Lessons Learned from Active Clinical and Laboratory Surveillance during the Sheep Pox Virus Outbreak in Spain, 2022–2023
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Inspection
2.2. Sampling
2.3. Preparation and Processing of Samples
2.3.1. Oral Swabs
2.3.2. Peripheral Blood
2.3.3. Scabs from Skin Lesion
2.4. Nucleic Acid Extraction
2.5. Capripox Virus Real-Time PCR
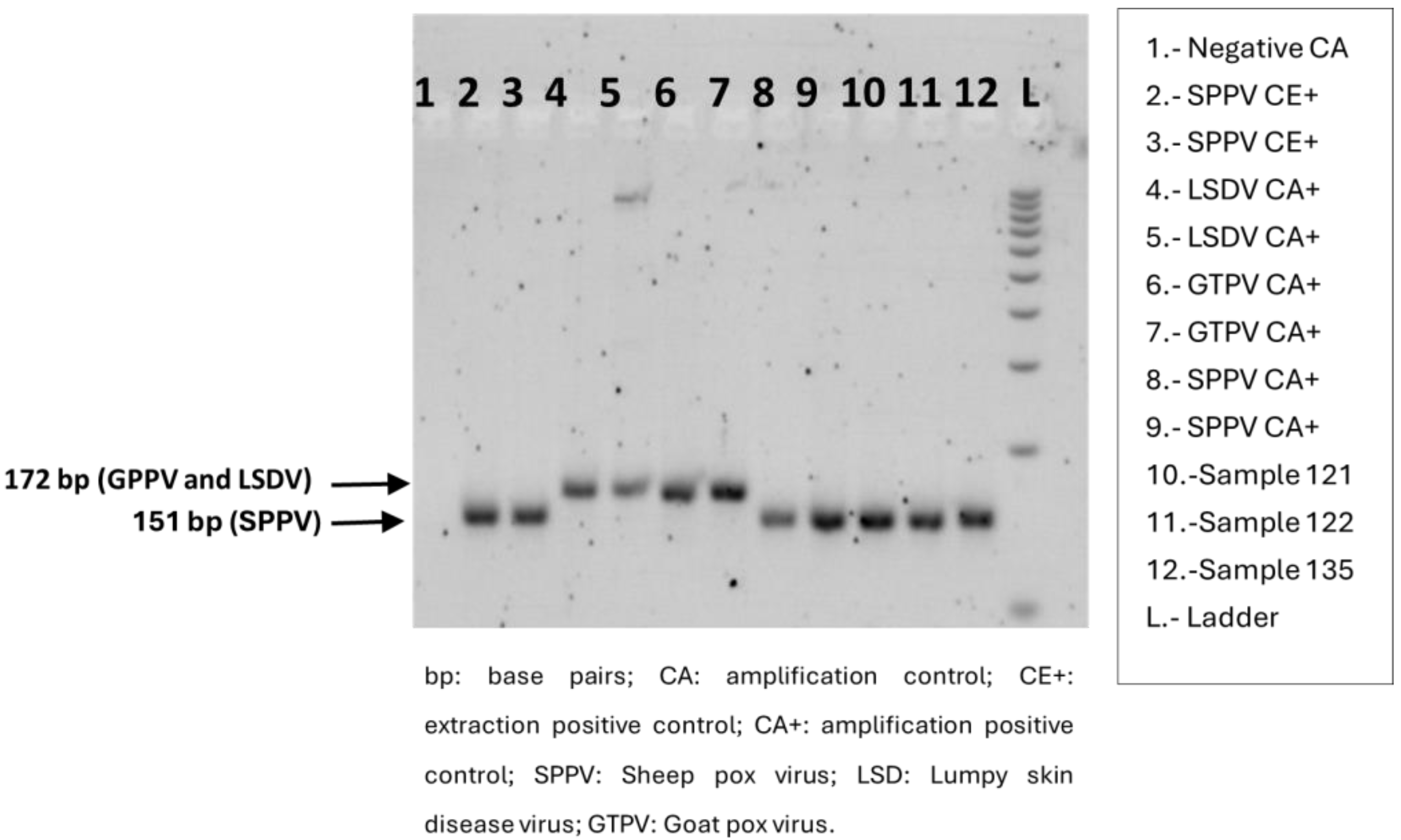
2.6. Sheep Pox Identification by Gel-Based PCR
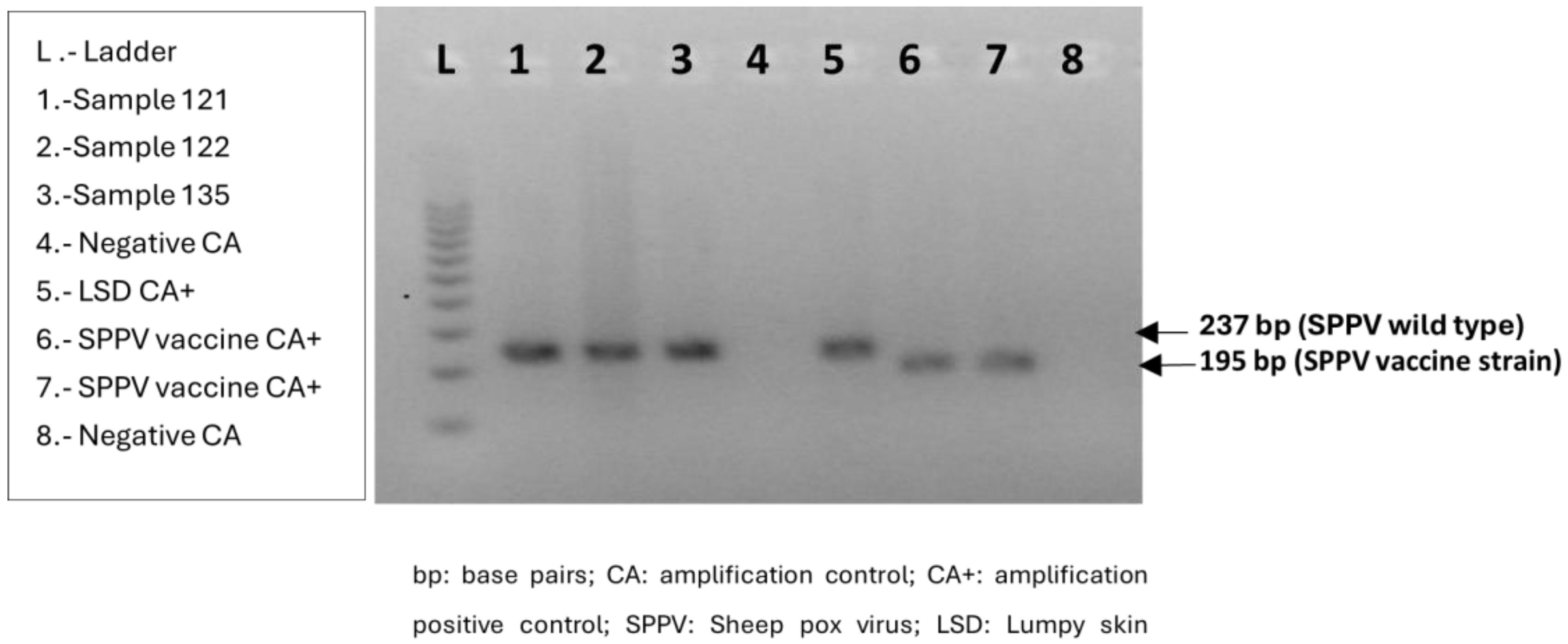
2.7. Differentiation between Field and Vaccine Strain Sheep Pox Virus
2.8. Detection of Specific Antibodies against Capripox Virus by Double Recognition ELISA
2.9. Statistical Analysis
2.10. Ethical Statement
3. Results
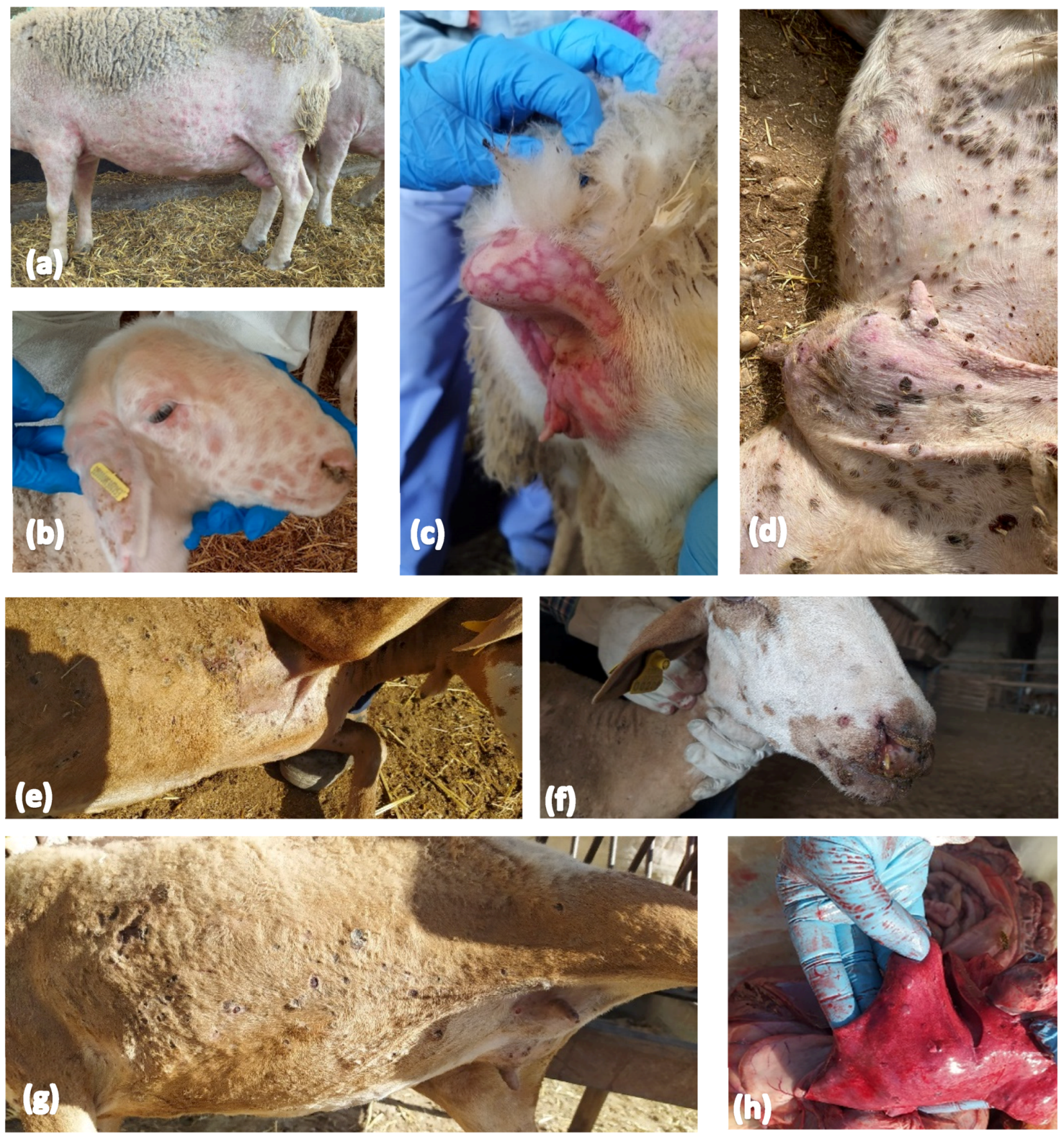
3.1. Confirmation of SPPV in the Index Case in Andalusia
3.2. Overall Clinical and Laboratory Surveillance in Sheep
3.3. Laboratory Surveillance in Positive Farms
| % (Positive/Total Samples) † | ||||||||
|---|---|---|---|---|---|---|---|---|
