Cross-Reactive Antibodies to the NS1 Protein of Omsk Hemorrhagic Fever Virus Are Absent in the Sera of Patients with Tick-Borne Encephalitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sera from Patients
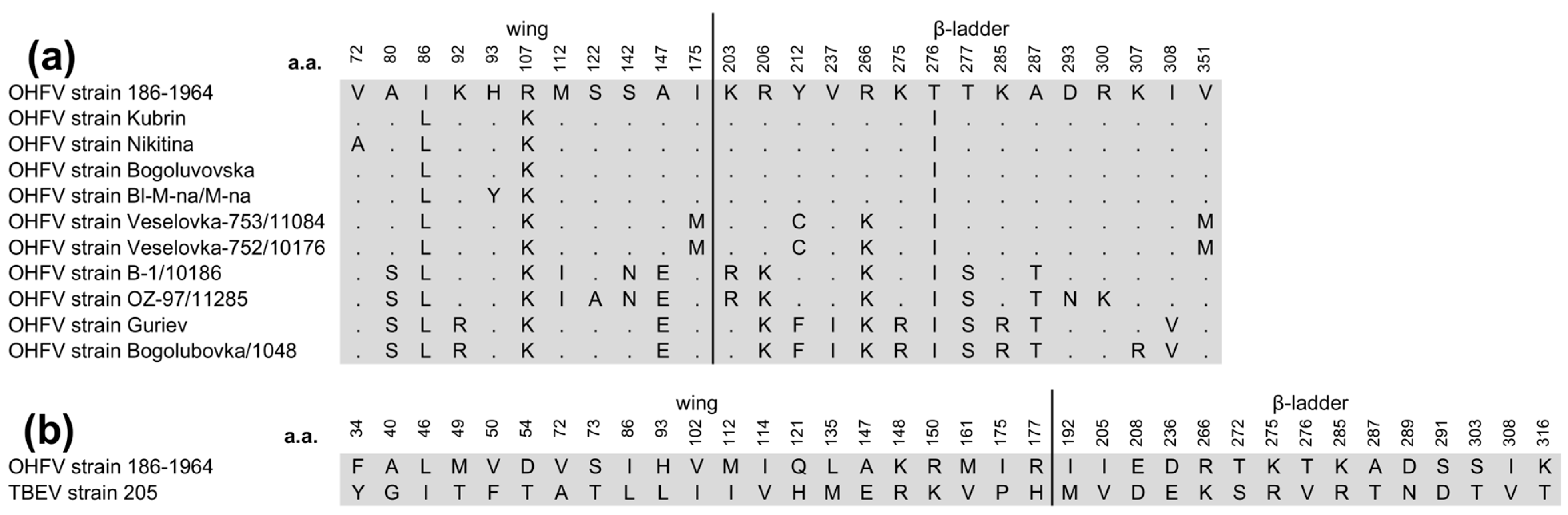
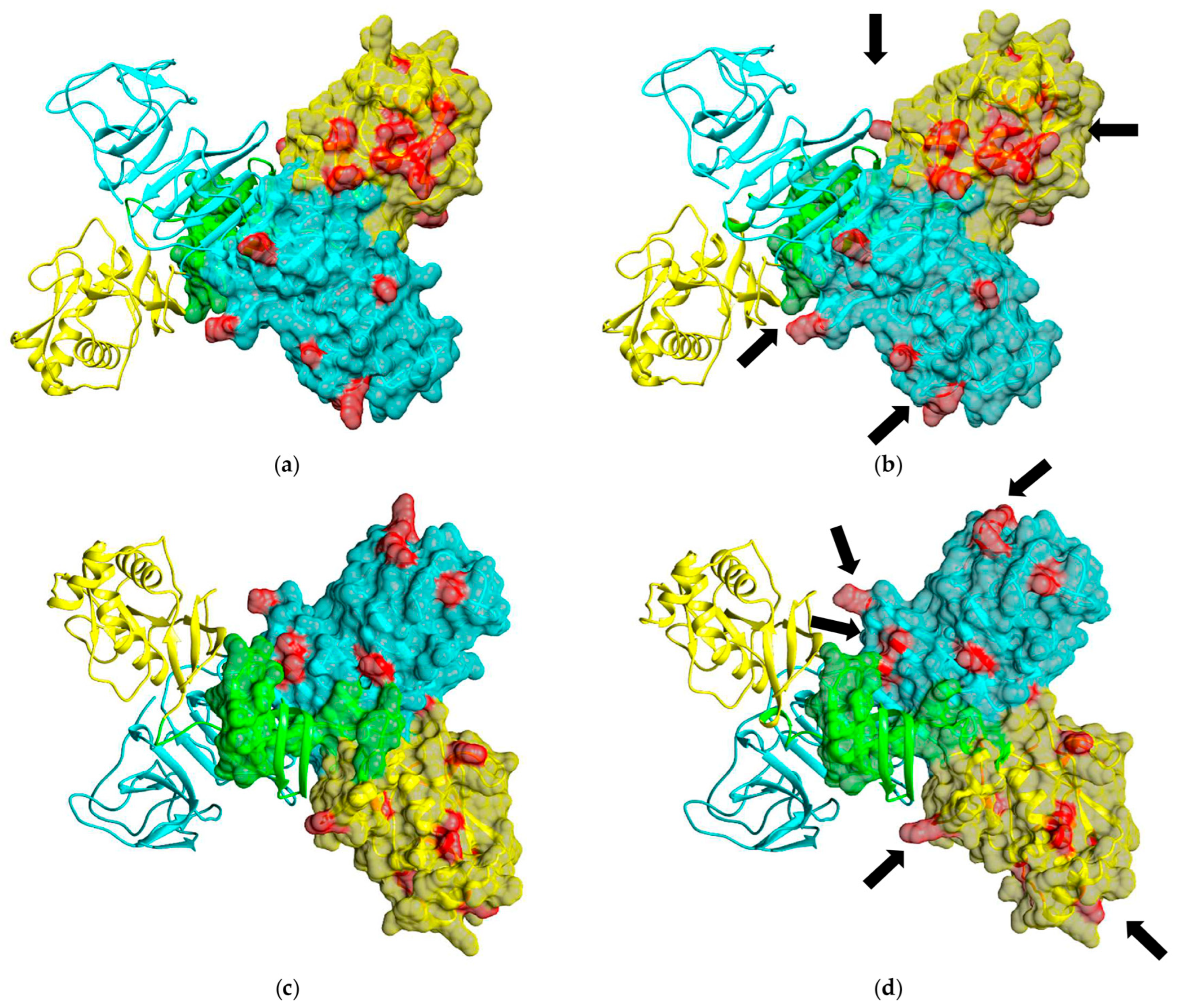
2.2. Phylogenetic Analyses of OHFV NS1 Gene and In Silico Structural Analysis of NS1 Protein of OHFV Strain 186–1964
2.3. The Construction of the Plasmid Encoding the OHFV NS1 Protein
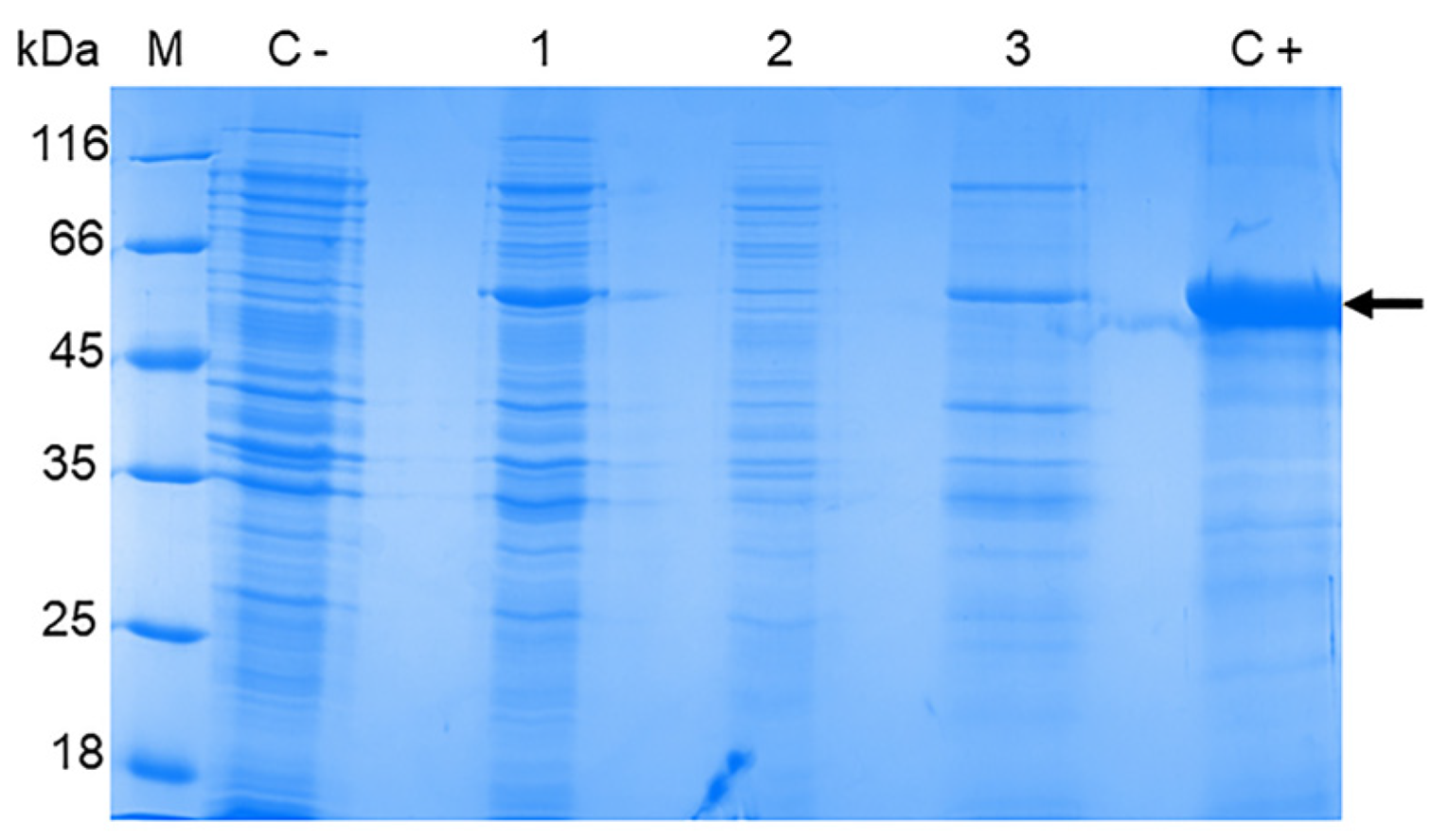
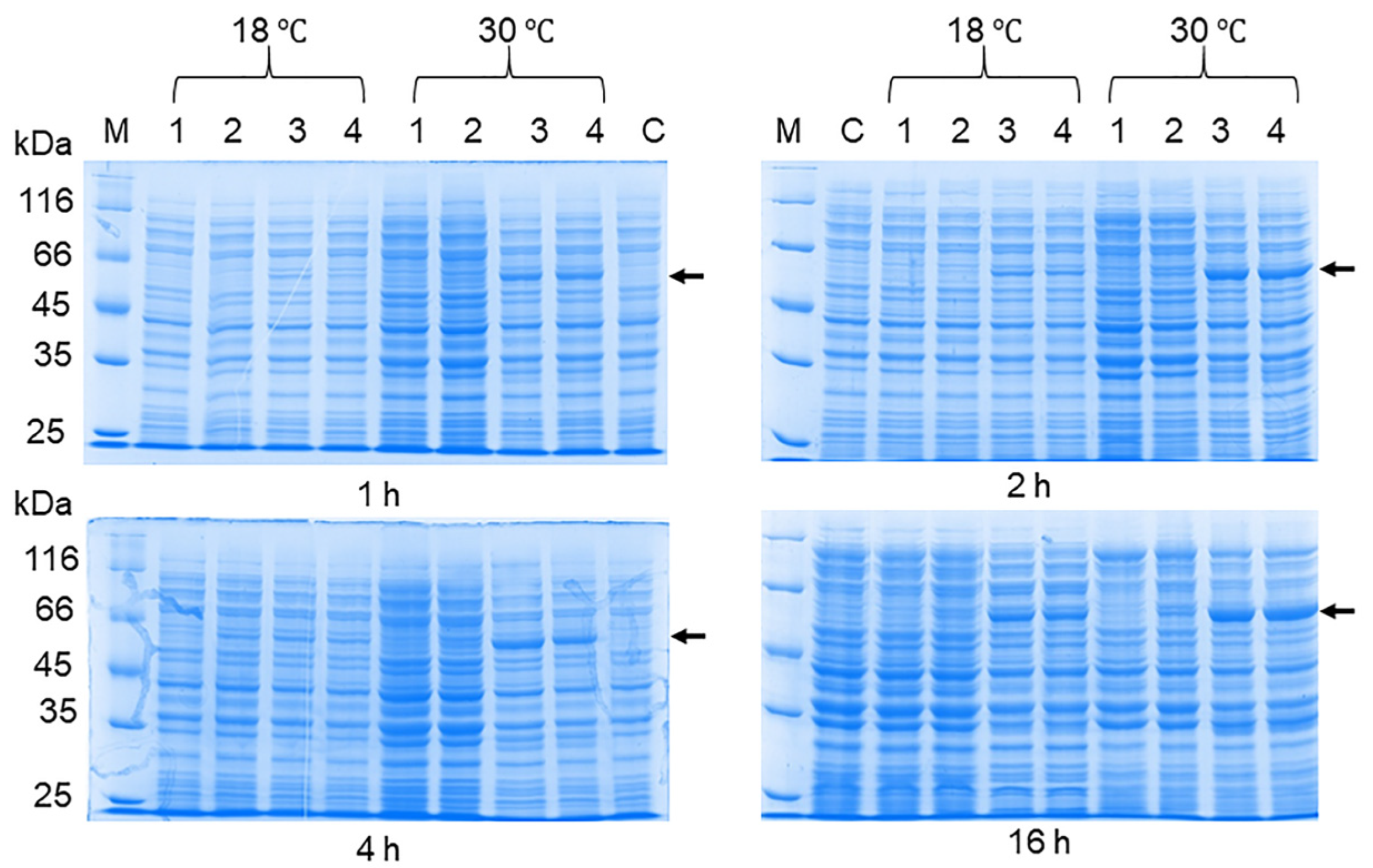
2.4. The Optimization of the OHFV NS1 Expression
2.5. The Purification of the Recombinant OHFV NS1 Protein
2.6. Western Blot Analysis
2.7. ELISA
2.8. Statistics
3. Results
3.1. The Phylogenetic Analysis and In Silico Modeling of the OHFV NS1 Protein
3.2. The Production of the OHFV NS1 Protein in E. coli Cells
3.3. OHFV NS1 Protein Isolation and Purification
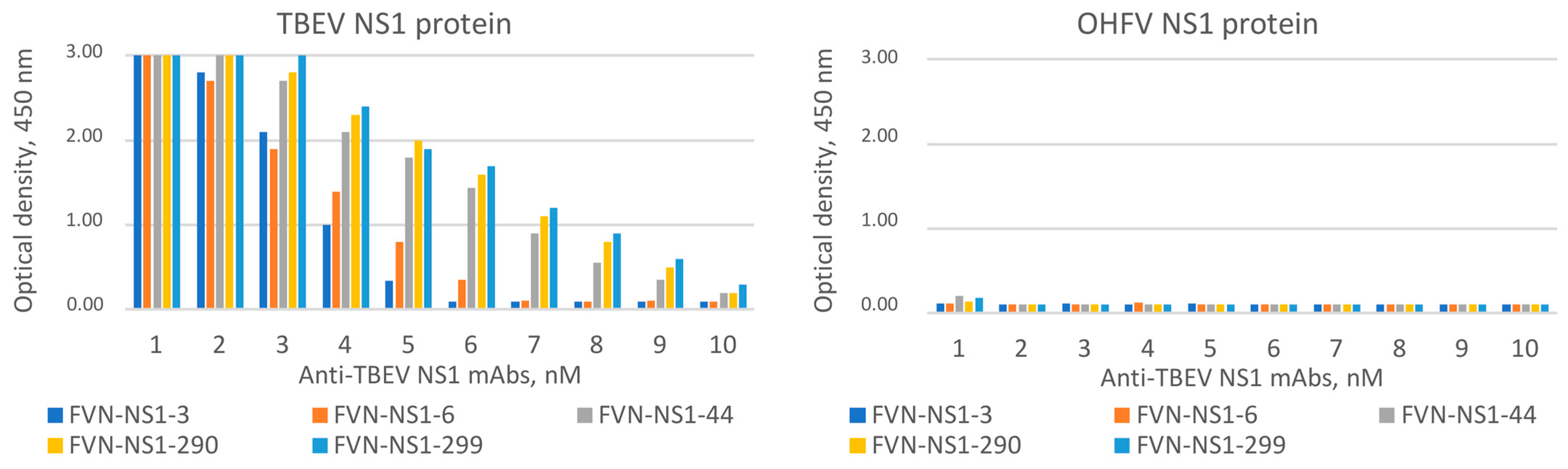
3.4. The Evaluation of the Antigenic Properties of the Recombinant OHFV NS1 Protein
3.5. The Examination of Sera from Patients with TBE for Cross-Reactivity with the OHFV NS1 Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Růžek, D.; Yakimenko, V.V.; Karan, L.S.; Tkachev, S.E. Omsk haemorrhagic fever. Lancet 2010, 376, 2104–2113. