Prevalence and Genetic Diversity of Bat Hepatitis B Viruses in Bat Species Living in Gabon
Abstract
1. Introduction
2. Materials and Methods
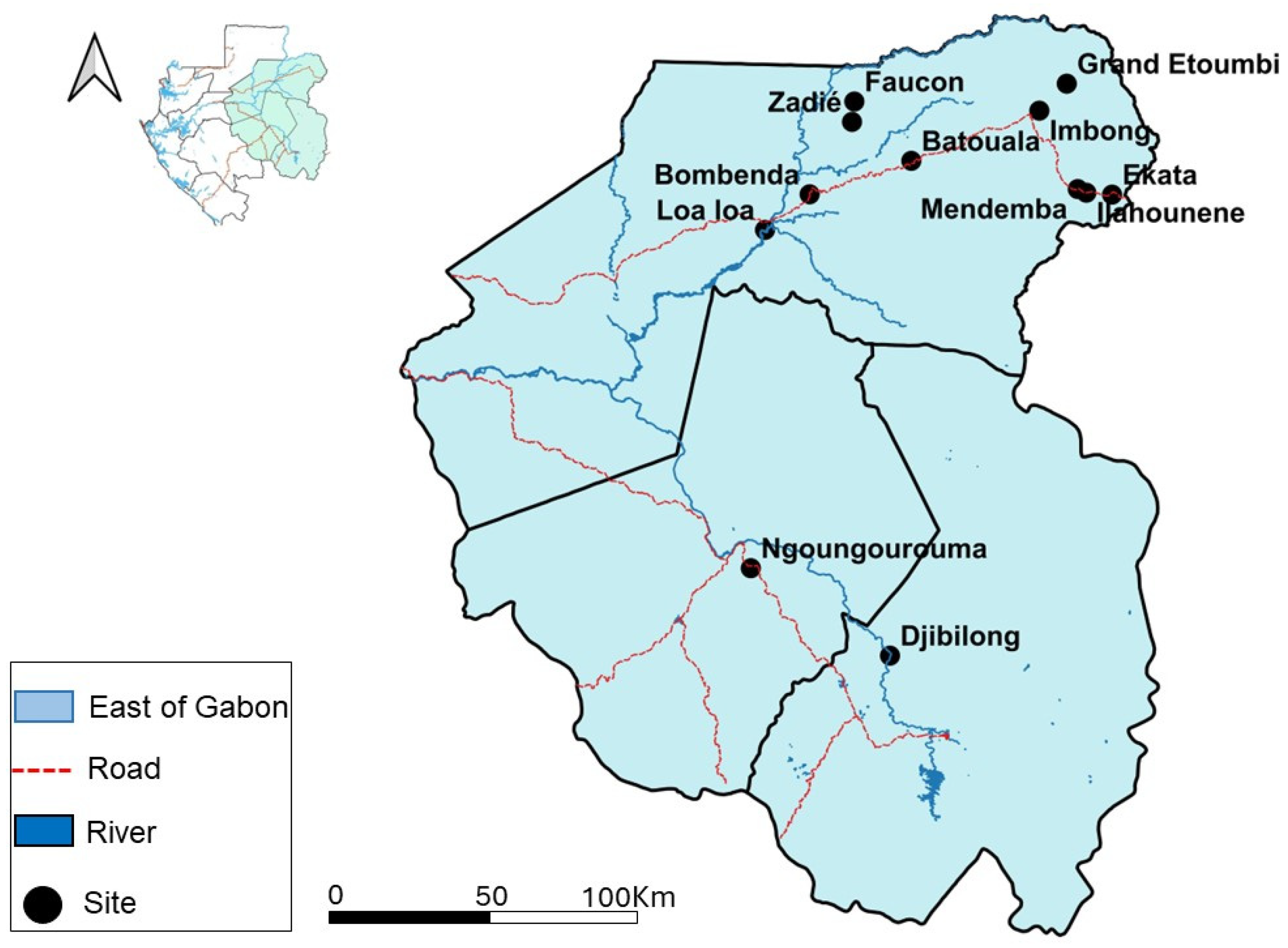
2.1. Study Area and Sample Origins
2.2. Sample Preparation and DNA Extraction
2.3. BtHBV PCR Amplification and Sequencing
2.4. Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Studied Population
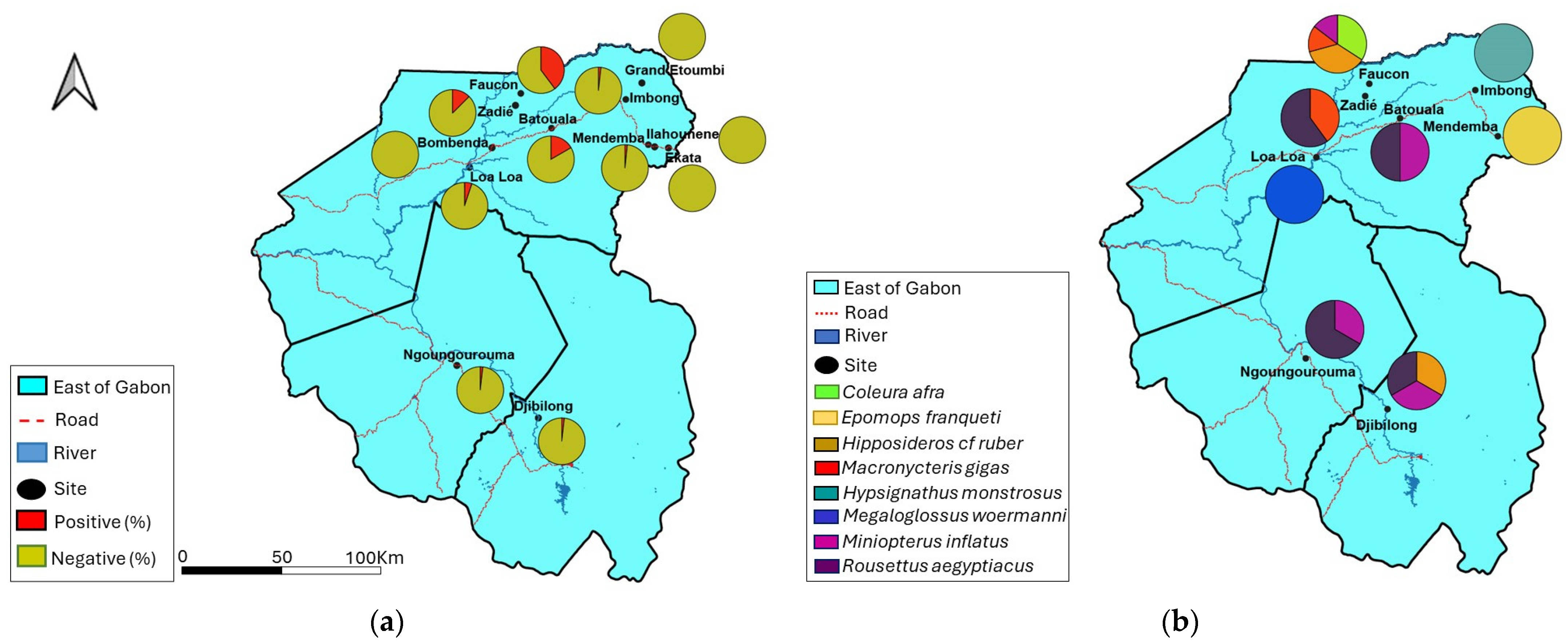
3.2. Detection of BtHBV in Bats
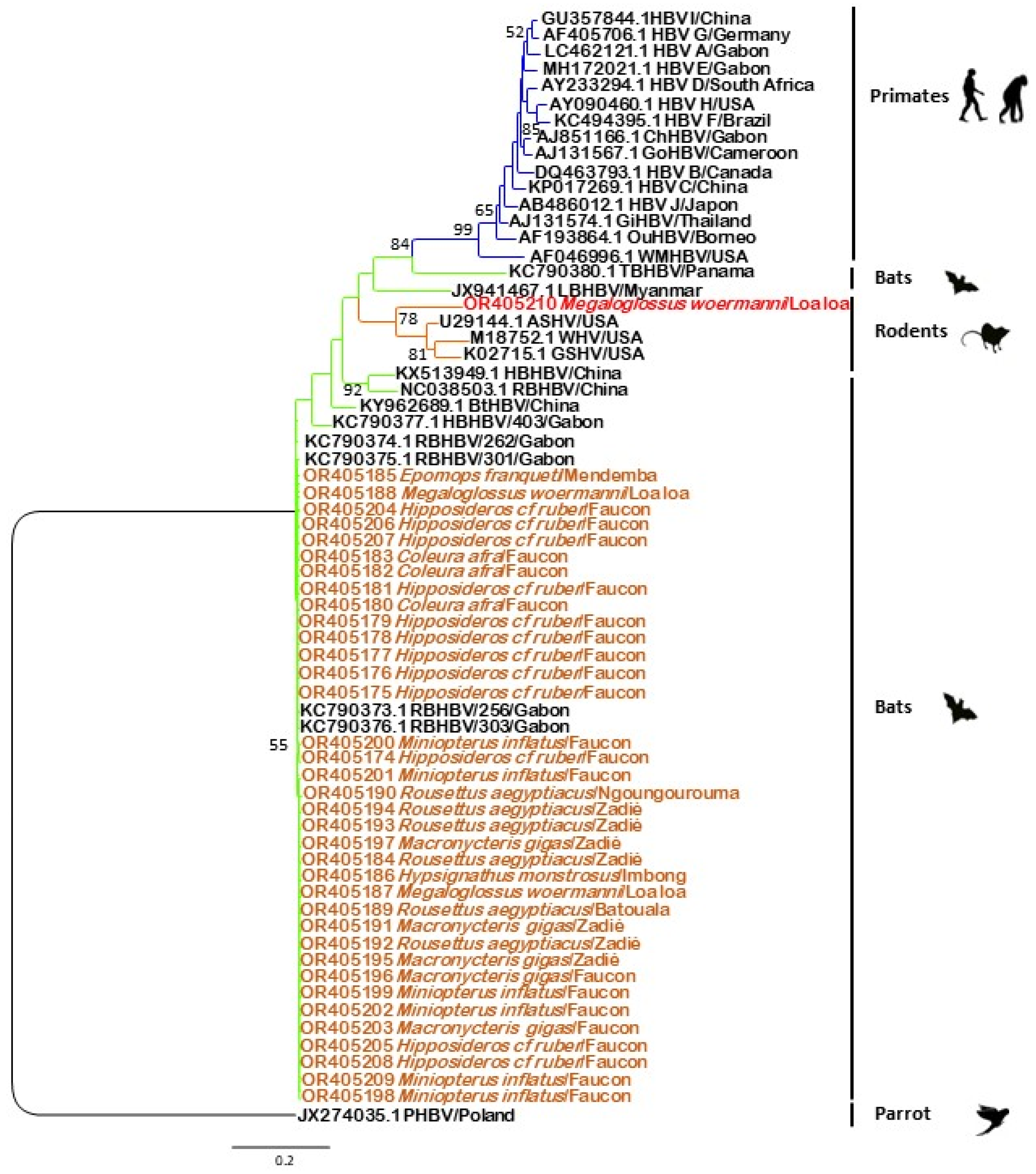
3.3. Phylogenetic Analysis
3.4. Factors Favoring BtHBV Positivity in Bats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Health Sector Strategy on Viral Hepatitis 2016–2021. [Internet]. Global Hepatitis Programme Department of HIV/AIDS. 2016. Available online: https://www.who.int/publications/i/item/WHO-HIV-2016.06 (accessed on 4 July 2022).
- Stanaway, J.D.; Flaxman, A.D.; Naghavi, M.; Fitzmaurice, C.; Vos, T.; Abubakar, I.; Abu-Raddad, L.J.; Assadi, R.; Bhala, N.; Cowie, B.; et al. The global burden of viral hepatitis from 1990 to 2013: Findings from the Global Burden of Disease Study 2013. Lancet 2016, 388, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Feschotte, C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010, 8, e1000495. [Google Scholar] [CrossRef] [PubMed]
- Paraskevis, D.; Magiorkinis, G.; Magiorkinis, E.; Ho, S.Y.; Belshaw, R.; Allain, J.-P.; Hatzakis, A. Dating the origin and dispersal of hepatitis B virus infection in humans and primates. Hepatology 2013, 57, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Testut, P.; Renard, C.A.; Terradillos, O.; Vitvitski-Trepo, L.; Tekaia, F.; Degott, C.; Blake, J.; Boyer, B.; Buendia, M.A. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J. Virol. 1996, 70, 4210–4219. [Google Scholar] [CrossRef] [PubMed]
- Marion, P.L.; Oshiro, L.S.; Regnery, D.C.; Scullard, G.H.; Robinson, W.S. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc. Natl. Acad. Sci. USA 1980, 77, 2941–2945. [Google Scholar] [CrossRef] [PubMed]
- Summers, J.; Smolec, J.M.; Snyder, R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc. Natl. Acad. Sci. USA 1978, 75, 4533–4537. [Google Scholar] [CrossRef]
- Starkman, S.; MacDonald, D.; Lewis, J.; Holmes, E.; Simmonds, P. Geographic and species association of hepatitis B virus genotypes in non-human primates. Virology 2003, 314, 381–393. [Google Scholar] [CrossRef]
