Abstract
After the first phase of the COVID-19 pandemic in Europe, a new highly pathogenic variant of echovirus 11 (E11) was detected. The aim of this study was to analyze the genetic diversity of Polish E11 environmental and clinical strains circulating between 2017 and 2023 as well as compare them with E11 strains isolated from severe neonatal sepsis cases reported in Europe between 2022 and 2023. Additionally, the study explores the effectiveness of environmental monitoring in tracking the spread of new variants. For this purpose, the complete sequences of the VP1 capsid protein gene were determined for 266 E11 strains isolated in Poland from 2017 to 2023, and phylogenetic analysis was performed. In the years 2017–2023, a significant increase in the detection of E11 strains was observed in both environmental and clinical samples in Poland. The Polish E11 strains represented three different genotypes, C3, D5 and E, and were characterized by a high diversity. In Poland, the intensive circulation of the new variant E11, responsible for severe neonatal infections with a high mortality in Europe, was detected in the years 2022–2023. This investigation demonstrates the important role of environmental surveillance in the tracking of enteroviruses circulation, especially in settings with limited clinical surveillance.
1. Introduction
Echovirus 11 (E11) is one of the most common enteroviruses detected in both clinical and environmental samples worldwide [1,2,3,4,5,6]. It belongs to the B species of the Enterovirus genus of the Picornaviridae family. Though E11 is known mainly for mild or asymptomatic infections, it is responsible for severe syndromes, characterized by high rates of morbidity and mortality [7]. The wide range of diseases encompass aseptic meningitis, encephalitis, acute flaccid paralysis (AFP), upper respiratory tract infections, hand, foot and mouth disease (HFMD), acute gastroenteritis, myocarditis and uveitis [8,9,10,11,12]. In particular, this virus can cause severe systemic infection in newborns, including acute hepatitis with coagulopathy, which is one of the most fatal complications of severe neonatal enterovirus infections [13,14]. As with other enteroviruses, E11 has the ability to cross the placenta and infect the fetus, leading to heart disease and fetal death [15,16,17]. People with primary immunodeficiency are also at risk of chronic infection caused by E11 [18]. This virus has an epidemic pattern of circulation and has been frequently identified as the causal agent of outbreaks that occur irregularly and often last for several years [1]. Similarly, E11 has been associated with nosocomial infections and numerous outbreaks in neonatal nurseries [19,20,21].
The infection outbreaks and silent circulation within the population can be tracked through environmental surveillance. Testing sewage samples allows for the detection of a wider spectrum of virus variants circulating in the population compared to those isolated from patients. Specifically, raw sewage contains viruses shed from both asymptomatic and symptomatic individuals. Moreover, it was proven that the pathogens detected in sewage were highly correlated with clinical cases and disease outbreaks [5,22,23].
The World Health Organization (WHO) and the European Center for Disease Prevention and Control (ECDC) have reported an increasing number of severe neonatal infections associated with E11 in 2022–2023 [24,25,26,27,28]. E11 was found in neonatal patients with severe sepsis, complicated by liver failure and neurological or myocardial involvement. The lethality rate of severe neonatal E11 infection was very high. Consequently, a new variant of E11 was reported from France, Croatia, Italy, Spain, Sweden and the United Kingdom of Great Britain and Northern Ireland [29,30].
Mutation and recombination play crucial roles in the formation of new genetic variants of E11 with novel biological properties associated with changes in tissue tropism, the evasion of host immunity, increased virulence and pathogenesis and the emergence of disease outbreaks. VP1 is the most immunodominant structural protein of E11, and antigenic variants are selected in the presence of antibodies to it. Molecular methods based on the amplification and sequencing of the VP1 coding region (1D) are used for the molecular characterization of strains and epidemiological studies. E11 strains circulating worldwide are classified into six genotypes (A–F), and genotypes A, C, and D are divided into A1–5, C1–4 and D1–5 [31,32].
The aim of this study was to examine the range of genetic variation within the Polish environmental and clinical E11 strains circulating between 2017 and 2023 and to compare them with E11 strains isolated from severe cases of neonatal sepsis reported in 2022 and 2023 in Europe. The phylogenetic relationship with other worldwide circulating strains was also analyzed. For this purpose, the complete sequences of the VP1 capsid protein gene were determined for E11 strains isolated in Poland from 2017 to 2023. Furthermore, this study addresses the utility of environmental surveillance for tracking the circulation of new variants of viruses.
2. Materials and Methods
2.1. Viruses
In total, 266 isolates of E11 were obtained from the collection of the National Institute of Public Health, NIH-NRI in Poland. Two hundred fifty-five of them were isolated from sewage samples collected in 2017–2023, and eleven were isolated from clinical cases (aseptic meningitis, neonatal sepsis) in 2018–2019 and 2022–2023. The environmental strains were isolated from sewage samples collected in five Polish cities at different time intervals: Warsaw (2017–2023, one to eight collection sites), Lublin (2022–2023, one to eight collection sites), Rzeszow (2022–2023, one collection site), Krakow (2023, one collection site), and Gdansk (2023, one collection site). The frequency of sampling varied during the study period, with collections occurring once a week from July 2017 to March 2020, once a quarter in 2021, and twice a month in 2022–2023. Sewage samples were concentrated according to the protocol described earlier [33] and inoculated onto two cell lines, L20B and RD.
The isolates of E11 were propagated in RD (rhabdomyosarcoma) cell cultures according to the instructions described in Polio Laboratory Manual, 4th edition, 2004, World Health Organization [34]. RD cells were maintained in MEM (Minimum Essential Medium) supplemented with 10% FBS (Fetal Bovine Serum).
2.2. RNA Isolation and RT-PCR
Viral RNA was extracted from the cell culture supernatant using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The complete VP1 coding region was amplified by reverse transcription PCR using Superscript III (Invitrogen, Carlsbad, CA, USA), specific primers and PCR cycling times and temperature, as previously described [35]. The amplified products were analyzed in 1.5% agarose gels, GelRed-stained and examined under a UV DNA transilluminator.
2.3. Sequencing and Sequence Analysis
The PCR products were processed in a cycle sequencing reaction with BigDye 3.1 (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. The product of the sequencing reaction was run in an automated genetic analyzer (Applied Biosystems, Foster City, CA, USA, model 3730). The resulting sequences were manually edited using BioEdit version 7.1.9 and examined in terms of the closest homologue sequences using BLAST software, version BLAST+ 2.15.0 (http://www.ncbi.nlm.nih.gov/BLAST/, accessed on 1 March 2024). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version X [36]. The sequences of isolated strains were aligned with the reference strains and a selection of sequences from worldwide strains. In total, 344 VP1 sequences were included in the phylogenetic analysis, including the 266 sequences generated in this study. A phylogenetic tree was computed using the neighbor-joining method with a bootstrap of 1000 replicates. All sequences obtained in this study were submitted to GenBank and were assigned accession numbers from PP534556 to PP534821.
In total, 150 VP1 sequences were included in the haplotype analysis, along with the 132 Polish sequences from 2018 to 2023, 17 European sequences from 2022 to 2023 and 1 sequence from the USA (2022). Median-joining network analysis was performed using PopART (Population Analysis with Reticulate Trees) software version 1.7 based on multisequence aligned haplotype data and traits data representing the geographical distribution [37].