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Li, L.; Dick, D.; Shope, R.E.; Feldmann, H.; Barrett, A.D.; Holbrook, M.R. Analysis of the complete genome of the tick-borne flavivirus Omsk hemorrhagic fever virus. Virology 2003, 313, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Karan, L.S.; Ciccozzi, M.; Yakimenko, V.V.; Presti, A.L.; Cella, E.; Zehender, G.; Rezza, G.; Platonov, A.E. The deduced evolution history of Omsk hemorrhagic fever virus. J. Med. Virol. 2014, 86, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.; Shin, A.; Tukhanova, N.; Turebekov, N.; Nurmakhanov, T.; Sutyagin, V.; Berdibekov, A.; Maikanov, N.; Lezdinsh, I.; Shapiyeva, Z.; et al. First Indications of Omsk Haemorrhagic Fever Virus beyond Russia. Viruses 2022, 14, 754. [Google Scholar] [CrossRef] [PubMed]
- Dias, B.P.; Barbosa, C.C.; Ferreira, C.S.; Mayra Soares Alves Dos Santos, S.; Arrieta, O.A.P.; Malta, W.C.; Gomes, M.L.M.D.; Alves ESilva, M.; Fonseca, J.M.; Borges, L.P.; et al. Challenges in Direct Detection of Flaviviruses: A Review. Pathogens 2023, 12, 643. [Google Scholar] [CrossRef] [PubMed]
- Chidumayo, N.N.; Yoshii, K.; Kariwa, H. Evaluation of the European tick-borne encephalitis vaccine against Omsk hemorrhagic fever virus. Microbiol. Immunol. 