Methodology for the Differential Classification of Dengue and Chikungunya According to the PAHO 2022 Diagnostic Guide
Abstract
:1. Introduction
2. Background
2.1. Machine Learning in the Differential Classification of Arboviruses
2.2. Quality Metrics for Model Evaluation
Confusion Matrix
- positive observation.
- Negative (N): The observation is not positive; that is, it is negative.
- True Positive (TP): The model correctly predicted the positive class.
- True Negative (TN): The model correctly predicts the negative class.
- False Positive (FP): Known as a type 1 error, it occurs when the model incorrectly predicts the positive class when it is of the negative class.
- False Negative (FN): Known as a type 2 error, it occurs when the model incorrectly predicts the negative class when it is of the positive class.
- Accuracy: This metric indicates the proportion of correct predictions made by a model relative to the total predictions [39].
- Precision: This is known as the positive predictive value and is the ratio of relevant instances to retrieved instances [39].
- Sensitivity: The rate of hits or the true positive rate (TPR) was calculated as the ratio of the total number of instances retrieved. This quality metric provides an answer to the real positives that are correctly identified [40].
- Specificity: This metric is known as the true negative rate (TNR), and it evaluates the proportion of true negatives that are correctly identified. This is the counterpart to the sensitivity [40].
- F1 Score: This metric is known as the harmonic mean of precision and recall, and is a measure that takes into account all the results of the confusion matrix, allowing a metric of the precision and robustness of the model [39].
2.3. Linear Interpolation
2.4. Synthesis of a Guide for the Diagnosis and Treatment of Dengue, Chikungunya, and Zika in the American Region
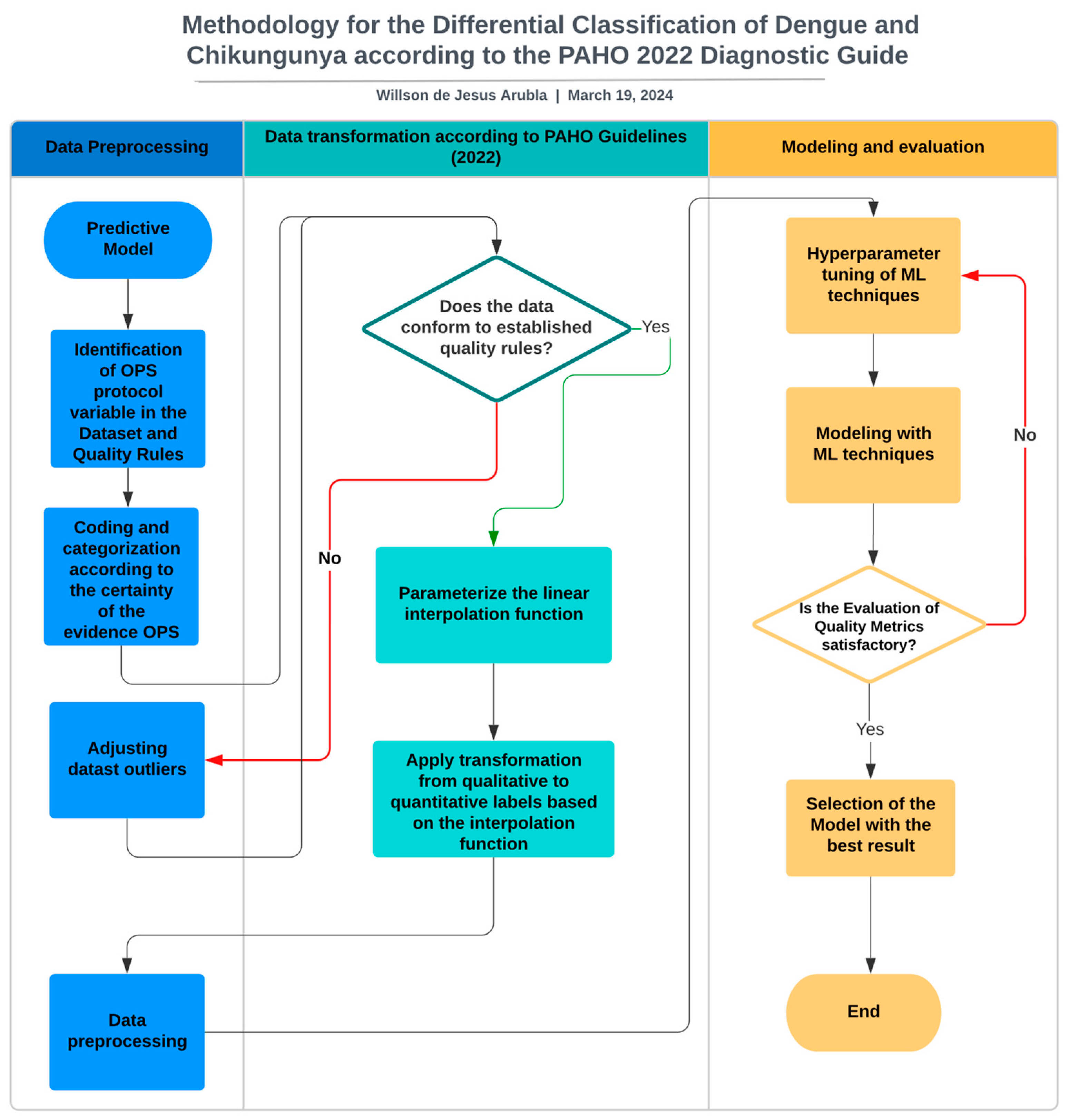
3. Materials and Methods
3.1. Identification of the PAHO Protocol Variables in the Dataset and Quality Rules
3.1.1. Dataset Selection
Age Rule
Rules for the Course of Symptoms of the Disease
3.2. Coding and Categorization According to the Certainty of the Evidence from the PAHO
3.3. Adjusting Datast Outliers
3.4. Parameterise the Linear Interpolation Function
- = the interpolated value for day x.
- = the value of the day to be interpolated.
- = the range of day values ranging from 0 to 12.
- , = values of the established range High, Moderate, Low, and Very Low towards which the interpolation of days is desired.