- Bonvicino, C.R.; Moreira, M.A.; Soares, M.A. Hepatitis B virus lineages in mammalian hosts: Potential for bidirectional cross-species transmission. World J. Gastroenterol. 2014, 20, 7665–7674. [Google Scholar] [CrossRef]
- He, B.; Fan, Q.; Yang, F.; Hu, T.; Qiu, W.; Feng, Y.; Li, Z.; Li, Y.; Zhang, F.; Guo, H.; et al. Hepatitis Virus in Long-Fingered Bats, Myanmar. Emerg Infect. Dis. 2013, 19, 638–640. [Google Scholar] [CrossRef]
- Drexler, J.F.; Geipel, A.; König, A.; Corman, V.M.; van Riel, D.; Leijten, L.M.; Bremer, C.M.; Rasche, A.; Cottontail, V.M.; Maganga, G.D.; et al. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. USA 2013, 110, 16151–16156. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, Q.; Wang, H.; Yao, X. Severe zoonotic viruses carried by different species of bats and their regional distribution. Clin. Microbiol. Infect. 2023. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.M.; Kumulungui, B.; Pourrut, X.; Rouquet, P.; Hassanin, A.; Yaba, P.; Délicat, A.; Paweska, J.T.; Gonzalez, J.-P.; Swanepoel, R. Fruit bats as reservoirs of Ebola virus. Nature 2005, 438, 575–576. [Google Scholar] [CrossRef] [PubMed]
- Maganga, G.D.; Bourgarel, M.; Ebang Ella, G.; Drexler, J.F.; Gonzalez, J.P.; Drosten, C.; Leroy, E.M. Is marburg virus enzootic in Gabon? J. Infect. Dis. 2011, 204 (Suppl. 3), S800–S803. [Google Scholar] [CrossRef] [PubMed]
- Halpin, K.; Young, P.L.; Field, H.E.; Mackenzie, J.S. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [CrossRef] [PubMed]
- Yob, J.M.; Field, H.; Rashdi, A.M.; Morrissy, C.; van der Heide, B.; Rota, P.; bin Adzhar, A.; White, J.; Daniels, P.; Jamaluddin, A.; et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001, 7, 439–441. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Drosten, C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014, 101, 45–56. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Sharma, A.; Ahmad Farouk, I.; Lal, S.K. COVID-19: A review on the novel coronavirus disease evolution, transmission, detection, control and prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef]
- Shi, Z. Bat and virus. Protein Cell 2010, 1, 109–114. [Google Scholar] [CrossRef]