2014, 58, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Orlinger, K.K.; Hofmeister, Y.; Fritz, R.; Holzer, G.W.; Falkner, F.G.; Unger, B.; Loew-Baselli, A.; Poellabauer, E.M.; Ehrlich, H.J.; Barrett, P.N.; et al. A tick-borne encephalitis virus vaccine based on the European prototype strain induces broadly reactive cross-neutralizing antibodies in humans. J. Infect. Dis. 2011, 203, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Ternovoi, V.A.; Kurzhukov, G.P.; Sokolov, Y.V.; Ivanov, G.Y.; Ivanisenko, V.A.; Loktev, A.V.; Ryder, R.W.; Netesov, S.V.; Loktev, V.B. Tick-Borne Encephalitis with Hemorrhagic Syndrome, Novosibirsk Region, Russia, 1999. Emerg. Infect. Dis. 2003, 9, 743–746. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.Y.; Leow, C.Y.; Abdul Majeed, A.B.; Leow, C.H. Flavivirus infection—A review of immunopathogenesis, immunological response, and immunodiagnosis. Virus Res. 2019, 274, 197770. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus NS1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Crooks, A.J.; Lee, J.M.; Easterbrook, L.M.; Timofeev, A.V.; Stephenson, J.R. The NS1 protein of tick-borne encephalitis virus forms multimeric species upon secretion from the host cell. J. Gen. Virol. 1994, 75, 3453–3460. [Google Scholar] [CrossRef] [PubMed]