| Outbreak | Period of Surveillance | % Morbidity (Affected/Census) | % Mortality (Death/Census) | Type of Clinical Signs | EDTA Blood | Oral Swab | Scab | Serum |
| 2022/3 | Epidemiological link | 0.1 (1/890) | 0.0 (0/890) | Mild | n.c. | 100 (3/3) | n.c. | n.c. |
| 2022/6 | Active | 0.4 (1/227) | 0.0 (0/227) | Mild | n.c. | 100 (1/1) | n.c. | n.c. |
| 2022/7 | Active | 0.4 (8/1.877) | 0.0 (0/1.877) | Mild | n.c. | 100 (2/2) | n.c. | n.c. |
| 2022/8 | Active | 0.2 (15/5.075) | 0.0 (0/5.075) | Mild | n.c. | 100 (2/2) | n.c. | n.c. |
| 2022/4 | Active | 2.3 (182/7.654) | 0.0 (0/7.654) | Mild | n.c. | 100 (5/5) | n.c. | n.c. |
| 2022/9 | Active | 2.5 (15/591) | 0.0 (0/591) | Mild | n.c. | 100 (2/2) | n.c. | n.c. |
| 2022/15 | Active | 0.02 (2/7.354) | 0.0 (0/7.354) | Mild | n.c. | 100 (2/2) | n.c. | n.c. |
| Re-visited | no data | no data | Scabs | 41.6 (25/60) | 80 (48/60) | 100 (20/20) | 10 (6/60) | |
| 2022/16 | Active | 0.2 (10/3.591) | 0.0 (0/3.591) | Mild | n.c. | 100 (2/2) | n.c. | n.c. |
| 2022/22 | Active | 0.9 (15/1.519) | 0.3 (5/1.519) | Mild | n.c. | 100 (9/9) | n.c. | n.c. |