3.5. The Transformation from Qualitative to Quantitative Labels Was Applied Based on the Interpolation Function
| Algorithm 1 The Transformation from Qualitative to Quantitative Labels Was Applied Based on the Interpolation Function |
| Define x, x0, x1, y0, y1 as real numbers Define x_min = 0, x_max = 12, cero = 0 as constants Define índice = 0 as an integer Define Very_low_value_0 = 0.0, Very_low_value_1 = 0.25, Low_value_0 = 0.26, Low_value_1 = 0.50, Moderate_value_0 = 0.51, Moderate_value_1 = 0.75, High_value_0 = 0.76, High_value_1 = 1 as constants Function LinearInterpolation (x, x0, x1, y0, y1): y = y0 + ((x − x0) * (y1 − y0)) / (x1 − x0) Return y For each row in the dataset dataset1: For each index and element in the row until the second-to-last element: If the last element of the row is “chikungunya” and the current element is “yes”: Apply the interpolation function Assign the interpolated value to the element Else, if the last element of the row is “chikungunya” and the current element is “no”: Assign the value of cero to the element Else, if the last element of the row is “dengue” and the current element is “yes”: Apply the interpolation function Assign the interpolated value to the element Else, if the last element of the row is “dengue” and the current element is “no”: Assign the value of cero to the element End for End for End function |
3.6. Data Preprocessing
3.7. Hyperparameter Tuning of ML Techniques
3.8. Modelling with ML Techniques
3.9. Selection of the Model with the Best Result
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNITED NATIONS Sustainable Development Goals. Available online: https://www.un.org/sustainabledevelopment/ (accessed on 13 March 2024).
- Lambrechts, L.; Scott, T.W.; Gubler, D.J. Consequences of the Expanding Global Distribution of Aedes Albopictus for Dengue Virus Transmission. PLoS Neglected Trop. Dis. 2010, 4, e646. [Google Scholar] [CrossRef] [PubMed]
- Chaw, J.K.; Chaw, S.H.; Quah, C.H.; Sahrani, S.; Ang, M.C.; Zhao, Y.; Ting, T.T. A Predictive Analytics Model Using Machine Learning Algorithms to Estimate the Risk of Shock Development among Dengue Patients. Healthc. Anal. 2024, 5, 100290. [Google Scholar] [CrossRef]
- PAHO/WHO Epidemiological Update—Dengue, Chikungunya and Zika—10 June 2023—PAHO/WHO | Pan American Health Organization. Available online: https://www.paho.org/en/documents/epidemiological-update-dengue-chikungunya-and-zika-10-june-2023 (accessed on 13 March 2024).