- Carpio, K.L.; Barrett, A.D.T. Flavivirus NS1 and Its Potential in Vaccine Development. Vaccines 2021, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Oceguera, L.F.; Patiris, P.J.; Chiles, R.E.; Busch, M.P.; Tobler, L.H.; Hanson, C.V. Flavivirus Serology by Western Blot Analysis. Am. J. Trop. Med. Hyg. 2007, 77, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Lorch, M.S.; Collado, M.S.; Argüelles, M.H.; Rota, R.P.; Spinsanti, L.I.; Lozano, M.E.; Goñi, S.E. Production of recombinant NS1 protein and its possible use in encephalitic flavivirus differential diagnosis. Protein Expr. Purif. 2019, 153, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Igolkina, Y.; Rar, V.; Krasnova, E.; Filimonova, E.; Tikunov, A.; Epikhina, T.; Tikunova, N. Occurrence and clinical manifestations of tick-borne rickettsioses in Western Siberia: First Russian cases of Rickettsia aeschlimannii and Rickettsia slovaca infections. Ticks Tick-Borne Dis. 2022, 13, 101927. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Matveev, A.; Pyankov, O.; Khlusevich, Y.; Tyazhelkova, O.; Emelyanova, L.; Timofeeva, A.; Shipovalov, A.; Chechushkov, A.; Zaitseva, N.; Kudrov, G.; et al. Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19. Int. J. Mol. Sci. 2023, 24, 10799. [Google Scholar] [CrossRef] [PubMed]
- Andrey, M.; Yana, K.; Olga, G.; Bogdana, K.; Sergey, T.; Lyudmila, E.; Nina, T. Tick-borne encephalitis nonstructural protein NS1 expressed in E. coli retains immunological properties of the native protein. Protein Expr. Purif. 2022, 191, 106031. [Google Scholar] [CrossRef] [PubMed]