3. Results
3.1. E11 Detection during 2017–2023
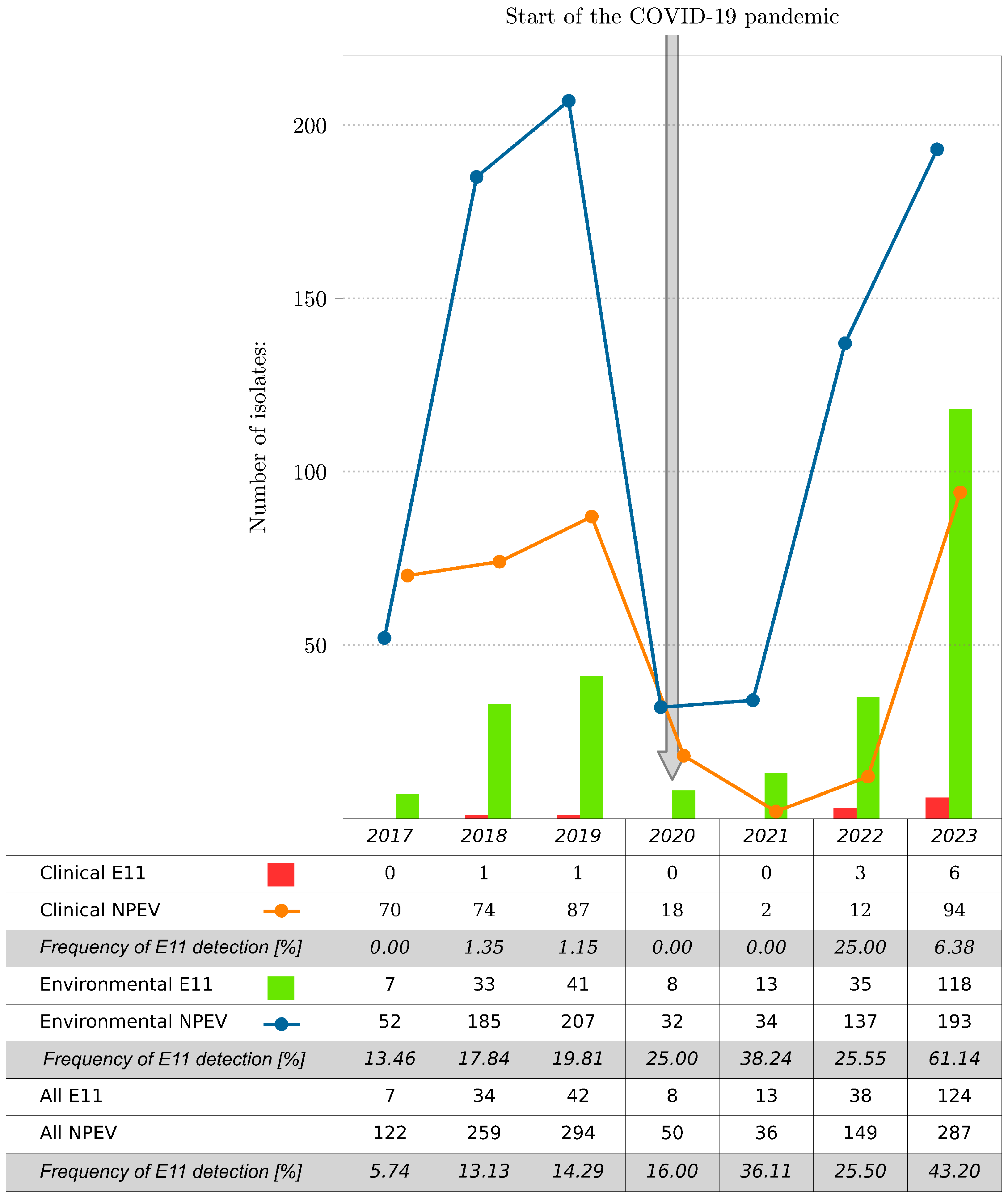
In total, 1197 NPEV isolates were collected at the National Institute of Public Health, NIH-NRI (NIPH NIH-NRI) in Poland, including 266 E11 isolates (266/1197; 22.22%), during 2017–2023. Most of the E11 came from environmental samples. The environmental strains of E11 accounted for 30.36% of the total NPEV isolated from sewage samples (255/840); the clinical strains of E11 represented only 3.08% of the total NPEV isolated from clinical samples (11/357). E11 was detected during each of the reported years. The number of isolated E11 isolates varied in particular years (Figure 1). The lowest number of E11 isolates, both clinical and environmental, was observed in 2017, and the highest was observed in 2023. The number of E11 isolates increased significantly from 2017 (n = 7) to 2023 (n = 124), representing 5.74% and 43.20% of the total number of isolates detected for the respective years. NPEV isolations, including E11, during the COVID-19 pandemic, especially during 2020 and 2021, were considerably lower than those after and before the pandemic. E11 was not detected in clinical samples during 2020 and 2021 at NIPH NIH-NRI, suggesting that a reduced transmission of E11 occurred during the intense phases of non-pharmaceutical interventions and social mitigations of COVID-19 in Poland.
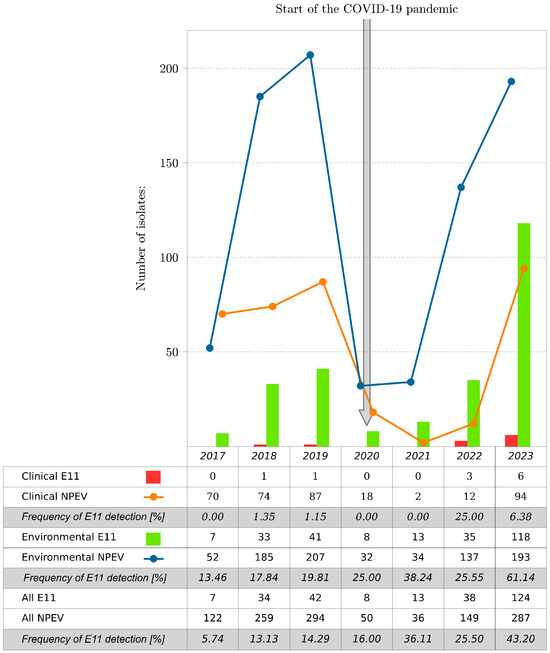
Figure 1.
Number of E11 and all NPEV isolates by year, Poland, 2017–2023. The frequency of E11 detection is shown as the percentage of E11 to NPEV per year.
3.2. Geographic and Temporal Distribution of E11 Strains
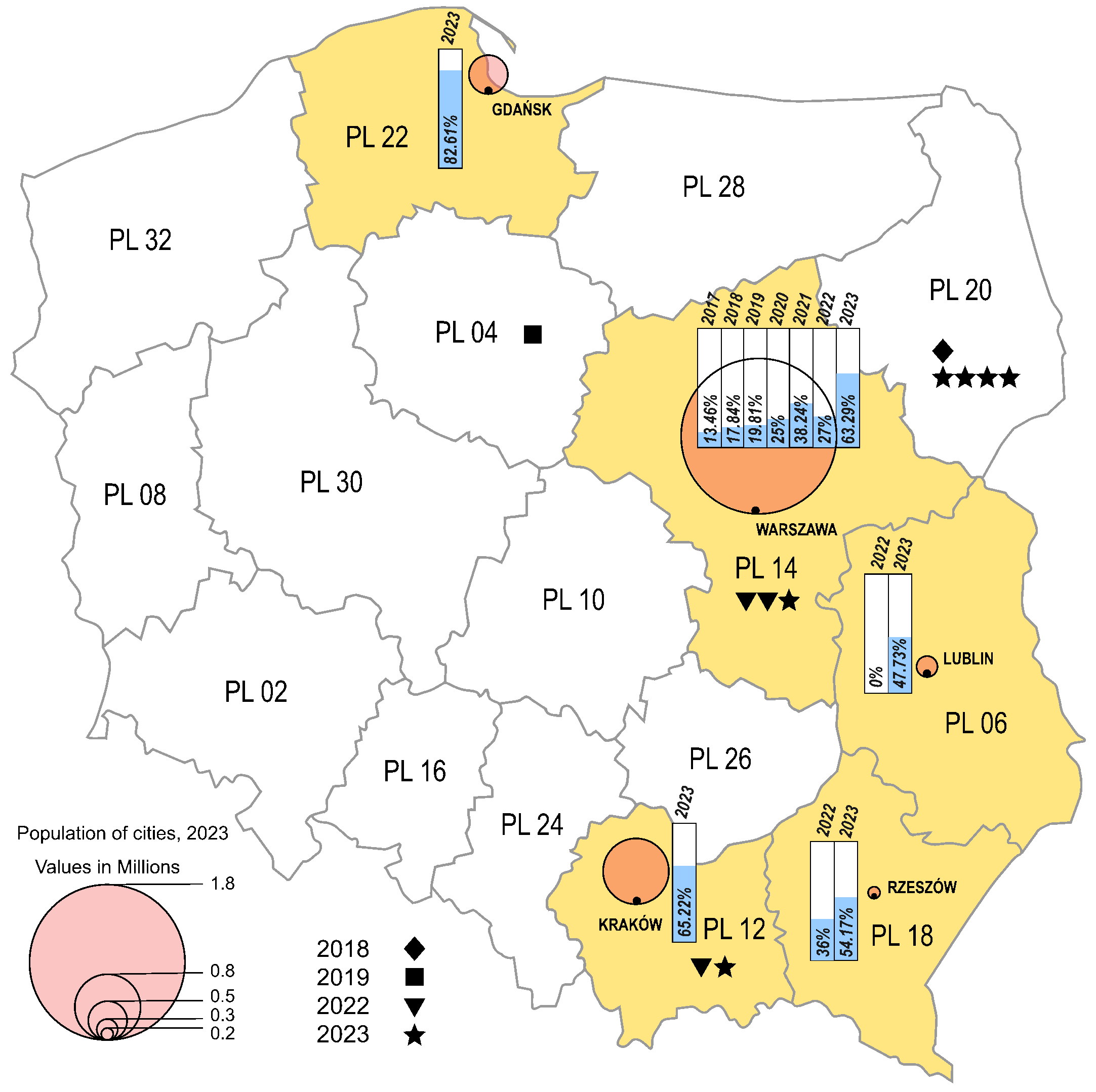
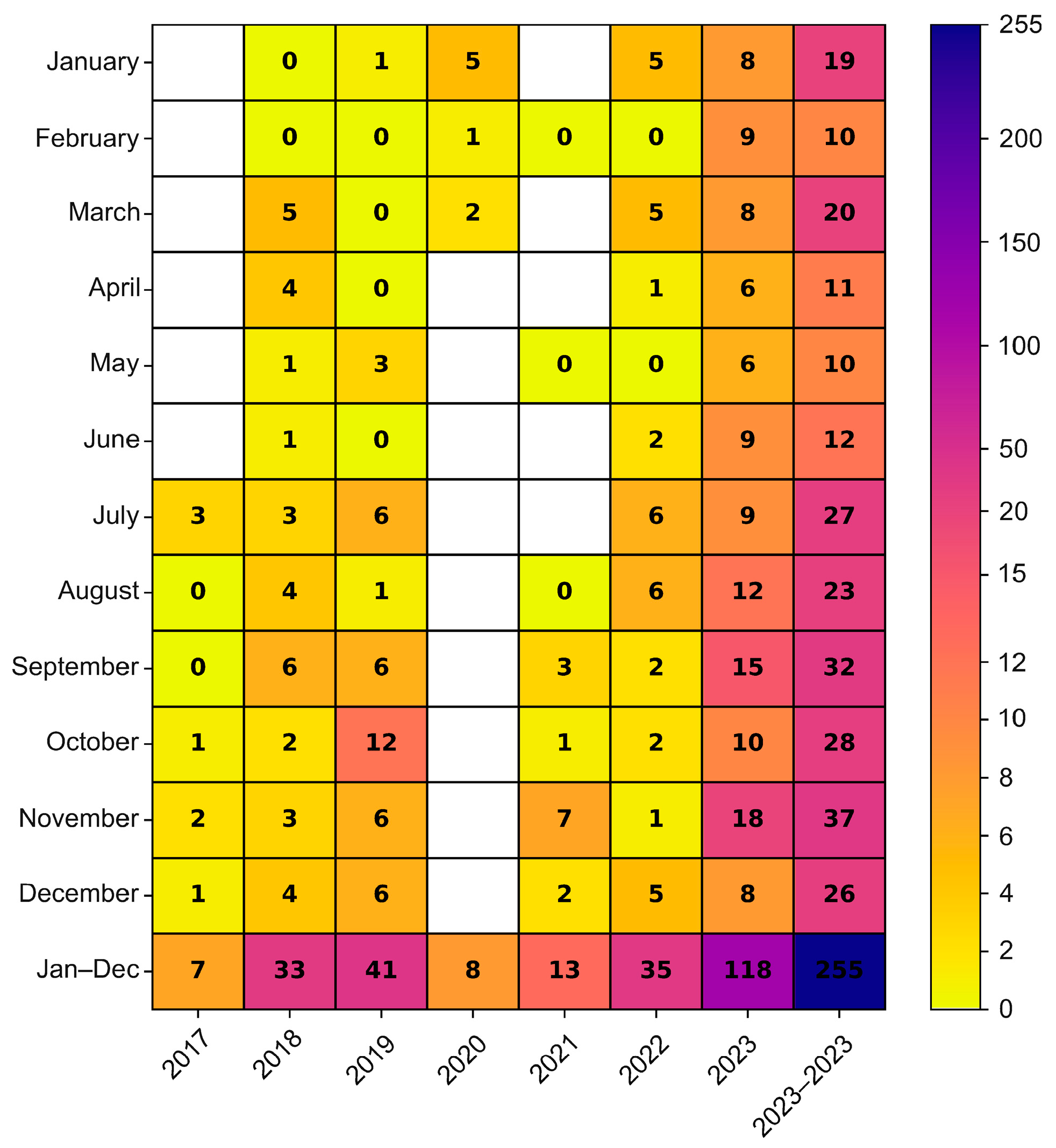