| Re-visited | no data | no data | Mild | 62.5 (5/8) | n.c. | n.c. | 37.5 (3/8) | |
| 2022/23 | Active | 3.6 (30/820) | 1.8 (15/820) | Mild | n.c. | 100 (9/9) | n.c. | n.c. |
| 2023/1 * | Reinforced | 3.6 (50/1.359) | 0.0 (0/1.359) | Mild | n.c. | 88.8 (8/9) | n.c. | n.c. |
| 2023/2 | Reinforced | 0.0 (0/3.544) | 0.0 (0/3.544) | No | n.c. | 5–24 (7/145) ** | n.c. | n.c. |
| Revisited | 0.02 (1/3.544) | 0.0 (0/3.544) | Mild | n.c. | 63.8 (23/36) | n.c. | n.c. | |
| 2023/3 | Reinforced | 5.9 (480/8.100) | 0.06 (5/8.100) | Mild | n.c. | 20–100 (30/147) ** | n.c. | n.c. |
| 2023/4 | Reinforced | 0.0 (0/1.216) | 0.0 (0/1.216) | No | n.c. | 16–79 (9/57) ** | n.c. | n.c. |
| Revisited | 0.3 (4/1.216) | 0.0 (0/1.216) | Mild | n.c. | 20 (12/60) | n.c. | 0 (0/60) | |
| 2023/5 | Reinforced | 0.3 (5/1.410) | 0.0 (0/1.410) | Mild | n.c. | 26.6 (16/60) | n.c. | n.c. |
| 2023/6 * | Reinforced | 0.03 (1/3.142) | 0.0 (0/3.142) | Mild | n.c. | 100 (2/2) | n.c. | n.c. |
| Revisited | no data | no data | Mild | n.c. | 1.3–6.8 (2/145) ** | n.c. | n.c. | |