- WHO Dengue- Global Situation. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON498 (accessed on 13 March 2024).
- Rigau-Pérez, J.G.; Clark, G.G.; Gubler, D.J.; Reiter, P.; Sanders, E.J.; Vorndam, A.V. Dengue and Dengue Haemorrhagic Fever. Lancet 1998, 352, 971–977. [Google Scholar] [CrossRef] [PubMed]
- PAHO Síntesis de evidencia: Directrices para el diagnóstico y el tratamiento del dengue, el chikunguña y el zika en la Región de las Américas. Rev. Panam. Salud Pública 2022, 46, 1. [CrossRef]
- Rico-Mendoza, A.; Porras-Ramírez, A.; Chang, A.; Encinales, L.; Lynch, R. Co-Circulation of Dengue, Chikungunya, and Zika Viruses in Colombia from 2008 to 2018. Rev. Panam. Salud Pública 2019, 43, 1. [Google Scholar] [CrossRef] [PubMed]
- Villamil-Gómez, W.E.; Rodríguez-Morales, A.J.; Uribe-García, A.M.; González-Arismendy, E.; Castellanos, J.E.; Calvo, E.P.; Álvarez-Mon, M.; Musso, D. Zika, Dengue, and Chikungunya Co-infection in a Pregnant Woman from Colombia. Int. J. Infect. Dis. 2016, 51, 135–138. [Google Scholar] [CrossRef]
- Caicedo, D.M.; Méndez, A.C.; Tovar, J.R.; Osorio, L.; Caicedo, D.M.; Méndez, A.C.; Tovar, J.R.; Osorio, L. Desarrollo de algoritmos clínicos para el diagnóstico del dengue en Colombia. Biomédica 2019, 39, 170–185. [Google Scholar] [CrossRef]
- Carlos, M.A.; Nogueira, M.; Machado, R.J. Analysis of Dengue Outbreaks Using Big Data Analytics and Social Networks. In Proceedings of the 2017 4th International Conference on Systems and Informatics (ICSAI), Hangzhou, China, 11–13 November 2017; IEEE: Hangzhou, China, 2017; pp. 1592–1597. [Google Scholar]
- Manogaran, G.; Lopez, D. A Gaussian Process Based Big Data Processing Framework in Cluster Computing Environment. Clust. Comput. 2018, 21, 189–204. [Google Scholar] [CrossRef]
- Noorbakhsh-Sabet, N.; Zand, R.; Zhang, Y.; Abedi, V. Artificial Intelligence Transforms the Future of Health Care. Am. J. Med. 2019, 132, 795–801. [Google Scholar] [CrossRef]
- Wiljer, D.; Hakim, Z. Developing an Artificial Intelligence–Enabled Health Care Practice: Rewiring Health Care Professions for Better Care. J. Med. Imaging Radiat. Sci. 2019, 50, S8–S14. [Google Scholar] [CrossRef] [PubMed]
- Bharambe, A.; Chandorkar, A.A.; Kalbande, D. A Deep Learning Approach for Dengue Tweet Classification. In Proceedings of the 2021 Third International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 2–4 September 2021; IEEE: Coimbatore, India, 2021; pp. 1043–1047. [Google Scholar]
- Khotimah, P.H.; Fachrur Rozie, A.; Nugraheni, E.; Arisal, A.; Suwarningsih, W.; Purwarianti, A. Deep Learning for Dengue Fever Event Detection Using Online News. In Proceedings of the 2020 International Conference on Radar, Antenna, Microwave, Electronics, and Telecommunications (ICRAMET), Virtual, 18–20 November 2020; IEEE: Tangerang, Indonesia, 2020; pp. 261–266. [Google Scholar]
- Gambhir, S.; Sanjay, K.M.; Jaypee, Y.K. The Diagnosis of Dengue Disease: An Evaluation of Three Machine Learning Approaches. Int. J. Healthc. Inf. Syst. Inform. 2018, 13, 1–9. [Google Scholar] [CrossRef]
- Acosta Torres, J.; Oller Meneses, L.; Sokol, N.; Balado Sardiñas, R.; Montero Díaz, D.; Balado Sansón, R.; Sardiñas Arce, M.E. Técnica Árboles de Decisión Aplicada al Método Clínico En El Diagnóstico Del Dengue. Rev. Cuba. Pediatría 2016, 88, 441–453. [Google Scholar]