The environmental strains were isolated from sewage samples collected in five Polish cities, in different time intervals (Warsaw 2017–2023, Lublin 2022–2023, Rzeszow 2022–2023, Krakow 2023, Gdansk 2023), while the clinical isolates came from four different regions from 2018 to 2019 and from 2022 to 2023 (Masovian Voivodeship, Lesser Poland Voivodeship, Kuyavian-Pomeranian Voivodeship, Podlaskie Voivodeship) (Figure 2). In the cities where environmental samples were collected, an increasing trend of E11 detection was observed in subsequent years of the study. In 2023, environmental E11 isolates represented more than 40%, while in 2017–2022, they represented less than 40% of the total number of NPEV detected in a given year at each collection site. In the individual sampling cities, E11 constituted 0 (Lublin, 2022) to 82.61% (Gdansk, 2023) of the total NPEV obtained. The monthly distribution of environmental E11 is shown in Figure 3. E11 isolates were frequently isolated from environmental samples in 2023 and obtained throughout the year. In the years 2017–2023, E11 was most often isolated in the summer and autumn months (from July to December), peaking in November (Figure 3).
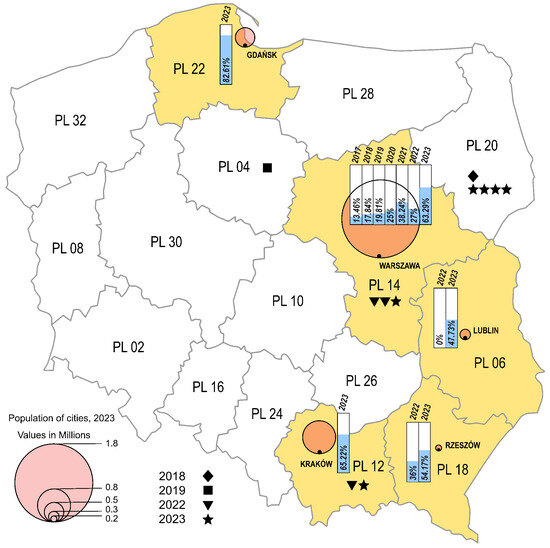
Figure 2.
Frequency of E11 detection in Polish regions, 2017–2023. The voivodeships in which environmental surveillance was carried out are marked in yellow. Each city monitored by environmental surveillance is represented by a circle, with the size proportional to the population of the city (Warsaw—1.8 million; Krakow—0.8 million; Gdansk—0.5 million; Lublin—0.3 million; Rzeszów—0.2 million). Symbols (star, triangle, square, diamond) mark cases of E11 infections. The percentage of environmental E11-positive samples is shown in the bar charts.
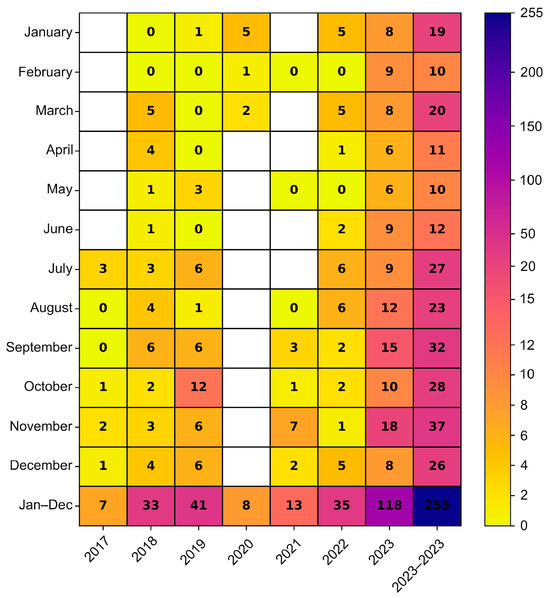
Figure 3.