- N’dilimabaka, N.; Pambou, J.B.M.; Maganga, G.D.; Mve-Ella, O.L.B.; Simo, H.D.; Boundenga, L.; Ngoubangoye, B.; Leroy, E.M. Absence of Mammarenavirus RNA among their Natural Rodent and Potential other Reservoirs in Wildlife in Gabon. Open Access J. Biomed Sci. 2022, 4, 1697–1703. [Google Scholar] [CrossRef]
- Maganga, G.D.; Pinto, A.; Mombo, I.M.; Madjitobaye, M.; Beyeme, A.M.M.; Boundenga, L.; Gouilh, M.A.; N’dilimabaka, N.; Drexler, J.F.; Drosten, C.; et al. Genetic diversity and ecology of coronaviruses hosted by cave-dwelling bats in Gabon. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Paterson, B.D.; Webala, P.W. Keys to the bats (Mammalia: Chiroptera) of East Africa. Fieldiana Life Earth Sci. 2012, 2012, 1–60. [Google Scholar] [CrossRef]
- Maganga, G.D.; Bourgarel, M.; Vallo, P.; Dallo, T.D.; Ngoagouni, C.; Drexler, J.F.; Drosten, C.; Nakouné, E.R.; Leroy, E.M.; Morand, S. Bat distribution size or shape as determinant of viral richness in african bats. PLoS ONE 2014, 9, e100172. [Google Scholar] [CrossRef] [PubMed]
- Kurbanov, F.; Tanaka, Y.; Mizokami, M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol. Res. 2010, 40, 14–30. [Google Scholar] [CrossRef]
- Norder, H.; Hammas, B.; Lofdahl, S.; Courouce, A.-M.; Magnius, L.O. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J. Gen. Virol. 1992, 73, 1201–1208. [Google Scholar] [CrossRef]
- Ding, H.; Liu, B.; Zhao, C.; Yang, J.; Yan, C.; Yan, L.; Zhuang, H.; Li, T. Amino acid similarities and divergences in the small surface proteins of genotype C hepatitis B viruses between nucleos(t)ide analogue-naïve and lamivudine-treated patients with chronic hepatitis B. Antivir. Res. 2013, 102, 29–34. [Google Scholar] [CrossRef]
- Liu, B.-M.; Li, T.; Xu, J.; Li, X.-G.; Dong, J.-P.; Yan, P.; Yang, J.-X.; Yan, L.; Gao, Z.-Y.; Li, W.-P.; et al. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naïve Chinese patients. Antivir. Res. 2010, 85, 512–519. [Google Scholar] [CrossRef]
- N’Dilimabaka, N.; Mavoungou, D.K.; Soami, V.; Kombila, L.B.; Mouguiama, R.M.; Mondjo, A.; Pambou, J.B.M.; Ngoma, J.F.; Ovengue, F.C.; Alilangori, T.P.; et al. Molecular analyses of human rabies virus associated with encephalitis in two children in Gabon. IJID Reg. 2022, 2, 180–183. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Herve, M. Package ‘RVAideMemoire’. 2023. Available online: https://cran.opencpu.org/web/packages/RVAideMemoire/RVAideMemoire.pdf (accessed on 22 December 2023).