- Arrubla-Hoyos, W.; Seveiche-Maury, Z.; Saeed, K.; Gómez, J.E.G.; De-La-Hoz-Franco, E. Comparison of Classical Machine Learning and Ensemble Techniques in the Context of Dengue Severity Prediction. In Proceedings of the 2023 IEEE Colombian Caribbean Conference (C3), Barranquilla, Colombia, 22–25 November 2023; pp. 1–5. [Google Scholar]
- Tanner, L.; Schreiber, M.; Low, J.G.; Ong, A.; Tolfvenstam, T.; Lai, Y.L.; Ng, L.C.; Leo, Y.S.; Thi Puong, L.; Vasudevan, S.G.; et al. Decision Tree Algorithms Predict the Diagnosis and Outcome of Dengue Fever in the Early Phase of Illness. PLoS Neglected Trop. Dis. 2008, 2, e196. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.-S.; Weng, T.-C.; Wang, J.-D.; Han, H.-C.; Cheng, H.-C.; Yang, C.-C.; Yu, C.-H.; Liu, Y.-J.; Hu, C.H.; Huang, C.-Y.; et al. Comparing Machine Learning with Case-Control Models to Identify Confirmed Dengue Cases. PLoS Neglected Trop. Dis. 2020, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Fathima, S.A.; Hundewale, N. Comparitive Analysis of Machine Learning Techniques for Classification of Arbovirus. In Proceedings of the 2012 IEEE-EMBS International Conference on Biomedical and Health Informatics, Hong Kong, China, 5–7 January 2012; IEEE: Hong Kong, China, 2012; pp. 376–379. [Google Scholar]
- Sajana, T.; Navya, M.; Gayathri, Y.; Reshma, N. Classification of Dengue Using Machine Learning Techniques. Int. J. Eng. Technol. 2018, 7, 212–218. [Google Scholar] [CrossRef]
- Sanjudevi, D.; Savitha, D. Dengue Fever Prediction Using Classification Techniques. Int. Res. J. Eng. Technol. (IRJET) 2019, 6, 558–563. [Google Scholar]
- Potts, J.A.; Gibbons, R.V.; Rothman, A.L.; Srikiatkhachorn, A.; Thomas, S.J.; Supradish, P.; Lemon, S.C.; Libraty, D.H.; Green, S.; Kalayanarooj, S. Prediction of Dengue Disease Severity among Pediatric Thai Patients Using Early Clinical Laboratory Indicators. PLoS Neglected Trop. Dis. 2010, 4, e769. [Google Scholar] [CrossRef] [PubMed]
- Phakhounthong, K.; Chaovalit, P.; Jittamala, P.; Blacksell, S.D.; Carter, M.J.; Turner, P.; Chheng, K.; Sona, S.; Kumar, V.; Day, N.P.J.; et al. Predicting the Severity of Dengue Fever in Children on Admission Based on Clinical Features and Laboratory Indicators: Application of Classification Tree Analysis. BMC Pediatr. 2018, 18, 109. [Google Scholar] [CrossRef]
- Faisal, T.; Ibrahim, F.; Taib, M.N. A Noninvasive Intelligent Approach for Predicting the Risk in Dengue Patients. Expert Syst. Appl. 2010, 37, 2175–2181. [Google Scholar] [CrossRef]
- Hossain, M.S.; Sultana, Z.; Nahar, L.; Andersson, K. An Intelligent System to Diagnose Chikungunya under Uncertainty. J. Wirel. Mob. Netw. Ubiquitous Comput. Dependable Appl. 2019, 10, 37–54. [Google Scholar]
- Veiga, R.V.; Schuler-Faccini, L.; França, G.V.; Andrade, R.F.; Teixeira, M.G.; Costa, L.C.; Paixão, E.S.; Costa, M. da C.N.; Barreto, M.L.; Oliveira, J.F.; et al. Classification Algorithm for Congenital Zika Syndrome: Characterizations, Diagnosis and Validation. Sci. Rep. 2021, 11, 6770. [Google Scholar] [CrossRef] [PubMed]
- da Silva Neto, S.R.; Tabosa Oliveira, T.; Teixeira, I.V.; Aguiar de Oliveira, S.B.; Souza Sampaio, V.; Lynn, T.; Endo, P.T. Machine Learning and Deep Learning Techniques to Support Clinical Diagnosis of Arboviral Diseases: A Systematic Review. PLoS Negl. Trop. Dis. 2022, 16, e0010061. [Google Scholar] [CrossRef] [PubMed]