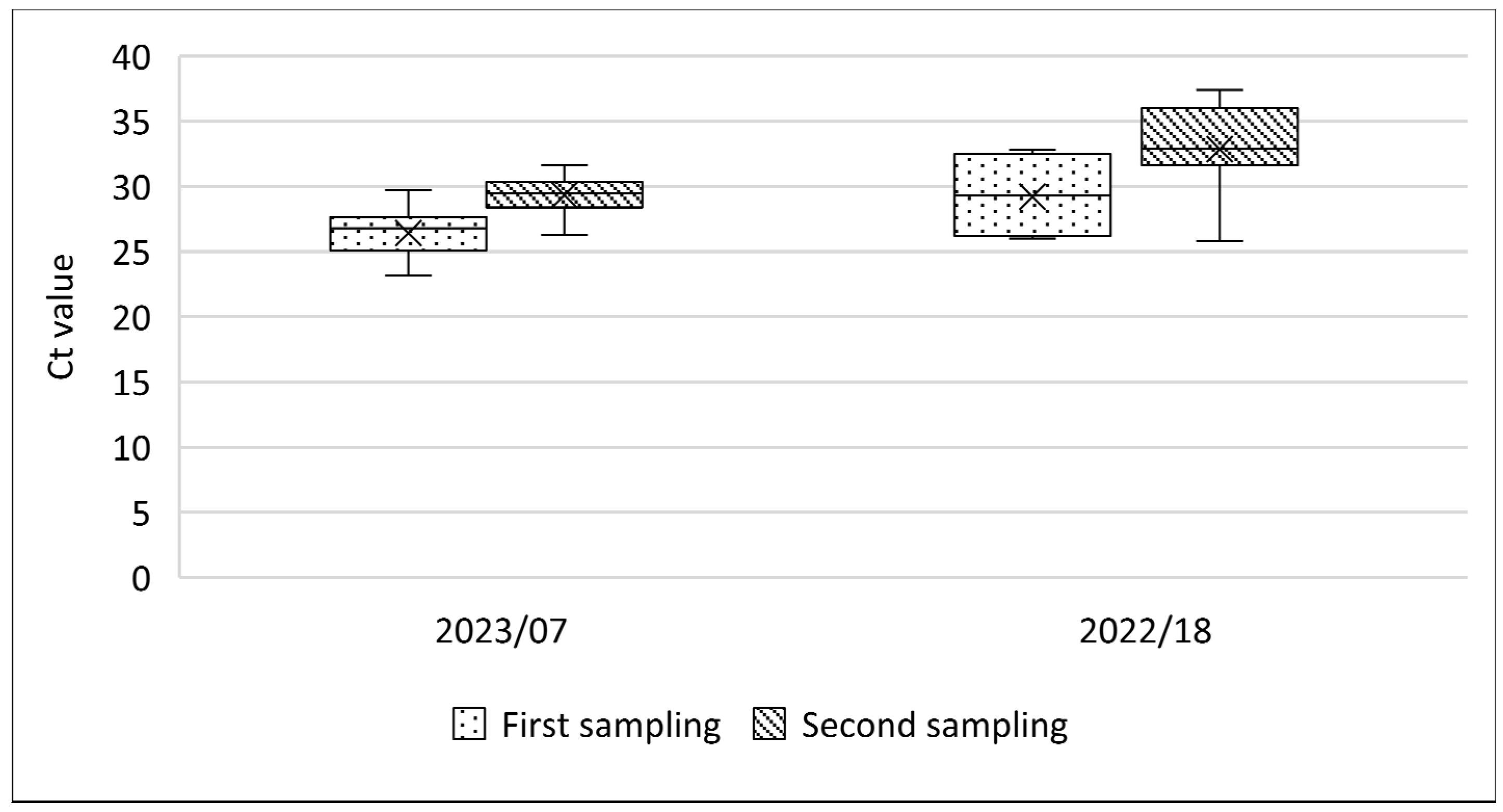
| 2023/7 * | Active | 6.2 (21/334) | 0.5 (2/334) | Mild | n.c. | 100 (21/21) | n.c. | n.c. |
| Revisited | no data | no data | Scabs | 45 (27/60) | 98.3 (59/60) | 100 (16/16) | 16.6 (10/60) | |
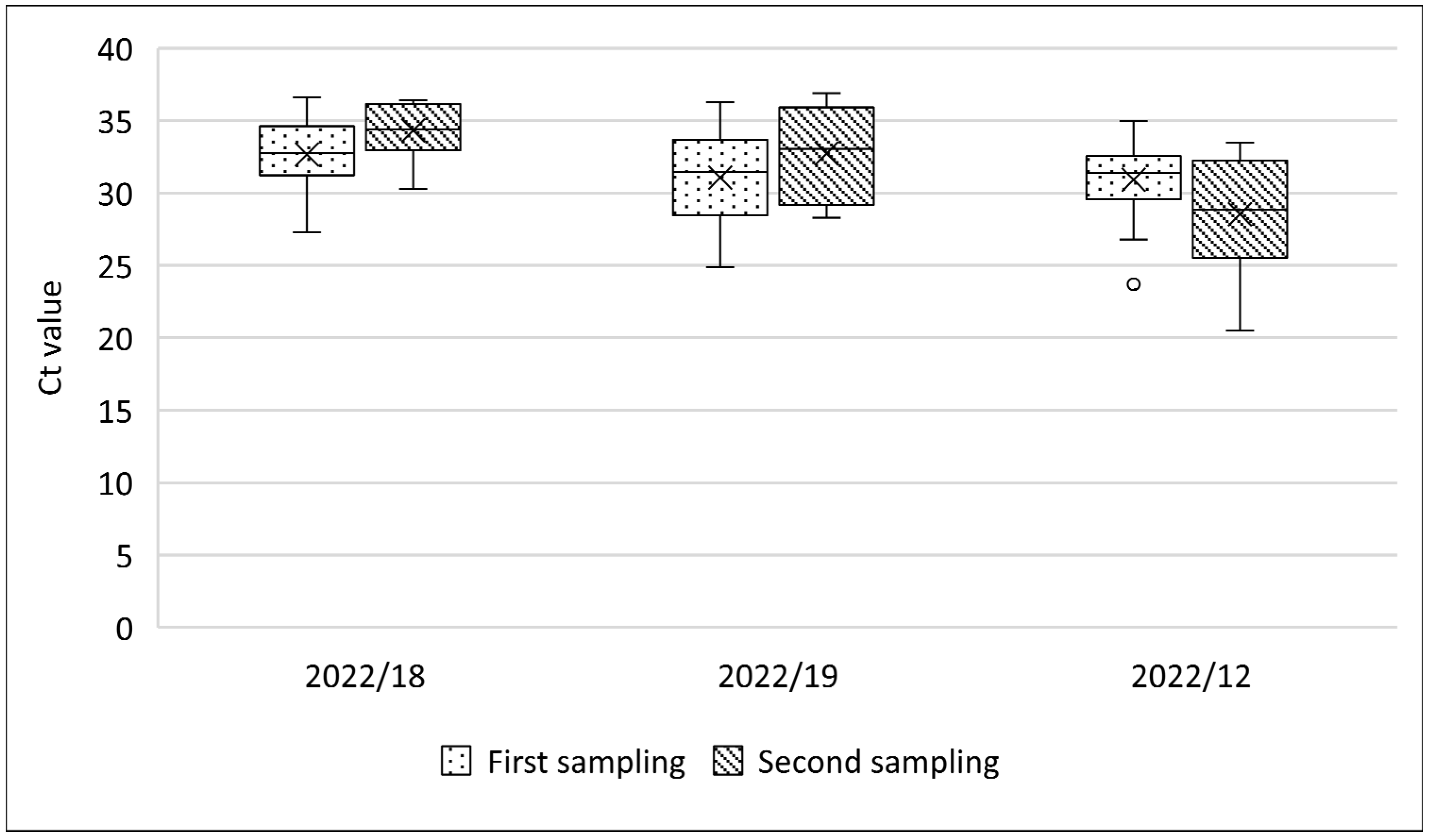
3.4. Evolution of SPPV Infection in Affected Farms
3.5. SPPV Detection in Goat Samples
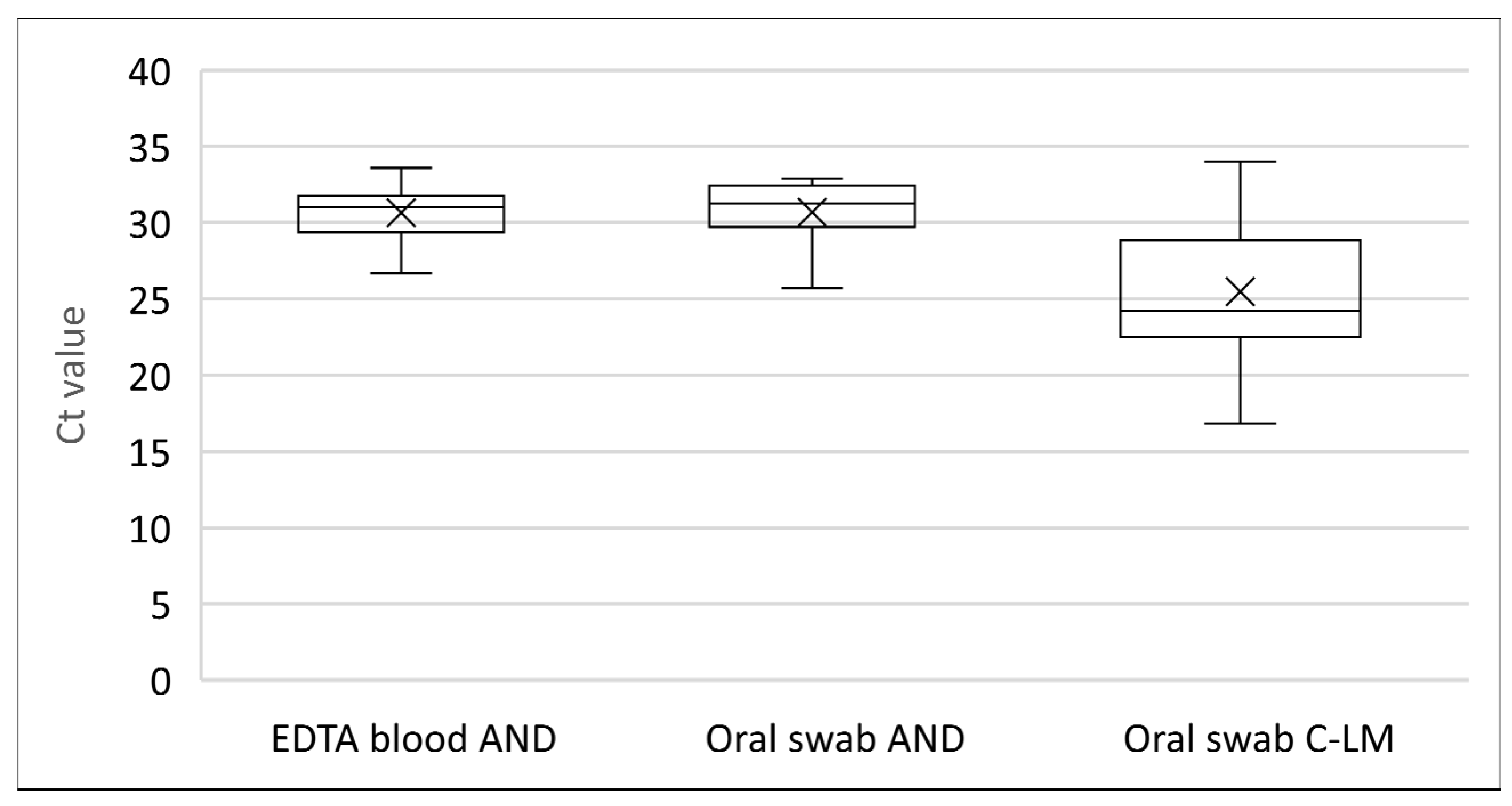
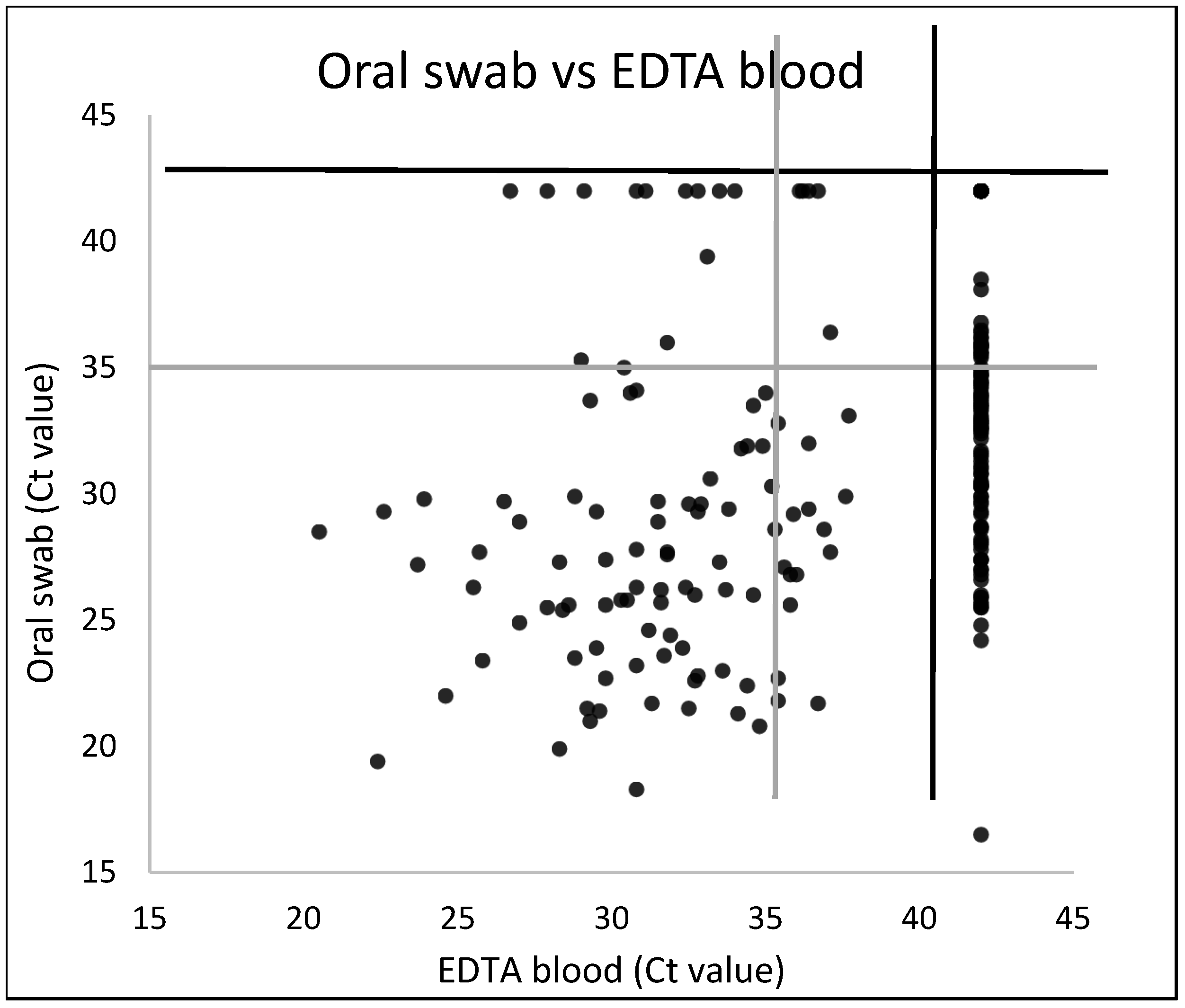
3.6. Comparative SPPV Detection in Blood and Oral Swabs Early after Infection
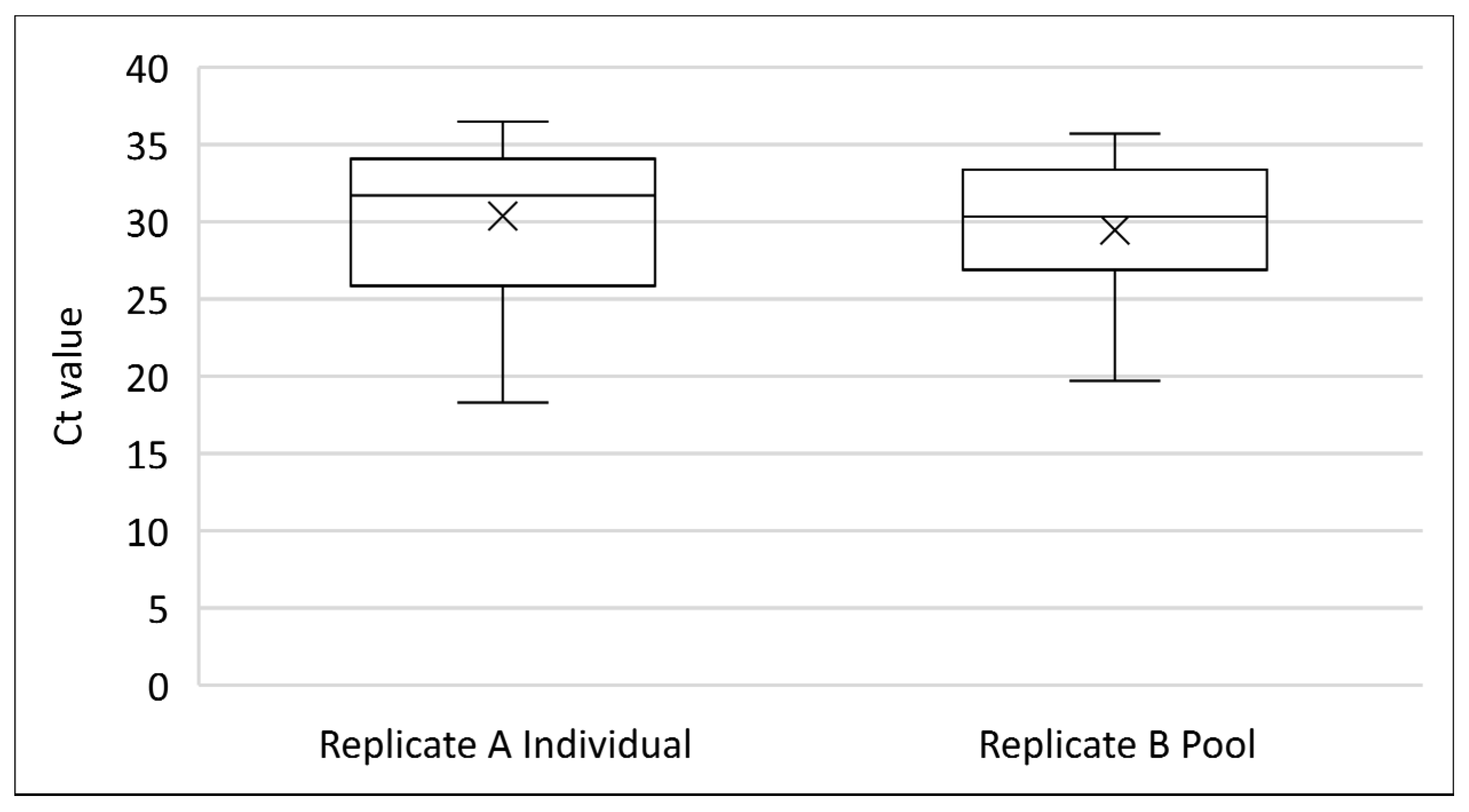
3.7. Effect of Oral Swab Pooling on Sensitivity
3.8. Laboratory Testing during Repopulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WOAH. Sheep pox and goat pox. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; WOAH: Paris, France, 2024; Chapter 3.8.12; Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.08.12_S_POX_G_POX.pdf (accessed on 10 January 2024).