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67. [Google Scholar] [CrossRef]
- Akaike, H. A New Look at the Statistical Model Identification. IEEE Trans. Automat Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Practical Use of the Information-Theoretic Approach. In Model Selection and Inference; Springer: New York, NY, USA, 1998. [Google Scholar]
- Burnham, K.P.; Anderson, D.R. Multimodel inference: Understanding AIC and BIC in model selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Rasche, A.; Souza, B.F.d.C.D.; Drexler, J.F. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr. Opin. Virol. 2016, 16, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Hiller, T.; Rasche, A.; Brändel, S.D.; König, A.; Jeworowski, L.; O’mara, M.T.; Cottontail, V.; Page, R.A.; Glebe, D.; Drexler, J.F.; et al. Host Biology and Anthropogenic Factors Affect Hepadnavirus Infection in a Neotropical Bat. Ecohealth 2019, 16, 82–94. [Google Scholar] [CrossRef]
- Van Nguyen, D.; Van Nguyen, C.; Bonsall, D.; Ngo, T.T.; Carrique-Mas, J.; Pham, A.H.; Bryant, J.E.; Thwaites, G.; Baker, S.; Woolhouse, M.; et al. Detection and characterization of homologues of human hepatitis viruses and pegiviruses in rodents and bats in vietnam. Viruses 2018, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.-Y.; Lin, X.-D.; Hao, Z.-Y.; Chen, X.-N.; Wang, Z.-X.; Wang, M.-R.; Wu, J.; Wang, H.-W.; Zhao, G.; Ma, R.Z.; et al. Extensive diversity and evolution of hepadnaviruses in bats in China. Virology 2018, 514, 88–97. [Google Scholar] [CrossRef]
- Lei, S.-C.; Xiao, X.; Liu, J.-W.; Han, H.-J.; Gong, X.-Q.; Zhao, M.; Wang, L.-J.; Qin, X.-R.; Yu, X.-J. High prevalence and genetic diversity of hepatitis B viruses in insectivorous bats from China. Acta Trop. 2019, 199, 105130. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.-L.; Li, W.; Zhu, Y.; Ge, X.-Y.; Zhang, L.-B.; Zhang, Y.-Z.; Bock, C.-T.; Shi, Z.-L. Detection and genome characterization of four novel bat hepadnaviruses and a hepevirus in China. Virol. J. 2017, 14, 1–10. [Google Scholar] [CrossRef]
- He, B.; Zhang, F.; Xia, L.; Hu, T.; Chen, G.; Qiu, W.; Fan, Q.; Feng, Y.; Guo, H.; Tu, C. Identification of a novel orthohepadnavirus in pomona roundleaf bats in China. Arch. Virol. 2015, 160, 335–337. [Google Scholar] [CrossRef]