Monthly distribution of E11 isolated from environmental samples in Poland, 2017–2023. The number of environmental Polish E11 isolates detected by months was given in cells.
3.3. Clinical Characteristics of E11 Cases
Between 2017 and 2023, 11 E11 clinical isolates were identified in NIPH NIH-NRI (Table 1). Two isolates were obtained in 2018–2019, and the remaining nine were obtained in 2022–2023. Of all the clinical isolates from 2023, more than half were associated with infection in infants and toddlers. The isolates were generally obtained from patients with symptoms of aseptic meningitis. Most of the mentioned cases had a mild course, except one case of systemic infection with hepatitis. Cerebrospinal fluid (CSF) examination showed pleocytosis with white blood cells (WBC) ranging from 7 to 263 cells/µL in Polish E11 infection cases. Female patients (6/11, 54.55%) and infants <3 months of age (6/11, 54.55%) represented more than half of the reported cases.

Table 1.
Clinical characteristics of E11 cases, Poland, 2018–2023.
3.4. Phylogenetic Analysis of E11 Strains
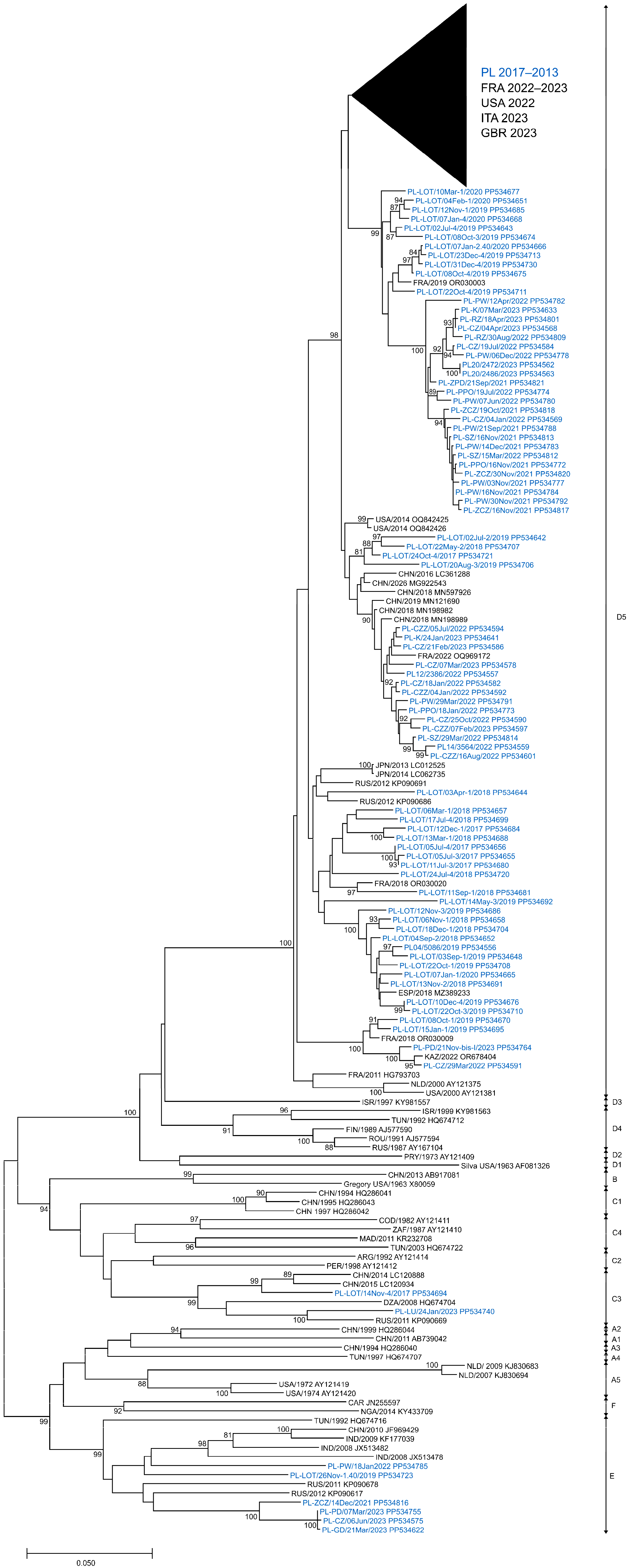
In this analysis, a total of 266 E11 isolates derived from clinical and environmental samples, collected between 2017 and 2023 in Poland, were entered. To evaluate the phylogenetic relationships among circulating E11 in Poland, full-length VP1 sequences (876 bp) of 266 isolated strains were investigated. The presented phylogenetic tree (Figure 4) was constructed based on 282 sequences, with 204 generated in this study. Sequences that were identical or highly similar were not included. The study isolates were grouped into three different genotypes, C3, D5 and E (Figure 4), according to the nomenclature proposed by Oberste et al. and Li et al. [31,32]. All clinical isolates (n = 11) and most environmental isolates (n = 244) were clustered in genotype D5. Nine environmental isolates from 2019 and 2021–2023 were placed in genotype E together with isolates from Eurasia and Africa (1992–2012). Two environmental isolates from 2017 and 2023 were placed in genotype C3 along with isolates from China (2014–2015), Russia (2011) and Algeria (2008). Sequences of clinical isolates from 2022 to 2023 were grouped together with those from France and Italy from 2022 to 2023.
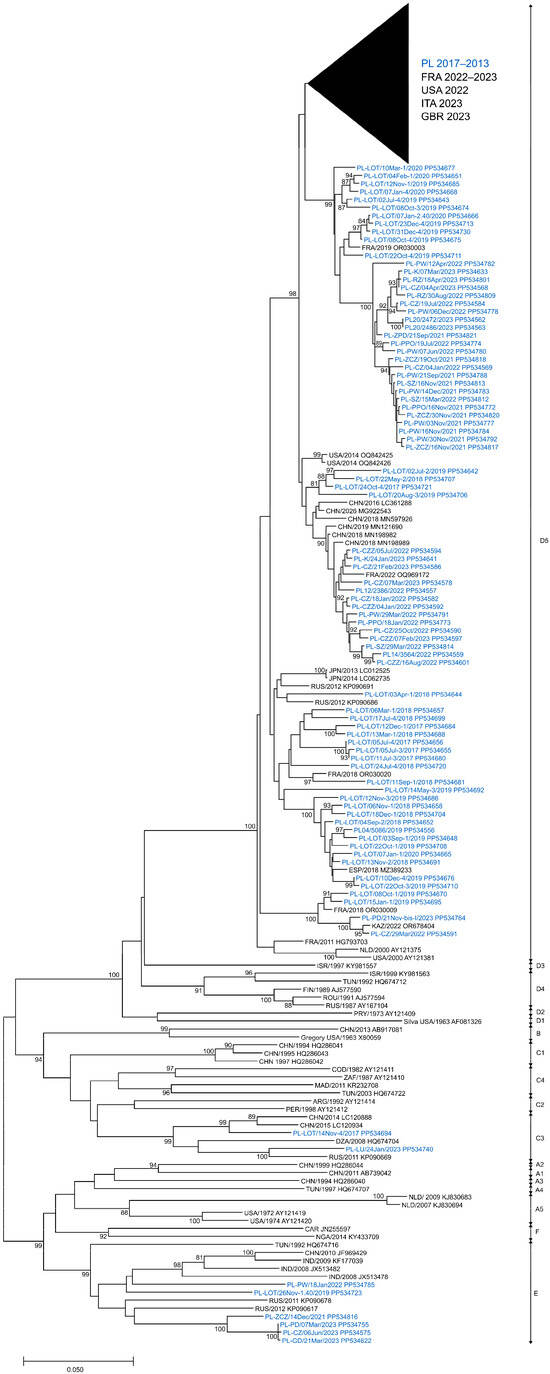
Figure 4.
Phylogenetic tree of echovirus 11 complete 1D sequences from clinical and environmental samples in Poland (2017–2023) and other countries (1963–2023). The tree was constructed by the neighbor-joining method and evaluated with 1000 bootstrap pseudoreplicates. Only bootstrap values ≥ 80% are indicated. Genetic distances were calculated with the Kimura 2-parameter algorithm. Analyses were conducted in MEGA version X. Sequence identifiers consist of a country abbreviation, a year of detection and an accession number.