- Akey, D.L.; Brown, W.C.; Jose, J.; Kuhn, R.J.; Smith, J.L. Structure-guided insights on the role of NS1 in flavivirus infection. BioEssays 2015, 37, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 Structures Reveal Surfaces for Associations with Membranes and the Immune System. Science 2014, 343, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Edeling, M.A.; Diamond, M.S.; Fremont, D.H. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. USA 2014, 111, 4285–4290. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, S.Y.; Mazurina, E.A.; Yakimenko, V.V. Molecular variability and genetic structure of Omsk hemorrhagic fever virus, based on analysis of the complete genome sequences. Ticks Tick-Borne Dis. 2021, 12, 101627. [Google Scholar] [CrossRef] [PubMed]
- Steinhagen, K.; Probst, C.; Radzimski, C.; Schmidt-Chanasit, J.; Emmerich, P.; van Esbroeck, M.; Schinkel, J.; Grobusch, M.P.; Goorhuis, A.; Warnecke, J.M.; et al. Serodiagnosis of Zika virus (ZIKV) infections by a novel NS1-based ELISA devoid of cross-reactivity with dengue virus antibodies: A multicohort study of assay performance, 2015 to 2016. Eurosurveillance 2016, 21, 30426. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, C.; Joguet, G.; Mengelle, C.; Chapuy-Regaud, S.; Pavili, L.; Prisant, N.; Izopet, J.; Bujan, L.; Mansuy, J.M. Kinetics of anti-ZIKV antibodies after Zika infection using two commercial enzyme-linked immunoassays. Diagn. Microbiol. Infect. Dis. 2018, 90, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Lustig, Y.; Zelena, H.; Venturi, G.; Van Esbroeck, M.; Rothe, C.; Perret, C.; Koren, R.; Katz-Likvornik, S.; Mendelson, E.; Schwartz, E. Sensitivity and Kinetics of an NS1-Based Zika Virus Enzyme-Linked Immunosorbent Assay in Zika Virus-Infected Travelers from Israel, the Czech Republic, Italy, Belgium, Germany, and Chile. J. Clin. Microbiol. 2017, 55, 1894–1901. [Google Scholar] [CrossRef] [PubMed]
- Kadkhoda, K.; Gretchen, A.; Racano, A. Evaluation of a commercially available Zika virus IgM ELISA: Specificity in focus. Diagn. Microbiol. Infect. Dis. 2017, 88, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, I.; Bin, H.; Schlezinger, S.; Schwartz, E. NS1 antigen testing for the diagnosis of dengue in returned Israeli travelers. J. Med. Virol. 2014, 86, 2005–2010. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Tonry, J.; Hall, R.A.; Williams, B.; Palacios, G.; Ashok, M.S.; Jabado, O.; Clark, D.; Tesh, R.B.; Briese, T.; et al. NS1 protein secretion during the acute phase of West Nile virus infection. J. Virol. 