- Yang, L.; Wu, J.; Hu, T.; Qin, S.; Deng, B.; Liu, J.; Zhang, F.; He, B.; Tu, C. Genetic diversity of bat orthohepadnaviruses in China and a proposed new nomenclature. Infect. Genet. Evol. 2018, 63, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Rougeron, V.; Suquet, E.; Maganga, G.D.; Jiolle, D.; Mombo, I.M.; Bourgarel, M.; Motsch, P.; Arnathau, C.; Durand, P.; Drexler, F.; et al. Characterization and phylogenetic analysis of new bat astroviruses detected in Gabon, Central Africa. Acta Virol. 2016, 60, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Breed, A.C.; Field, H.E.; Smith, C.S.; Edmonston, J.; Meers, J. Bats without borders: Long-distance movements and implications for disease risk management. Ecohealth 2010, 7, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Willoughby, A.R.; Phelps, K.L.; PREDICT Consortium; Olival, K.J. A Comparative analysis of viral richness and viral sharing in cave-roosting bats. Diversity 2017, 9, 35. [Google Scholar] [CrossRef]
- Turmelle, A.S.; Olival, K.J. Correlates of viral richness in bats (Order Chiroptera). Ecohealth 2009, 6, 522–539. [Google Scholar] [CrossRef] [PubMed]
- Mickleburgh, S.; Waylen, K.; Racey, P. Bats as bushmeat: A global review. Oryx 2009, 43, 217–234. [Google Scholar] [CrossRef]
- Olivero, J.; Fa, J.E.; Farfán, M.; Márquez, A.L.; Real, R.; Juste, F.J.; Leendertz, S.A.; Nasi, R. Human activities link fruit bat presence to Ebola virus disease outbreaks. Mammal Rev. 2020, 50, 1–10. [Google Scholar] [CrossRef]
- Wright, G.S. Hipposideros caffer (Chiroptera: Hipposideridae). Mamm. Species 2005, 845, 1–9. [Google Scholar] [CrossRef][Green Version]
- Jacquet, S.; Pons, J.-B.; De Bernardo, A.; Ngoubangoye, B.; Cosset, F.-L.; Régis, C.; Etienne, L.; Pontier, D. Evolution of Hepatitis B Virus Receptor NTCP Reveals Differential Pathogenicities and Species Specificities of Hepadnaviruses in Primates, Rodents, and Bats. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef]
- Schountz, T.; Baker, M.L.; Butler, J.; Munster, V. Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Front. Immunol. 2017, 8, 1098. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, B.; Hou, J.; Sun, J.; Hao, R.; Xiang, K.; Yan, L.; Zhang, J.; Zhuang, H.; Li, T. Naturally occurring deletions/insertions in HBV core promoter tend to decrease in hepatitis B e antigen-positive chronic hepatitis B patients during antiviral therapy. Antivir Ther. 2015, 20, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, L.; Chen, Y.; Liu, A.; Wang, L.; Xu, P.; Liu, Y.; Li, L.; Meng, F. Integrating nested PCR with high-throughput sequencing to characterize mutations of HBV genome in low viral load samples. Medicine 2017, 96, e7588. [Google Scholar] [CrossRef] [PubMed]