- Tabosa de Oliveira, T.; da Silva Neto, S.R.; Teixeira, I.V.; Aguiar de Oliveira, S.B.; de Almeida Rodrigues, M.G.; Sampaio, V.S.; Endo, P.T. A Comparative Study of Machine Learning Techniques for Multiclass Classification of Arboviral Diseases. Front. Trop. Dis. 2022, 2, 769968. [Google Scholar] [CrossRef]
- Medeiros Neto, L.; Rogerio da Silva Neto, S.; Endo, P.T. A Comparative Analysis of Converters of Tabular Data into Image for the Classification of Arboviruses Using Convolutional Neural Networks. PLoS ONE 2023, 18, e0295598. [Google Scholar] [CrossRef] [PubMed]
- Tchapet Njafa, J.-P.; Nana Engo, S.G. Quantum Associative Memory with Linear and Nonlinear Algorithms for the Diagnosis of Some Tropical Diseases. Neural. Netw. 2018, 97, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Quijada, C.; Gomez-Marquez, J.; Hamad-Schifferli, K. Repurposing Old Antibodies for New Diseases by Exploiting Cross-Reactivity and Multicolored Nanoparticles. ACS Nano 2020, 14, 6626–6635. [Google Scholar] [CrossRef]
- Braga, O.; Albuquerque, G.; Oliveira, M.; Monteiro, O. Intelligent Solution for Classification of Diseases Transmitted by Vector Aedes Aegypti. In Proceedings of the Euro American Conference on Telematics and Information Systems, Fortaleza Brazil, 12–15 November 2018; ACM: Fortaleza, Brazil, 2018; pp. 1–5. [Google Scholar]
- Iqbal, N.; Islam, M. Machine Learning for Dengue Outbreak Prediction: A Performance Evaluation of Different Prominent Classifiers. Informatica 2019, 43, 363–371. [Google Scholar] [CrossRef]
- Blackmist Evaluación de los Resultados de los Experimentos de Aprendizaje Automático Automatizado—Azure Machine Learning. Available online: https://learn.microsoft.com/es-es/azure/machine-learning/how-to-understand-automated-ml (accessed on 23 October 2022).
- Narayanasamy, S.K.; Elçi, A. An Effective Prediction Model for Online Course Dropout Rate. Int. J. Distance Educ. Technol. (IJDET) 2020, 18, 94–110. [Google Scholar] [CrossRef]
- Hicks, S.A.; Strümke, I.; Thambawita, V.; Hammou, M.; Riegler, M.A.; Halvorsen, P.; Parasa, S. On Evaluation Metrics for Medical Applications of Artificial Intelligence. Sci. Rep. 2022, 12, 5979. [Google Scholar] [CrossRef]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for Multiclass Classification: An Overview. arXiv 2020, arXiv:2008.05756. [Google Scholar]
- Swasnita, S.; Suparti, S.; Sugito, S. Perhitungan Suku Bunga Efektif Untuk Penentuan Alternatif Pembiayaan Kendaraan Motor Pada Leasing Dan Bank Dengan Metode Interpolasi Linier (Studi Kasus Harga Sepeda Motor Honda Beat Injeksi Terdaftar Bulan September 2014). J. Gaussian 2015, 4, 403–412. [Google Scholar]
- Fu-bin, P.; Yu-bo, Y.; Jian-fei, J. The Influences of Message Jitter on Linear Interpolation for Electronic Transformer Data Synchronization. In Proceedings of the TENCON 2015-2015 IEEE Region 10 Conference, Macao, China, 1–4 November 2015; pp. 1–5. [Google Scholar]
- Veracierta, J.G.P. La Interpolación Lineal En La Distribución t: Valores y Errores. SABER. Rev. Multidiscip. Cons. Investig. Univ. Oriente 2009, 21, 261–268. [Google Scholar]
- Al Amin, I.H.; Lusiana, V.; Hartono, B. Pencarian Lintasan Pada Collision Detection Menggunakan Pendekatan Interpolasi Linier. Seminar Nasional Teknologi Informasi dan Aplikasi Komputer SINTAK 2018, 2, 57–61. Available online: https://www.unisbank.ac.id/ojs/index.php/sintak/article/view/6513 (accessed on 14 November 2018).