- Buller, R.M.; Arif, B.M.; Black, D.N.; Dumbell, K.R.; Esposito, J.J.; Lefkowitz, E.J.; Moss, B.; Mercer, A.A.; Moyer, R.W.; Skinner, M.A.; et al. Family Poxviridae. In Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses; Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A., Fauquet, C.M., Eds.; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Babiuk, S.; Bowden, T.R.; Boyle, D.B.; Wallace, D.B.; Kitching, R.P. Capripoxviruses: An emerging worldwide threat to sheep, goats and cattle. Transbound. Emerg. Dis. 2008, 55, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Hosamani, M.; Nandi, S.; Mondal, B.; Singh, R.K.; Rasool, T.J.; Bandyopadhyay, S.K.A. Vero cell-attenuated Goatpox virus provides protection against virulent virus challenge. Acta Virol. 2003, 48, 15–21. [Google Scholar]
- Balinsky, C.A.; Delhon, G.; Smoliga, G.; Prarat, M.; French, R.A.; Geary, S.J.; Rock, D.L.; Rodriguez, L.L. Rapid preclinical detection of sheeppox virus by a real-time PCR assay. J. Clin. Microbiol. 2008, 46, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.M.; Chu, Y.F.; Wu, G.H.; Zhao, Z.X.; Li, J.; Zhu, H.X.; Zhang, Q. An outbreak of sheep pox associated with goat poxvirus in Gansu province of China. Vet. Microbiol. 2012, 156, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Encyclopaedia of Virology, 2nd ed.; Granoff, A., Webster, R.G., Eds.; Elsevier Science: Amsterdam, The Netherlands, 1999; ISBN 9780080547978. [Google Scholar]
- Tuppurainen, E.S.M.; Venter, E.H.; Shisler, J.L.; Gari, G.; Mekonnen, G.A.; Juleff, N.; Lyons, N.A.; De Clercq, K.; Upton, C.; Bowden, T.R.; et al. Review: Capripoxvirus Diseases: Current Status and Opportunities for Control. Transbound. Emerg. Dis. 2017, 64, 729–745. [Google Scholar] [CrossRef] [PubMed]
- European Union. Regulation (EU) 2016/429 of the European Parliament and of the Council of 9 March 2016 on Transmissible Animal Diseases and Amending and Repealing Certain Acts in the Area of Animal Health (‘Animal Health Law’). Official Journal of the European Union-L84.31.3.2016. 1 November 2016. Available online: http://data.europa.eu/eli/reg/2016/429/oj (accessed on 10 January 2024).