| Variables | Number of Samples n = 859 (%) | Haut-Ogooué n = 167 (%) | Ogooue-Ivindo n = 528 (%) | Ogooué-Lolo n = 164 (%) |
|---|---|---|---|---|
| Age | ||||
| J | 26 (3) | 5 (3) | 15 (2.8) | 6 (3.7) |
| SA | 149 (17.4) | 12 (7.2) | 62 (11.8) | 75 (45.7) |
| A | 682 (79.4) | 149 (89.2) | 450 (85.2) | 83 (50.6) |
| Unknown | 2 (0.2) | 1 (0.6) | 1 (0.2) | - |
| Sex | ||||
| Males | 413 (48.1) | 112 (67.1) | 238 (45.1) | 63 (38.4) |
| Females | 444 (51.7) | 55 (32.9) | 289 (54.7) | 100 (61) |
| Unknown | 2 (0.2) | - | 1 (0.2) | 1 (0.6) |
| Species | ||||
| Coleura afra | 18 (2.1) | - | 18 (3.4) | - |
| Eidolon helvum | 2 (0.2) | - | 2 (0.4) | - |
| Epomops franqueti | 168 (19.6) | - | 168 (31.9) | - |
| Hipposideros cf ruber | 30 (3.5) | 5 (3) | 25 (4.7) | - |
| Macronycterisgigas | 44 (5.1) | - | 40 (7.6) | 4 (2.4) |
| Hypsignathus monstrosus | 5 (0.6) | - | 5 (0.9) | - |
| Megaloglossus woermanni | 107 (12.5) | - | 107 (20.3) | - |
| Miniopterus inflatus | 198 (23) | 154 (92.2) | 40 (7.6) | 4 (2.4) |
| Myonycteris torquata | 10 (1.2) | - | 10 (1.9) | - |
| Neoromicia tenuipinnis | 5 (0.6) | - | 5 (0.9) | - |
| Rousettus aegyptiacus | 241 (28) | 8 (4.8) | 77 (14.6) | 156 (95.2) |
| Unidentified | 31 (3.6) | - | 31 (5.8) | - |
| Diet | ||||
| Insectivorous | 295 (34.3) | 159 (95.2) | 128 (24.2) | 8 (4.9) |
| Frugivorous | 426 (49.6) | 8 (4.8) | 262 (49.6) | 156 (95.1) |
| Nectarivorous | 107 (12.5) | - | 107 (20.3) | - |
| Unidentified | 31 (3.6) | - | 31 (5.9) | - |
| Collection sites | ||||
| Caves | ||||
| Djibilong | 167 (31.7) | 167 (100) | - | - |
| Batouala | 12 (2.3) | - | 12 (6.2) | - |
| Faucon | 103 (19.6) | - | 103 (52.8) | - |
| Zadié | 80 (15.2) | - | 80 (41) | - |
| Ngoungourouma | 164 (31.2) | - | - | 164 (100) |
| Total | 526 (100) | 167 | 195 | 164 |
| Villages | ||||
| Bombenda | 10 (3) | - | 10 (3) | - |
| Ekata | 34 (10.2) | - | 34 (10.2) | - |
| Grand Etoumbi | 48 (14.4) | - | 48 14.4) | - |
| Ilahounene | 68 (20.4) | - | 68 (20.4) | - |