In general, nucleotide sequence divergence in pairwise comparisons between Polish E11 isolates ranged from 0.0% to 25.7% (0.0–12.7% aa divergence) (Table 2) and depended on the year of detection. In 2020, it varied from 0.0% to 8.3% (0.0–2.7% aa divergence) and in 2023, it varied from 0.0% to 25.3% (0.0–12.3% aa divergence) when the three genotypes were detected. The viruses of E showed the highest nucleotide divergence (0.0–15.1%), and D5 showed the smallest (0–9.9%). Furthermore, the nucleotide diversity of environmental isolates was significantly higher than that of clinical E11. Polish E11 strains showed 75.6–81.2% nucleotide and 87.3–94.2% amino acid similarity with the prototype strain—Gregory.

Table 2.
Nucleotide and amino acid divergence of E11 isolates detected in Poland (2017–2023), based on the full-length VP1 sequence analysis.
The complete VP1 coding sequence of E11 consisted of 292 aa, and 69.18% of aa sites (202/292) were conserved between the Polish strains. A significant number of aa substitutions were observed; 90 sites of 292 aa residues had been changed between isolates. In most polymorphic sites, amino acid substitutions were not associated with clustering (49/90, 54.44%). Forty-one sites showed specific aa conservation for genotypes, and most of them were located in the amino and carboxyterminal region of the VP1 protein or in loops (Table 3). The conservation of aa signatures specific to these genotypes was incomplete, showing a return to the original state at locations 23, 84 and 292 in the C3 genotype and at locations 84, 245, 271 and 280 in the E genotype.

Table 3.
Amino acid consensus of VP1 in Polish E11; strains of differential genotypes were compared with the prototype strain—Gregory.
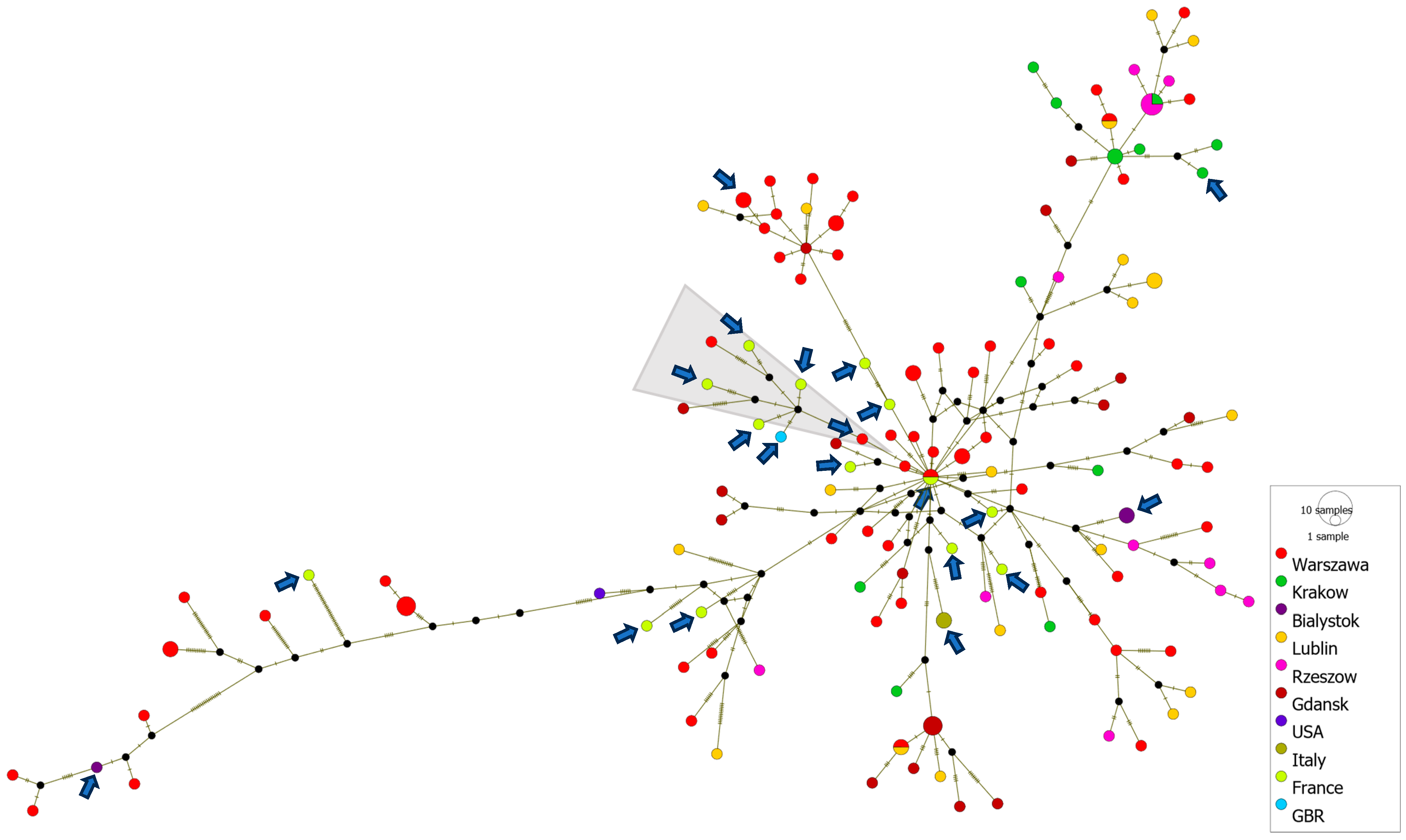
The generation of haplotype networks is a widely used methodology for analyzing and visualizing the relationships between sequences from different geographic destinations. A cluster of severe neonatal cases associated with a new variant of E-11, which contains, among others, sequences from France (2022–2023), Italy (2023) and Poland (2017–2023), was analyzed using PopArt software version 1.7. A total of 150 D5 genotype sequences were subjected to the median-joining haplotype network analysis. The analysis showed a close relationship of Polish environmental and clinical isolates with clinical cases from France, Italy and Great Britain, associated with severe cases of neonatal infections in Europe. The haplotype analysis also revealed that most of the strains from France were separated by as few as 0–6 nucleotide substitutions in the VP1 gene from Polish environmental or clinical strains. The French clinical isolate from 2022 shares an identical VP1 gene sequence with the Polish environmental isolate from the same year. Additionally, the Polish clinical strain (PL14/4786/2022, PP534560), isolated from a severe case of hepatitis and multisystemic disease, clusters with clinical strains from France and Great Britain associated with multiorgan failure, neonatal sepsis and sudden infant death, as well as with Polish environmental strains from Gdansk and Warsaw (depicted by a grey triangle in Figure 5).
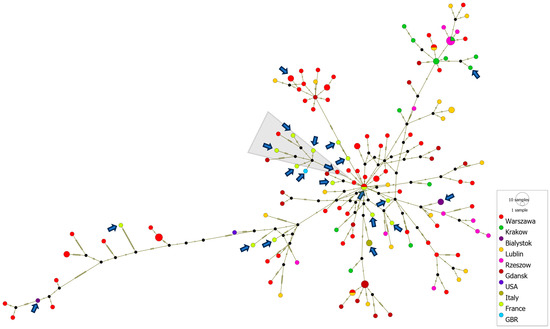
Figure 5.
Median-joining haplotype network of 150 D5 genotype sequences constructed in PopArt. The analysis is based on entire VP1 gene sequences (876 nt) from clinical and environmental isolates collected in Poland from 2017 to 2023, as well as from other countries during 2022–2023. Each circle represents a unique haplotype, and its size reflects the number of strains expressing that haplotype. Crosshatches indicate the number of nucleotide differences between haplotypes. Color codes denote the geographic location of the strains. Clinical isolates are marked with arrows. The group of strains associated with severe E11 infections is marked with a gray triangle.