- Yan, X.; Enhua, X. ARIMA and Multiple Regression Additive Models for PM2. 5 Based on Linear Interpolation. In Proceedings of the 2020 International Conference on Big Data & Artificial Intelligence & Software Engineering (ICBASE), Bangkok, Thailand, 30 October–1 November 2020; pp. 266–269. [Google Scholar]
- Sujito; Gumilar, L.; Hadi, R.R.; Rodhi Faiz, M.; Syafriyudin; Nugroho, Z.S. Analysis Comparison of Linear Interpolation and Quadratic Interpolation Methods for Forecasting a Growth Total of Electricity Customers in Kotawaringin Barat Regency at 2022-2025 Years. In Proceedings of the 2022 International Electronics Symposium (IES), Surabaya, Indonesia, 9–11 August 2022; pp. 73–78. [Google Scholar]
- World Health Organization. Global Report on Ageism; World Health Organization: Geneva, Switzerland, 2021; ISBN 978-92-4-001686-6. [Google Scholar]
- WHO Dengue y Dengue Grave. Available online: https://www.who.int/es/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 3 October 2021).
- Pan American Health Organization; Espinal, M.A.; World Health Organization. Dengue: Guías para la Atención de Enfermos en la Región de las Américas; World Health Organization: Geneva, Switzerland, 2016; ISBN 978-92-75-31890-4. [Google Scholar]
- Staples, J.E.; Breiman, R.F.; Powers, A.M. Chikungunya Fever: An Epidemiological Review of a Re-Emerging Infectious Disease. Clin. Infect. Dis. 2009, 49, 942–948. [Google Scholar] [CrossRef] [PubMed]
- OMS Chikungunya. Available online: https://www.who.int/es/news-room/fact-sheets/detail/chikungunya (accessed on 7 March 2024).




| Certainty in the Evidence According to the PAHO Guidelines (2022) | Meaning |
|---|---|
High     | Further studies are unlikely to change the confidence in the estimated result. |
Moderate     | New studies could have a significant impact on the confidence in the result. |
Low     | New studies have a high probability of significantly impacting the confidence in the estimated result, potentially modifying it. |
Very Low     | The level of certainty regarding any estimated results is very low. |
 equals 25% and
equals 25% and  0%.
0%.| Certainty in the Evidence According to the PAHO Guidelines (2022) | Manifestations of Dengue | Manifestations of Chikunguña | Manifestations of Zika |
|---|---|---|---|
High     | Thrombocytopenia Progressive increase of haematocrit Leukopenia | Arthralgias | Pruritus |
Moderate     | Anorexia or hyporexia Vomiting Abdominal pain Shaking chills Haemorrhages (includes bleeding on the skin, mucous membranes or both) | Rash Conjunctivitis Arthritis Myalgia or bone pain | Rash Conjunctivitis |
Low     | Retroocular pain Hepatomegaly Headache Diarrhoea Dysgeusia Cough Elevation of transaminases Tourniquet test positive | Bleeding (includes bleeding on the skin or mucous membranes) | Lymphadenopathy Pharyngitis/odynophagia |
Very Low     | − | − | − |
 equals 25% and
equals 25% and  0%.
0%.| Variable | Certainty in the Evidence According to the PAHO Guidelines (2022) | |
|---|---|---|
| Demonstrations in Dengue | Demonstrations in Chikungunya | |
| Myalgia | − | Moderate |
| Headache | Low | − |
| Exanthema | − | Moderate |
| Threw up | Low | − |
| Conjunctivitis | − | Moderate |
| Arthritis | − | Moderate |
| Arthralgia | − | High |
| Laco (symptom—tourniquet test) | Low | − |
| Retroocular pain | Low | − |
| Certainty in the Evidence According to the PAHO Guidelines (2022) | Meaning | Quantitative Value |
|---|---|---|
High     | Further studies are unlikely to change the confidence in the estimated result. | 0.76–1 |
Moderate     | New studies could have a significant impact on the confidence in the result. | 0.51–0.75 |
Low     | New studies have a high probability of significantly impacting the confidence in the estimated result, potentially modifying it. | 0.26–0.50 |
Very Low     | The level of certainty regarding any estimated results is very low. | 0–0.25 |
 equals 25% and
equals 25% and  0%.