| Imbong | 55 (16.5) | - | 55 (16.5) | - |
| Loa loa | 58 (17.4) | - | 58 (17.4) | - |
| Mendemba | 60 (18.1) | - | 60 (18.1) | - |
| Total | 333 (100) | - | 333 | - |
| Season | ||||
| Dry season | 460 (53.6) | - | 460 (87.1) | - |
| Rainy season | 399 (46.4) | 167 (100) | 68 (12.9) | 164 (100) |
| Positive PCR (%) | ||||||
|---|---|---|---|---|---|---|
| Variables | Number of Samples n = 859(%) | Haut-Ogooué n = 167(%) | Ogooue-Ivindo n = 528(%) | Ogooué-Lolo n = 164(%) | Total Positive PCR (%) | Fisher Test/Chi Test (p-Value) |
| Age | ||||||
| J | 26 | 0/5 | 0/15 | 0/6 | 0/26 | |
| SA | 149 | 0/12 | 8/62 (12.9) | 1/75 (1.3) | 9/149 (6) | |
| A | 682 | 3/149 (2) | 50/450 (11.1) | 2/83 (2.4) | 55/682 (8) | 0.397 |
| Unknown | 2 | 0/1 | 0/1 | - | 0/2 (0) | |
| Sex | ||||||
| Males | 413 | 1/112 (0.9) | 39/238 (16.4) | 3/63 (4.8) | 43/413 (10.4) | |
| Females | 444 | 2/55 (3.6) | 19/289 (6.6) | 0/100 | 21/444 (4.7) | 0.002432 ** |
| Unknown | 2 | - | 0/1 | 0/1 | 0/2 | |
| Species | ||||||
| Coleura afra | 18 | - | 14/18 (75) | - | 14/18 (75) | |
| Eidolon helvum | 2 | - | 0/2 | - | 0/2 | |
| Epomops franqueti | 168 | - | 1/168 (0.6) | - | 1/168 (0.6) | |
| Hipposideros cf ruber | 30 | 1/5 (20) | 15/25 (60) | - | 16/30 (53.3) | |
| Macronycterisgigas | 44 | - | 10/40 (25) | 0/4 | 10/44 (25) | |
| Hypsignathus monstrosus | 5 | - | 1/5 (20) | - | 1/5 (20) | |
| Megaloglossus woermanni | 107 | - | 3/107 (2.8) | - | 3/107 (2.8) | 0.0004998 *** |
| Miniopterus inflatus | 198 | 1/154 (0.6) | 7/40 (17.5) | 1/4 (25) | 9/198 (4.5) | |
| Myonycteris torquata | 10 | - | 0/10 | - | 0/10 | |
| Neoromicia tenuipinnis | 5 | - | 0/5 | - | 0/5 | |
| Rousettus aegyptiacus | 241 | 1/8 (12.5) | 7/77 (9.1) | 2/156 (1.3) | 10/241 (4.1) | |
| Unidentified | 31 | - | 0/31 | - | 0/31 | |
| Diet | ||||||
| Insectivorous | 295 | 2/159 (1.3) | 46/128 (35.9) | 1/8 (12.5) | 49/295 (16.6) | |
| Frugivorous | 426 | 1/8 (12.5) | 9/262 (0.8) | 2/156 (1.3) | 12/426 (2.8) | 4.312 × 1011 *** |
| Nectarivorous | 107 | - | 3/107 (2.8) | - | 3/107 (2.8) | |
| Unidentified | 31 | - | 0/31 | - | 0/31 | |
| Collection sites | ||||||
| Caves | ||||||
| Djibilong | 167 | 3/167 (1.8) | - | - | 3/167 (1.8) | |
| Batouala | 12 | - | 2/12 (16.7) | - | 2/12 (16.7) | |
| Faucon | 103 | - | 41/103 (39.8) | - | 41/103 (39.8) | |
