathogens 2022, 11, 1368. [Google Scholar] [CrossRef]
- Turkish Statistical Institute (TurkStat). Available online: https://data.tuik.gov.tr/Bulten/Index?p=Su-Urunleri-2022-49678 (accessed on 12 March 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).