0%.| Variable | Certainty in Evidence | Quantitative Weight Assignment | |
|---|---|---|---|
| Manifestations in Dengue | Demonstrations in Chikungunya | ||
| Myalgia | * | Moderate | 0.51–0.75 |
| Headache | Low | * | 0.26–0.50 |
| Exanthema | * | Moderate | 0.51–0.75 |
| Threw up | Low | * | 0.26–0.50 |
| Conjunctivitis | * | Moderate | 0.51–0.75 |
| Arthritis | * | Moderate | 0.51–0.75 |
| Arthralgia | * | High | 0.76–1 |
| Laco (symptom—tourniquet test) | Low | * | 0.26–0.50 |
| Retroocular pain | Low | * | 0.26–0.50 |
| Myalgia | Headache | Exanthema | Threw Up | Conjunctivitis | Arthritis | Arthralgia | Laco (Symptom—Tourniquet Test) | Retroocular Pain | |
|---|---|---|---|---|---|---|---|---|---|
| count | 11,448 | 11,448 | 11,448 | 11,448 | 11,448 | 11,448 | 11,448 | 11,448 | 11,448 |
| mean | 0.199 | 0.140 | 0.098 | 0.081 | 0.014 | 0.041 | 0.373 | 0.006 | 0.045 |
| std | 0.255 | 0.151 | 0.206 | 0.195 | 0.083 | 0.143 | 0.403 | 0.044 | 0.110 |
| min | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25% | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 50% | 0.078 | 0.078 | 0 | 0 | 0 | 0 | 0.078 | 0 | 0 |
| 75% | 0.53 | 0.32 | 0.078 | 0 | 0 | 0 | 0.835 | 0 | 0 |
| max | 0.75 | 0.44 | 0.75 | 0.69 | 0.75 | 1 | 0.44 | 0.44 | 0.44 |
| ML Technique | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Tree Decision | 98.5% | 99% | 99% | 99% |
| KNN | 81% | 81% | 81% | 81% |
| Neural Network | 98% | 98% | 98% | 98% |
| SVM | 98% | 97% | 97% | 97% |
| RF | 99% | 99% | 99% | 99% |
| Baggin | 98% | 98% | 98% | 98% |
| Boosting | 98% | 98% | 98% | 98% |
| Hard-voting | 99% | 99% | 99% | 99% |
| Soft-voting | 98% | 98% | 98% | 98% |
| Stacking | 99% | 99% | 99% | 99% |
| ML Technique | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Tree Decision | 75% | 75% | 75% | 75% |
| KNN | 68% | 68% | 68% | 68% |
| Neural Network | 77% | 77% | 77% | 77% |
| SVM | 73% | 73% | 73% | 73% |
| RF | 78% | 79% | 78% | 78% |
| Baggin | 77% | 77% | 77% | 77% |
| Boosting | 71% | 71% | 71% | 71% |
| Hard-voting | 79% | 79% | 79% | 79% |
| Soft-voting | 77% | 78% | 77% | 77% |
| Stacking | 79% | 79% | 79% | 79% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arrubla-Hoyos, W.; Gómez, J.G.; De-La-Hoz-Franco, E. Methodology for the Differential Classification of Dengue and Chikungunya According to the PAHO 2022 Diagnostic Guide. Viruses 2024, 16, 1088. https://doi.org/10.3390/v16071088
Arrubla-Hoyos W, Gómez JG, De-La-Hoz-Franco E. Methodology for the Differential Classification of Dengue and Chikungunya According to the PAHO 2022 Diagnostic Guide. Viruses. 2024; 16(7):1088. https://doi.org/10.3390/v16071088
Chicago/Turabian StyleArrubla-Hoyos, Wilson, Jorge Gómez Gómez, and Emiro De-La-Hoz-Franco. 2024. "Methodology for the Differential Classification of Dengue and Chikungunya According to the PAHO 2022 Diagnostic Guide" Viruses 16, no. 7: 1088. https://doi.org/10.3390/v16071088
APA StyleArrubla-Hoyos, W., Gómez, J. G., & De-La-Hoz-Franco, E. (2024). Methodology for the Differential Classification of Dengue and Chikungunya According to the PAHO 2022 Diagnostic Guide. Viruses, 16(7), 1088. https://doi.org/10.3390/v16071088