4. Discussion
In this study, we conducted a genetic comparison of E11 strains isolated from clinical samples with environmental E11 strains collected in Poland from 2017 to 2023. This research was prompted by the emergence of severe neonatal E11 infections with high mortality rates reported by the World Health Organization (WHO) and the European Center for Disease Prevention and Control (ECDC) in Europe during 2022–2023. Between 2022 and July 2023, WHO and ECDC reported 21 cases of severe sepsis caused by E11 infection, which were complicated by liver failure and neurological or myocardial involvement. These cases occurred in countries including France, Croatia, Italy, Spain, Sweden and the United Kingdom of Great Britain and Northern Ireland [24,25,26,27,28,29,30] and were related to the emergence of a new variant of E11. Our analysis revealed the intensive circulation of the same variant in Poland between 2022 and 2023. Both Polish environmental and clinical isolates were closely related to strains from France, Italy and Great Britain. The virus collection from the Department of Virology, NIPH NHI-NRI, comprised nine clinical isolates of E11 from 2022 to 2023, with only one isolate associated with a severe case of sepsis accompanied by hepatitis. Over half of these E11 infections (58%) were reported in infants under the age of three months old. This reiterates the previous observations that E11 infections were most commonly notified in children younger than 3 months old [2]. Furthermore, the number of E11 clinical isolates was significantly higher compared to those of the years before the COVID-19 pandemic (2017–2019; n = 2). Numerous authors are addressing the spread of viruses in the midst of and following the COVID-19 pandemic. The reduction in virus circulation during the COVID-19 pandemic is believed to have affected the severity of new infections due to the prolonged absence of ongoing natural exposure to viruses. Lower levels of population immunity, especially among younger children, may result in a greater prevalence of disease and potentially more severe infection when the virus circulation resumes. Research conducted in the USA showed a significant disruption in seasonal EV detections during the early phases of the COVID-19 pandemic and a rapid increase in detections during the summer and fall of 2022 [38]. Similar disturbances in EV circulation have been observed in the Netherlands during and after lockdown [39].
Generally, E11 is one of the most commonly reported kinds of NPEV in the world. Between 1970 and 2005, in the USA, E11 represented 11.4% of all reported isolates from clinical samples [1]. A similar situation occurred in China, where E11 accounted for 11.2% of all the detected NPEV from cerebrospinal fluid samples during 2018–2019 [3,40]. In Europe, in turn, E11 was among the ten most frequently reported types, representing 4% of all typed EV positive samples between 2015 and 2017 [2]. However, due to the fact that most enterovirus infections are generally asymptomatic or mildly symptomatic, it makes it difficult to trace infections through clinical surveillance. Therefore, environmental surveillance is a more effective method of detecting viruses that circulate silently.
Environmental studies reveal more clearly that E11 is among the most frequently detected enteroviruses across the world. Chinese environmental reports indicated the high frequency isolation of E11 from sewage samples in different time periods (26.4%—242/916, 2009–2012 [4], 26.3%—336/1279, 2013–2021 [5]). E11 was also among the most frequently isolated enteroviruses in environmental surveillance, for example, in the USA during 1994–2002 [41], in Russia during 2004–2017 [42], in India during 2007–2009 [43], in Italy during 2009–2015 [44] and in Japan during 2013–2016 and 2019–2021 [45,46]. Our study revealed a significantly higher presence of E11 in the environmental samples in 2022–2023. E11 represented more than 60% of all environmental NPEV strains collected in 2023 and was detected throughout the year, indicating a change in the seasonal pattern of circulation. In our previous environmental studies conducted in 2011, E11 was also the most prevalent enterovirus (26%, 28/107); however, by 2023, it had surpassed its previous level of frequency [6]. The research carried out in Sicily also showed a sudden increase in the detection of E11 in 2023 [47], which may indicate an increase in the circulation of E11 throughout Europe.
Epidemics and outbreaks caused by E11 are often accompanied by severe cases with high mortality. The epidemic in Hungary in 1989 is an example, where 13 fatal cases of hemorrhagic syndromes were recorded among 386 children suffering from E11-associated disease (fever, vomiting, diarrhea) [48]. The National Enterovirus Surveillance System in the United States showed that E11 was the most commonly isolated EV from newborns during 1983–2003, representing 14% of neonatal EV infections, 19% of which were fatal [49]. This information should be related to the fact that in a similar time period (1970–2005), E11 was the second most frequently detected enterovirus in the USA [1]. The intense circulation of E11 in Poland and throughout Europe during 2022–2023 may explain the occurrence of severe cases in newborns in many countries in Europe. The lack of specific maternal enterovirus antibodies, especially E11, can lead to an increased number of severe cases and a higher fatality rate of neonatal enterovirus infections. Some studies suggest that the level of neutralizing antibodies to E11 in cord sera is lower compared to that of other serotypes and also lower than in maternal sera, which may determine the severity of E11 infection in newborns [17,50].
In this study, phylogenetic analysis showed that E11 isolates from 2017 to 2023 belong to three genotypes, C3, D5 and E. Polish isolates of genotypes C3 and E were found only in sewage, while D5 isolates were detected in both sewage and human samples. The D5 genotype was predominant in Poland during 2017–2023 and was consistently detected over a 7-year research period. The close relationship between the Polish D5 strains and European clinical strains suggests sustained circulation in the region during the study period. Previous studies reported the detection of D5 on four different continents, indicating the cosmopolitan nature of this genotype [32]. Severe cases of multisystemic infection in newborns in Europe during 2022–2023 were linked to a new variant of this genotype [29]. The emergence of a new enterovirus variant may stem from either the introduction of a new virus from a geographically distinct area or the genetic evolution of an endemic virus population. Phylogenetic analysis suggests that the new E11 variant emerged from an endemic circulating population through evolutionary processes.
Our analysis showed a high nucleotide diversity (up to 25.7%) between Polish E11 strains and a high number of amino acid substitutions in VP1. Phylogenetic data from previous studies have shown a greater diversity range of E11, with variations of up to 27.6% [32]. The high variety and low genetic stability reported for E11 may facilitate the emergence of new variants. It is worth noting that such high variation was revealed in our study through the analysis of environmental strains isolated from sewage samples. Notably, environmental surveillance allowed for the detection of two genotypes (C3 and E) in Poland. The fact that those genotypes were not isolated from clinical cases suggests silent circulation in the Polish population. It could be stated that only a small amount of E11 circulating causes detectable symptomatic infections. Interestingly, the Chinese researchers also described the circulation of several E11 genotypes at the same time, albeit causing only sporadic cases [31]. The study from Russia showed that certain genotypes of E11 were more prevalent in sewage, compared to other genotypes like D5, which were isolated from both sewage and human samples [51].
There are certain limitations to the study. The primary issue is the insufficient data regarding the number of E11 cases in Poland following the initial phase of the COVID-19 outbreak. Due to the absence of clinical surveillance for NPEV in Poland, sporadic cases, including potentially severe ones, of E11 may have gone unnoticed. Environmental surveillance was not conducted with the same frequency throughout the study period, especially during 2020–2021, which was affected by the COVID-19 pandemic. This limitation could have led to an underestimated number of enterovirus isolates obtained. Moreover, environmental surveillance was primarily conducted in eastern Poland, potentially limiting the representativeness of the analyzed isolates for the entire country. Furthermore, the detection method, which involves isolation in cell cultures, can preferentially detect certain types of enteroviruses—for example, E11, which can multiply well in RD cells. Multiple studies confirmed that RD cells support more replication enteroviruses of species B and polioviruses compared to other species [52]. Other studies have reported that coxsackie B viruses are more frequently isolated from Hep-2 cells than from RD cells [40]. Therefore, it is possible that our research did not fully reflect the actual proportions of enteroviruses in the sewage. Moreover, the frequency of isolation of different serotypes depends on individual characteristics, such as different rates of multiplication or varying stability in sewage. This could result in a higher frequency of isolating certain enteroviruses, such as E11, compared to others. It should also be mentioned that the identification of enterovirus types has some limitations due to the methods and primers used [53]. Additionally, it is important to note that the phylogenetic analysis focused on a fragment encoding one viral protein rather than the entire genome, leaving it unclear whether recombination has occurred.
5. Conclusions
The research carried out on environmental samples indicated an intensive circulation of E11 in Poland in 2022–2023. Taking into account the close genetic relationship between isolates recovered in Poland and other European countries, we can conclude that the increase in infection events in Europe caused by E11, including the presence of severe cases, was related to the intensive circulation of the D5 genotype in the European population. Taking into account the scale of detection of E11 in environmental samples, the serious cases of infections reported in Europe probably constituted a small percentage of asymptomatic and mildly symptomatic infections.
This study contributes to a better understanding of the circulation patterns of E11 in humans and their possible implication in severe cases of multisystemic disease in neonates. Furthermore, the research conducted indicates the need for constant clinical enteroviral surveillance supplemented with environmental surveillance, which allows for a more complete assessment of the epidemiological situation.