- European Union. Commission Implementing Regulation (EU) 2018/1882 of 3 December 2018 on the Application of Certain Disease Prevention and Control Rules to Categories of Listed Diseases and Establishing a List of Species and Groups of Species Posing a Considerable Risk for the Spread of Those Listed Diseases. Official Journal of the European Union-L308.04.12.2018. 3 December 2018. Available online: http://data.europa.eu/eli/reg_impl/2018/1882/2024-02-01 (accessed on 15 February 2024).
- Bowden, T.R.; Babiuk, S.L.; Parkyn, G.R.; Copps, J.S.; Boyle, D.B. Capripoxvirus tissue tropism and shedding: A quantitative study in experimentally infected sheep and goats. Virology 2008, 371, 380–393. [Google Scholar] [CrossRef] [PubMed]
- Lamien, C.E.; Le Goff, C.; Silber, R.; Wallace, D.B.; Gulyaz, V.; Tuppurainen, E.; Madani, H.; Caufour, P.; Adam, T.; El Harrak, M.; et al. Use of the Capripoxvirus homologue of Vaccinia virus 30 kDa RNA polymerase subunit (RPO30) gene as a novel diagnostic and genotyping target: Development of a classical PCR method to differentiate Goat poxvirus from Sheep poxvirus. Vet. Microbiol. 2011, 149, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Haegeman, A.; Zro, K.; Sammin, D.; Vandenbussche, F.; Ennaji, M.M.; De Clercq, K. Investigation of a Possible Link between Vaccination and the 2010 Sheep Pox Epizootic in Morocco. Transbound. Emerg. Dis. 2016, 63, e278–e287. [Google Scholar] [CrossRef] [PubMed]
- Kitching, R.P.; Taylor, W.P. Clinical and antigenic relationship between isolates of sheep and goat pox viruses. Trop. Anim. Health Prod. 1985, 17, 64–74. [Google Scholar] [CrossRef]
- Mulatu, E.; Feyisa, A. Review: Lumpy Skin Disease. J. Vet. Sci. Technol. 2018, 9, 535. [Google Scholar] [CrossRef]
- Wolff, J.; Abd El Rahman, S.; King, J.; El-Beskawy, M.; Pohlmann, A.; Beer, M.; Hoffmann, B. Establishment of a Challenge Model for Sheeppox Virus Infection. Microorganisms 2020, 8, 2001. [Google Scholar] [CrossRef]
- Babiuk, S.; Wallace, D.B.; Smith, S.J.; Bowden, T.R.; Dalman, B.; Parkyn, G.; Copps, J.; Boyle, D.B. Detection of antibodies against capripoxviruses using an inactivated sheep pox virus ELISA. Transbound. Emerg. Dis. 2009, 56, 132–141. [Google Scholar] [CrossRef]
- Haegeman, A.; De Leeuw, I.; Mostin, L.; Van Campe, W.; Aerts, L.; Vastag, M.; De Clercq, K. An Immunoperoxidase Monolayer Assay (IPMA) for the detection of lumpy skin disease antibodies. J. Virol. Methods 2020, 277, 113800. [Google Scholar] [CrossRef] [PubMed]
- Bhanuprakash, V.; Venkatesan, G.; Balamurugan, V.; Hosamani, M.; Yogisharadhya, R.; Chauhan, R.S.; Pande, A.; Mondal, B.; Singh, R.K. Pox outbreaks in sheep and goats at Makhdoom (Uttar Pradesh), India: Evidence of sheeppox virus infection in goats. Transbound. Emerg. Dis. 2010, 57, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Santhamani, R.; Venkatesan, G.; Minhas, S.K.; Shivachandra, S.B.; Muthuchelvan, D.; Pandey, A.B.; Ramakrishnan, M.A. Detection and characterization of atypical capripoxviruses among small ruminants in India. Virus Genes 2015, 51, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Beard, P.M.; Sugar, S.; Bazarragchaa, E.; Gerelmaa, U.; Tserendorj, S.; Tuppurainen, E.; Sodnomdarjaa, R. A description of two outbreaks of capripoxvirus disease in Mongolia. Vet. Microbiol. 2010, 142, 427–431. [Google Scholar] [CrossRef] [PubMed]


| Period | Area | Number of Farms | Number of Samples | |||
|---|---|---|---|---|---|---|
| (Positive/Sampled) | Oral Swab | EDTA Blood | Scab | Serum | ||
| First period of Active surveillance | AND | 12/17 | 309 | 817 | 11 | 372 |
| C-LM | 10/11 † | 189 | 68 | 20 | 0 | |
| Reinforced active surveillance | AND | 0/49 | 3.772 | 0 | 0 | 0 |
| C-LM | 6/286 | 31.647 | 10 | 17 | 60 | |
| Second period of Active surveillance | AND | 0 | n.c. | n.c. | n.c. | n.c. |
| C-LM | 1/25 | 470 | 60 | 16 | 60 | |
| 29/388 | 36,387 | 955 | 64 | 492 | ||
| Period | Area | Number of Farms | Number of Samples | |||
|---|---|---|---|---|---|---|
| Sampled | Oral Swab | EDTA Blood | Scab | Serum | ||
| First period of Active surveillance | AND | 10 | 102 | 297 | 0 | 124 |
| C-LM | 0 | n.c. | n.c. | n.c. | n.c. | |
| Reinforced active surveillance | AND | 7 | 173 | 57 | 0 | 0 |
| C-LM | 12 | 1479 | 0 | 0 | 43 | |
| Second period of Active surveillance | AND | 0 | n.c. | n.c. | n.c. | n.c. |
| C-LM | 1 | 3 | 0 | 0 | 0 | |
| 30 | 1757 | 354 | 0 | 167 | ||