| Zadié | 80 | - | 10/80 (12.5) | - | 10/80 (12.5) | |
| Ngoungourouma | 164 | - | - | 3/164 (1.8) | 3/164 (1.8) | |
| Villages | 2.608 × 107 *** | |||||
| Bombenda | 10 | - | 0/10 | - | 0/10 | |
| Ekata | 34 | - | 0/34 | - | 0/34 | |
| Grand Etoumbi | 48 | - | 0/48 | - | 0/48 | |
| Ilahounene | 68 | - | 0/68 | - | 0/68 | |
| Imbong | 55 | - | 1/55 (1.8) | - | 1/55 (1.8) | |
| Loa loa | 58 | 3/58 (5.2) | - | 3/58 (5.2) | ||
| Mendemba | 60 | - | 1/60 (1.7) | - | 1/60 (1.7) | |
| Season | ||||||
| Dry season | 460 | - | 55/460 (12) | - | 55/460 (12) | 8.497 × 108 *** |
| Rainy season | 399 | 3/167 (1.8) | 3/68 (4.4) | 3/164 (1.8) | 9/399 (2.3) | |
| Model No. (Rank) | Fixed Effects | Df | ΔAIC | Akaike Weight | |||||
|---|---|---|---|---|---|---|---|---|---|
| Intercept | Age | Location | Season | Sex | Species | ||||
| 17 (1) | −3.747 | + | 14 | 0 | 0.216 | ||||
| 18 (2) | −3.513 | + | + | 17 | 0.28 | 0.187 | |||
| 19 (3) | −3.667 | + | + | 15 | 1.70 | 0.092 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koumba Mavoungou, D.S.; Bohou Kombila, L.; Longo Pendy, N.M.; Koumba Moukouama, S.E.; Lekana-Douki, S.E.; Maganga, G.D.; Leroy, E.M.; Aghokeng, A.F.; N’dilimabaka, N. Prevalence and Genetic Diversity of Bat Hepatitis B Viruses in Bat Species Living in Gabon. Viruses 2024, 16, 1015. https://doi.org/10.3390/v16071015
Koumba Mavoungou DS, Bohou Kombila L, Longo Pendy NM, Koumba Moukouama SE, Lekana-Douki SE, Maganga GD, Leroy EM, Aghokeng AF, N’dilimabaka N. Prevalence and Genetic Diversity of Bat Hepatitis B Viruses in Bat Species Living in Gabon. Viruses. 2024; 16(7):1015. https://doi.org/10.3390/v16071015
Chicago/Turabian StyleKoumba Mavoungou, Danielle S., Linda Bohou Kombila, Neil M. Longo Pendy, Schedy E. Koumba Moukouama, Sonia Etenna Lekana-Douki, Gaël D. Maganga, Eric M. Leroy, Avelin F. Aghokeng, and Nadine N’dilimabaka. 2024. "Prevalence and Genetic Diversity of Bat Hepatitis B Viruses in Bat Species Living in Gabon" Viruses 16, no. 7: 1015. https://doi.org/10.3390/v16071015
APA StyleKoumba Mavoungou, D. S., Bohou Kombila, L., Longo Pendy, N. M., Koumba Moukouama, S. E., Lekana-Douki, S. E., Maganga, G. D., Leroy, E. M., Aghokeng, A. F., & N’dilimabaka, N. (2024). Prevalence and Genetic Diversity of Bat Hepatitis B Viruses in Bat Species Living in Gabon. Viruses, 16(7), 1015. https://doi.org/10.3390/v16071015