Author Contributions
Conceptualization, M.W.; methodology, M.W., B.G., P.K., K.O., A.K. and K.T.; formal analysis, M.W., T.W. and K.T.; data curation, B.G., A.K., K.O. and P.K.; writing—original draft preparation, M.W.; writing—review and editing, B.G., K.T. and A.S.; visualization, T.W.; supervision, M.W.; funding acquisition, M.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute of Public Health NIH—National Research Institute, grant number 2BWBW/2021.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical review and approval were waived for the environmental samples used in the study, as these samples do not involve human or animal subjects and are thus exempt from the requirement for consent. Similarly, stool samples were collected and analyzed as part of routine surveillance for enteroviruses across various clinical sites and were therefore considered exempt from the requirement for consent. Additionally, ethical approval for the collection and use of clinical samples in Białystok was provided by the Bioethical Committee of the Medical University of Białystok (decision no. APK.002.186.2020, approved on 30 April 2020, and APK.002.423.2022, approved on 15 December 2022). Informed consent was obtained from all subjects recruited in Białystok.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Acknowledgments
We thank the employees of the municipal water and sewage companies from Warsaw, Krakow, Gdansk, Lublin and Rzeszow.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Khetsuriani, N.; Lamonte-Fowlkes, A.; Oberst, S.; Pallansch, M.A. Centers for Disease Control and Prevention. Enterovirus surveillance-United States, 1970–2005. MMWR Surveill. Summ. 2006, 55, 1–20. [Google Scholar] [PubMed]
- Bubba, L.; Broberg, E.K.; Jasir, A.; Simmonds, P.; Harvala, H. Enterovirus study collaborators. Circulation of non-polio enteroviruses in 24 EU and EEA countries between 2015 and 2017: A retrospective surveillance study. Lancet Infect. Dis. 2020, 20, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, M.; Xu, H.; Wang, T.; Liu, Y.; Yan, H.; Liu, P.; Qin, D.; Yang, Q. Analysis of enterovirus genotypes in the cerebrospinal fluid of children associated with aseptic meningitis in Liaocheng, China, from 2018 to 2019. BMC Infect. Dis. 2021, 21, 405. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Lu, J.; Zhang, Y.; Yoshida, H.; Guo, X.; Liu, L.; Li, H.; Zeng, H.; Fang, L.; Mo, Y.; et al. Prevalence of nonpolio enteroviruses in the sewage of Guangzhou city, China, from 2009 to 2012. Appl. Environ. Microbiol. 2013, 79, 7679–7683. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, S.; Zhang, Y.; Zhang, W.; Chen, M.; Li, C.; Guo, X.; Zhu, S.; Zeng, H.; Fang, L.; Ke, B.; et al. Prevalence of Non-Polio Enteroviruses in the Sewage of Guangzhou City, China, from 2013 to 2021. Microbiol. Spectr. 2023, 11, e0363222. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, M.; Ciąćka, A.; Witek, A.; Kuryk, Ł.; Żuk-Wasek, A. Environmental Surveillance of Non-polio Enteroviruses in Poland, 2011. Food Environ. Virol. 2015, 7, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.M.; Mulkey, S.B.; Campos, J.M.; DeBiasi, R.L. Laboratory diagnosis of CNS infections in children due to emerging and re-emerging neurotropic viruses. Pediatr. Res. 2024, 95, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S.; Forgie, S.; Robinson, J. Non-polio Enterovirus detection with acute flaccid paralysis: A systematic review. J. Med. Virol. 2018, 90, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Zhou, Y.; Zhu, Y.; Liu, Z.; Yang, F.; Yang, S.; Yu, Z.; Guo, C.; Ma, S. Molecular characterization of a new human echovirus 11 isolate associated with severe hand, foot and mouth disease in Yunnan, China, in 2010. Arch. Virol. 2015, 160, 2343–2347. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Lashkevich, V.A.; Koroleva, G.A.; Karganova, G.G. Phylogenetic and serological characterization of echovirus 11 and echovirus 19 strains causing uveitis. Arch. Virol. 2002, 147, 131–142. [Google Scholar] [CrossRef]
- Klein, J.O.; Lerner, A.M.; Finland, M. Acute gastroenteritis associated with ECHO virus, Type 11. Am. J. Med. Sci. 1960, 240, 749–753. [Google Scholar] [CrossRef]
- Szendrõi, A.; El-Sageyer, M.; Takács, M.; Mezey, I.; Berencsi, G. Nucleotide sequences and mutations of the 5′-nontranslated region (5′NTR) of natural isolates of an epidemic echovirus 11′ (prime). Arch. Virol. 2000, 145, 2575–2600. [Google Scholar] [CrossRef]
- Wang, C.; Yang, R.; Yang, F.; Han, Y.; Ren, Y.; Xiong, X.; Wang, X.; Bi, Y.; Li, L.; Qiu, Y.; et al. Echovirus 11 infection induces pyroptotic cell death by facilitating NLRP3 inflammasome activation. PLoS Pathog. 2022, 18, e1010787. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, H.; Tang, J.; He, Y.; Xiong, T.; Li, W.; Qu, Y.; Mu, D. Clinical characteristics of severe neonatal enterovirus infection: A systematic review. BMC Pediatr. 2021, 21, 127. [Google Scholar] [CrossRef] [PubMed]
- Tassin, M.; Martinovic, J.; Mirand, A.; Peigue-Lafeuille, H.; Picone, O.; Benachi, A.; Vauloup-Fellous, C. A case of congenital Echovirus 11 infection acquired early in pregnancy. J. Clin. Virol. 2014, 59, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.W.; Bendig, J.W.; Ossuetta, I. Vertical transmission of human echovirus 11 at the time of Bornholm disease in late pregnancy. Pediatr. Infect. Dis. J. 2005, 24, 88–89. [Google Scholar] [CrossRef]
- Hu, Y.L.; Lin, S.Y.; Lee, C.N.; Shih, J.C.; Cheng, A.L.; Chen, S.H.; Chang, L.Y.; Fang, C.T. Serostatus of echovirus 11, coxsackievirus B3 and enterovirus D68 in cord blood: The implication of severe newborn enterovirus infection. J. Microbiol. Immunol. Infect. 2023, 56, 766–771. [Google Scholar] [CrossRef]
- Laassri, M.; Zagorodnyaya, T.; Hassin-Baer, S.; Handsher, R.; Sofer, D.; Weil, M.; Karagiannis, K.; Simonyan, V.; Chumakov, K.; Shulman, L. Evolution of echovirus 11 in a chronically infected immunodeficient patient. PLoS Pathog. 2018, 14, e1006943. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.Y.; Chiu, C.H.; Huang, Y.C.; Chen, C.J.; Lien, R.; Chu, S.M.; Huang, C.G.; Tsao, K.C.; Shih, S.R.; Hsu, J.F. Investigation and successful control of an echovirus 11 outbreak in neonatal intensive care units. Pediatr. Neonatol. 2020, 61, 180–187. [Google Scholar] [CrossRef]
- Rueca, M.; Lanini, S.; Giombini, E.; Messina, F.; Castilletti, C.; Ippolito, G.; Capobianchi, M.R.; Valli, M.B. Detection of recombinant breakpoint in the genome of human enterovirus E11 strain associated with a fatal nosocomial outbreak. Virol. J. 2022, 19, 97. [Google Scholar] [CrossRef]
- Somekh, E.; Shohat, T.; Handsher, R.; Serour, F. An outbreak of echovirus 11 in a children’s home. Epidemiol. Infect. 2001, 126, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cui, N.; Wang, H.; Tao, Z.; Liu, Y.; Zhang, H.; Yoshida, H.; Song, Y.; Zhang, Y.; Song, L.; et al. Evaluating the prevalence and molecular epidemiology of echovirus 11 isolated from sewage in Shandong Province, China in 2010. Virus Genes 2012, 44, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Bubba, L.; Benschop, K.S.M.; Blomqvist, S.; Duizer, E.; Martin, J.; Shaw, A.G.; Bailly, J.L.; Rasmussen, L.D.; Baicus, A.; Fischer, T.K.; et al. Wastewater Surveillance in Europe for Non-Polio Enteroviruses and Beyond. Microorganisms 2023, 11, 2496. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Disease Outbreak News. Enterovirus-Echovirus 11 Infection in the European Region. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON474 (accessed on 7 July 2023).
- European Centre for Disease Prevention and Control. Epidemiological Update, Epidemiological Update: Echovirus 11 Infections in Neonates. Available online: https://www.ecdc.europa.eu/en/news-events/epidemiological-update-echovirus-11-infections-neonates (accessed on 19 July 2023).