2005, 79, 13924–13933. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.M.; Diamond, M.S. Defining the levels of secreted non-structural protein NS1 after West Nile virus infection in cell culture and mice. J. Med. Virol. 2008, 80, 547–556. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kravchuk, B.I.; Khlusevich, Y.A.; Chicherina, G.S.; Yakimenko, V.V.; Krasnova, E.I.; Tikunova, N.N.; Matveev, A.L. Cross-Reactive Antibodies to the NS1 Protein of Omsk Hemorrhagic Fever Virus Are Absent in the Sera of Patients with Tick-Borne Encephalitis. Viruses 2024, 16, 1032. https://doi.org/10.3390/v16071032
Kravchuk BI, Khlusevich YA, Chicherina GS, Yakimenko VV, Krasnova EI, Tikunova NN, Matveev AL. Cross-Reactive Antibodies to the NS1 Protein of Omsk Hemorrhagic Fever Virus Are Absent in the Sera of Patients with Tick-Borne Encephalitis. Viruses. 2024; 16(7):1032. https://doi.org/10.3390/v16071032
Chicago/Turabian StyleKravchuk, Bogdana I., Yana A. Khlusevich, Galina S. Chicherina, Valeriy V. Yakimenko, Elena I. Krasnova, Nina N. Tikunova, and Andrey L. Matveev. 2024. "Cross-Reactive Antibodies to the NS1 Protein of Omsk Hemorrhagic Fever Virus Are Absent in the Sera of Patients with Tick-Borne Encephalitis" Viruses 16, no. 7: 1032. https://doi.org/10.3390/v16071032