| % (Positive/Total Samples) † | ||||||||
|---|---|---|---|---|---|---|---|---|
| Outbreak | Period of Surveillance | % Morbidity (Affected/Census) | % Mortality (Death/Census) | Type of Clinical Signs | EDTA Blood | Oral Swab | Scab | Serum |
| 2022/1 * | Index case (passive) | 15.9 (50/314) | 9.5 (30/314) | scabs | 45 (27/60) | n.c. | 100 (27/27) | 40 (24/60) |
| 2022/2 * | Active | 0.5 (1/170) | 0.0 (0/170) | mild | 1.7 (1/58) | n.c. | n.c. | 1.7 (1/58) |
| 2022/5 * | Active | 0.5 (2/340) | 0.0 (0/340) | mild | 3.3 (2/59) | 100 (2/2) | n.c. | 0 (0/59) |
| 2022/10 * | Active | 2.7 (3/110) | 0.0 (0/110) | mild | 3.9 (2/51) | 100 (3/3) | n.c. | 0 (0/51) |
| 2022/11 * | Active | 1.2 (1/79) | 0.0 (0/79) | mild | 0 (0/38) | 100 (1/1) | n.c. | 0 (0/38) |
| 2022/12 * | Active | 1.0 (7/639) | 0.0 (0/639) | mild | 43 (26/60) | 100 (4/4) | n.c. | 1.6 (1/60) |
| Re-visited | no data | no data | mild | 76.1 (16/21) | 100 (21/21) | n.c. | 19 (4/21) | |
| 2022/13 * | Active | 26 (50/192) | 0.0 (0/192) | mild | 58.4 (31/53) | n.c. | n.c. | n.c. |
| 2022/14 | Active | 9.5 (4/42) | 0.0 (0/42) | mild | 44 (11/25) | n.c. | n.c. | n.c. |
| 2022/17 | Active | 0.2 (1/373) | 0.0 (0/373) | mild | 15 (9/60) | 76.9 (10/13) | n.c. | n.c. |
| 2022/18 * | Active | 15.4 (15/97) | 0.0 (0/97) | mild | 32 (16/50) | 97.6 (42/43) | n.c. | n.c. |
| Re-visited | no data | no data | scabs | 40 (24/60) | 51.6 (31/60) | 100 (9/9) | 16.6 (10/60) | |
| 2022/19 * | Active | 8.2 (30/364) | 0.0 (0/364) | mild | 47.7 (32/67) | 100 (7/7) | n.c. | n.c. |
| Re-visited | no data | no data | scabs | 36.6 (22/60) | 96.6 (58/60) | 100 (2/2) | 5 (3/60) | |
| 2022/20 * | Active | 9.7 (20/206) | 0.0 (0/206) | mild | 52 (26/50) | n.c. | n.c. | n.c. |
| 2022/21 * | Active | 0.6 (1/149) | 0.0 (0/149) | mild | 17.3 (9/52) | 40 (2/5) | n.c. | n.c. |
| EDTA Blood | |||
|---|---|---|---|
| Positive or Inconclusive | Negative | ||
| Oral swab | Positive or inconclusive | 91 | 97 |
| Negative | 13 | 28 | |
| % (Positive or Inconclusive/Total Samples) | ||||
|---|---|---|---|---|
| Outbreak | Interval to Repopulation (Days) * | Oral Swab | Serum | Environmental Swab |
| 2022/3 | 154 | 0 (0/60) | 0 (0/60) | n.c. |
| 2022/4 | 153 | 0 (0/60) | 0 (0/60) | n.c. |
| 2022/6 | 150 | 0 (0/60) | 0 (0/60) | n.c. |
| 2022/7 | 150 | 3.3% (2/60) | 0 (0/60) | n.c. |
| 156 | 3.5% (2/57) | n.c. | n.c. | |
| 167 | 8.7% (5/57) | 0 (0/57) | n.c. | |
| 182 | 12.2% (7/57) | 0 (0/57) | n.c. | |
| 202 | n.c. | 0 (0/56) | 10% (1/10) | |
| 2022/8 | 150 | 0 (0/60) | 0 (0/60) | n.c. |
| 2022/9 | 153 | 0 (0/60) | n.c. | n.c. |
| 2022/10 | 428 | 0 (0/31) | 0 (0/31) | n.c. |
| 2022/15 | 140 | 0 (0/60) | 0 (0/60) | n.c. |
| 2022/17 | 274 | 0 (0/08) | n.c. | n.c. |
| 2022/20 | 369 | 0 (0/37) | n.c. | n.c. |
| 2022/22 | 305 | 0 (0/47) | n.c. | n.c. |
| 2023/2 | 185 | 0 (0/43) | n.c. | n.c. |
| 2023/3 | 265 | 0 (0/91) | n.c. | n.c. |
| 2023/6 | 189 | 0 (0/94) | n.c. | n.c. |
| (16/942) | (0/561) | (1/10) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalba, R.; Haegeman, A.; Ruano, M.J.; Gómez, M.B.; Cano-Gómez, C.; López-Herranz, A.; Tejero-Cavero, J.; Capilla, J.; Bascuñan, M.V.; De Regge, N.; et al. Lessons Learned from Active Clinical and Laboratory Surveillance during the Sheep Pox Virus Outbreak in Spain, 2022–2023. Viruses 2024, 16, 1034. https://doi.org/10.3390/v16071034
Villalba R, Haegeman A, Ruano MJ, Gómez MB, Cano-Gómez C, López-Herranz A, Tejero-Cavero J, Capilla J, Bascuñan MV, De Regge N, et al. Lessons Learned from Active Clinical and Laboratory Surveillance during the Sheep Pox Virus Outbreak in Spain, 2022–2023. Viruses. 2024; 16(7):1034. https://doi.org/10.3390/v16071034
Chicago/Turabian StyleVillalba, Rubén, Andy Haegeman, María José Ruano, María Belén Gómez, Cristina Cano-Gómez, Ana López-Herranz, Jesús Tejero-Cavero, Jaime Capilla, María Victoria Bascuñan, Nick De Regge, and et al. 2024. "Lessons Learned from Active Clinical and Laboratory Surveillance during the Sheep Pox Virus Outbreak in Spain, 2022–2023" Viruses 16, no. 7: 1034. https://doi.org/10.3390/v16071034
APA StyleVillalba, R., Haegeman, A., Ruano, M. J., Gómez, M. B., Cano-Gómez, C., López-Herranz, A., Tejero-Cavero, J., Capilla, J., Bascuñan, M. V., De Regge, N., & Agüero, M. (2024). Lessons Learned from Active Clinical and Laboratory Surveillance during the Sheep Pox Virus Outbreak in Spain, 2022–2023. Viruses, 16(7), 1034. https://doi.org/10.3390/v16071034