- European Centre for Disease Control and Prevention. Communicable Disease Threat Report, Weekly Bulletin. 2023. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/communicable-disease-threats-report-16-jun-2023.pdf (accessed on 16 June 2023).
- World Health Organization. Disease Outbreak News. Enterovirus Infection—France 31 May 2023. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON469 (accessed on 31 May 2023).
- European Centre for Disease Control (ECDC). Echovirus 11 in Newly Born Twins: Case in Italy Shows Close Genetic Relation to Strains Found in France among Neonates. Available online: https://www.eurekalert.org/news-releases/992726 (accessed on 15 June 2023).
- Grapin, M.; Mirand, A.; Pinquier, D.; Basset, A.; Bendavid, M.; Bisseux, M.; Jeannoël, M.; Kireche, B.; Kossorotoff, M.; L’Honneur, A.S.; et al. Severe and fatal neonatal infections linked to a new variant of echovirus 11, France, July 2022 to April 2023. Eurosurveillance 2023, 28, 2300253. [Google Scholar] [CrossRef] [PubMed]
- Piralla, A.; Borghesi, A.; Di Comite, A.; Giardina, F.; Ferrari, G.; Zanette, S.; Figar, T.A.; Angelini, M.; Pisoni, C.; Pitrolo, A.M.G.; et al. Fulminant echovirus 11 hepatitis in male non-identical twins in northern Italy, April 2023. Eurosurveillance 2023, 28, 2300289. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yan, D.; Chen, L.; Zhang, Y.; Song, Y.; Zhu, S.; Ji, T.; Zhou, W.; Gan, F.; Wang, X.; et al. Multiple genotypes of Echovirus 11 circulated in mainland China between 1994 and 2017. Sci. Rep. 2019, 9, 10583. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Nix, W.A.; Kilpatrick, D.R.; Flemister, M.R.; Pallansch, M.A. Molecular epidemiology and type-specific detection of echovirus 11 isolates from the Americas, Europe, Africa, Australia, southern Asia and the Middle East. Virus Res. 2003, 91, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Zurbriggen, S.; Tobler, K.; Abril, C.; Diedrich, S.; Ackermann, M.; Pallansch, M.A.; Metzler, A. Isolation of sabin-like polioviruses from wastewater in a country using inactivated polio vaccine. Appl. Environ. Microbiol. 2008, 74, 5608–5614. [Google Scholar] [CrossRef] [PubMed]
- WHO Expanded Programme on Immunization & World Health Organization Division of Communicable Diseases. World Health Organization Manual for the Virological Investigation of Poliomyelitis. 1990. Available online: https://apps.who.int/iris/handle/10665/62186 (accessed on 3 June 2022).
- Leitch, E.C.; Harvala, H.; Robertson, I.; Ubillos, I.; Templeton, K.; Simmonds, P. Direct identification of human enterovirus serotypes in cerebrospinal fluid by amplification and sequencing of the VP1 region. J. Clin. Virol. 2009, 44, 119–124, Erratum in J. Clin. Virol. 2011, 51, 286. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full Feature Software for Haplotype Network Construction. Methods Evol. Ecol. 2015, 6, 1110–1116. Available online: https://popart.maths.otago.ac.nz/?s=PopART%3A+Full+Feature+Software+for+Haplotype+Network+Construction (accessed on 18 May 2020). [CrossRef]
- Lee, B.R.; Sasidharan, A.; Harrison, C.J.; Selvarangan, R. Disruption of seasonal enterovirus and parechovirus detections in the CSF and plasma of children during the COVID-19 pandemic. J. Clin. Virol. 2023, 160, 105381. [Google Scholar] [CrossRef] [PubMed]
- Forero, E.L.; Knoester, M.; Gard, L.; Ott, A.; Brandenburg, A.H.; McCall, M.B.B.; Niesters, H.G.M.; Van Leer-Buter, C. Changes in enterovirus epidemiology after easing of lockdown measures. J. Clin. Virol. 2023, 169, 105617. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cai, S.; Wu, X.; Zhang, Y.; Li, D.; Chen, Y.; Chen, Q.; Zhu, S.; Yan, D.; Xu, W.; et al. Analysis of the distribution characteristics of enterovirus types based on environmental surveillance from 2013 to 2021 in Fujian Province, China. Biosaf. Health 2023, 5, 240–249. [Google Scholar] [CrossRef]
- Sedmak, G.; Bina, D.; MacDonald, J. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from milwaukee, wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 2003, 69, 7181–7187. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, O.E.; Yarmolskaya, M.S.; Eremeeva, T.P.; Babkina, G.M.; Baykova, O.Y.; Akhmadishina, L.V.; Krasota, A.Y.; Kozlovskaya, L.I.; Lukashev, A.N. Environmental Surveillance for Poliovirus and Other Enteroviruses: Long-Term Experience in Moscow, Russian Federation, 2004–2017. Viruses 2019, 11, 424. [Google Scholar] [CrossRef]
- Tiwari, S.; Dhole, T.N. Assessment of enteroviruses from sewage water and clinical samples during eradication phase of polio in North India. Virol. J. 2018, 15, 157. [Google Scholar] [CrossRef]
- Delogu, R.; Battistone, A.; Buttinelli, G.; Fiore, S.; Fontana, S.; Amato, C.; Cristiano, K.; Gamper, S.; Simeoni, J.; Frate, R.; et al. Poliovirus and Other Enteroviruses from Environmental Surveillance in Italy, 2009–2015. Food Environ. Virol. 2018, 10, 333–342. [Google Scholar] [CrossRef]
- Ozawa, H.; Yoshida, H.; Usuku, S. Environmental Surveillance Can Dynamically Track Ecological Changes in Enteroviruses. Appl. Environ. Microbiol. 2019, 85, e01604-19. [Google Scholar] [CrossRef]
- Kitakawa, K.; Kitamura, K.; Yoshida, H. Monitoring Enteroviruses and SARS-CoV-2 in Wastewater Using the Polio Environmental Surveillance System in Japan. Appl. Environ. Microbiol. 2023, 89, e0185322. [Google Scholar] [CrossRef]
- Giammanco, G.M.; Filizzolo, C.; Pizzo, M.; Sanfilippo, G.L.; Cacioppo, F.; Bonura, F.; Fontana, S.; Buttinelli, G.; Stefanelli, P.; De Grazia, S. Detection of Echovirus 11 lineage 1 in wastewater samples in Sicily. Sci. Total Environ. 2024, 918, 170519. [Google Scholar] [CrossRef] [PubMed]
- El-Sageyer, M.M.; Szendröi, A.; Hütter, E.; Uj, M.; Szücs, G.; Mezey, I.; Tóth, I.; Kátai, A.; Kapiller, Z.; Páll, G.; et al. Characterisation of an echovirus type 11′ (prime) epidemic strain causing haemorrhagic syndrome in newborn babies in Hungary. Acta Virol. 1998, 42, 157–166. [Google Scholar] [PubMed]
- Khetsuriani, N.; Lamonte, A.; Oberste, M.S.; Pallansch, M. Neonatal enterovirus infections reported to the national enterovirus surveillance system in the United States, 1983–2003. Pediatr. Infect. Dis. J. 2006, 25, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.W.; Lukes, H.; Wells, B.; Thompson, L.; Low, D.E.; Cheang, M. Maternal and neonatal neutralizing antibody titers to selected enteroviruses. Pediatr. Infect. Dis. 1985, 4, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Yarmolskaya, M.S.; Shumilina, E.Y.; Ivanova, O.E.; Drexler, J.F.; Lukashev, A.N. Molecular epidemiology of echoviruses 11 and 30 in Russia: Different properties of genotypes within an enterovirus serotype. Infect. Genet. Evol. 2015, 30, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, M.; Sharif, S.; Klapsa, D.; Wilton, T.; Alam, M.M.; Fernandez-Garcia, M.D.; Rehman, L.; Mujtaba, G.; McAllister, G.; Harvala, H.; et al. Environmental Surveillance Reveals Complex Enterovirus Circulation Patterns in Human Populations. Open Forum Infect. Dis. 2018, 5, ofy250. [Google Scholar] [CrossRef]
- Liu, B. Universal PCR Primers Are Critical for Direct Sequencing-Based Enterovirus Genotyping. J. Clin. Microbiol. 2016, 55, 339–340. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).