A Truncated Isoform of Cyclin T1 Could Contribute to the Non-Permissive HIV-1 Phenotype of U937 Promonocytic Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cells
2.3. Immunoprecipitation and Western Blotting
2.4. RT-PCR
2.5. Mass Spectrometry
2.6. Treatments
2.7. Immunofluorescence and Confocal Microscopy
2.8. Transient Transfection and HIV-1-LTR Luciferase Assay
2.9. Statistical Analysis
3. Results
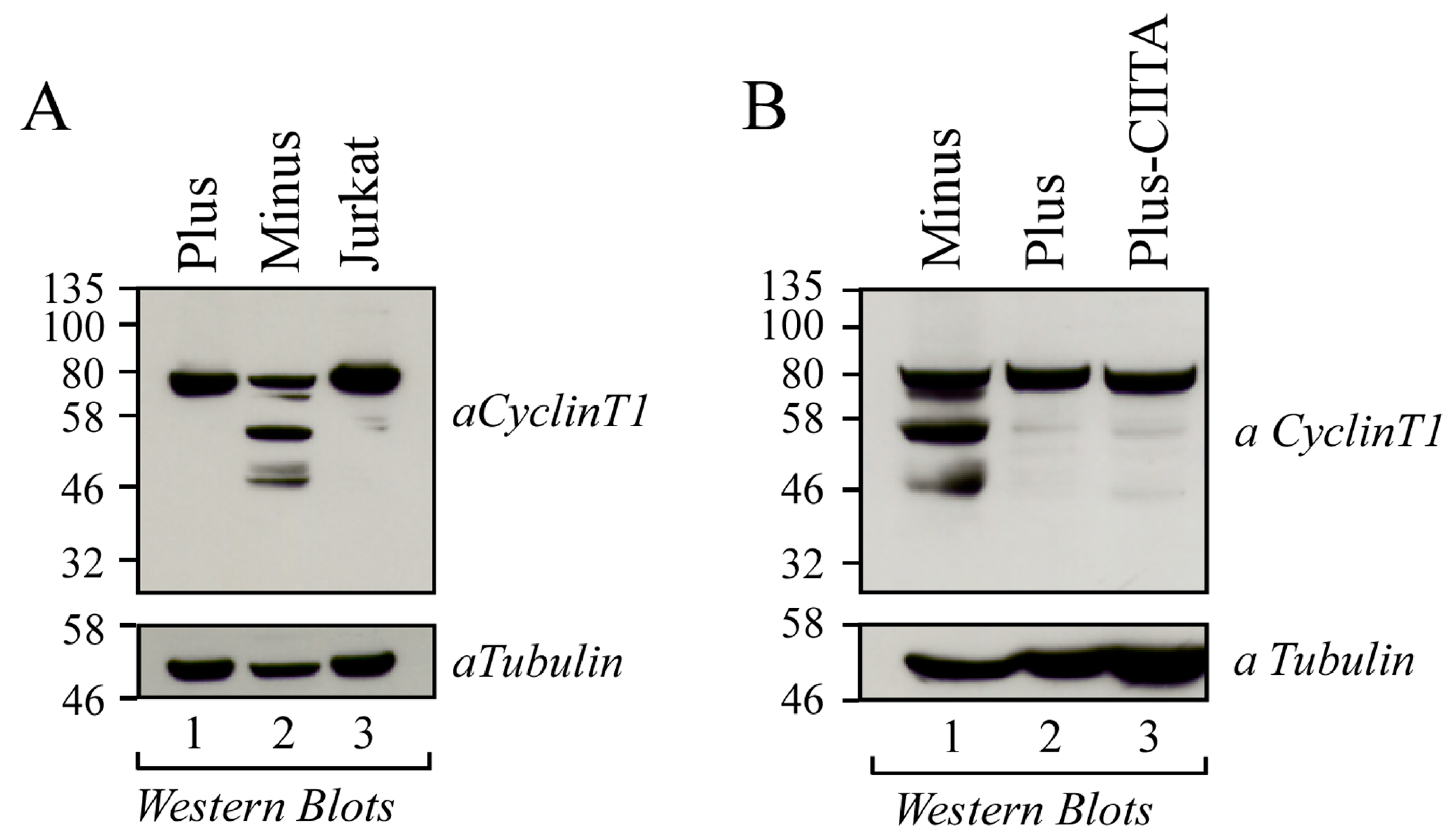
3.1. Cyclin T1 Is Mainly Expressed as a 50 kDa Protein in U937 Minus Cells
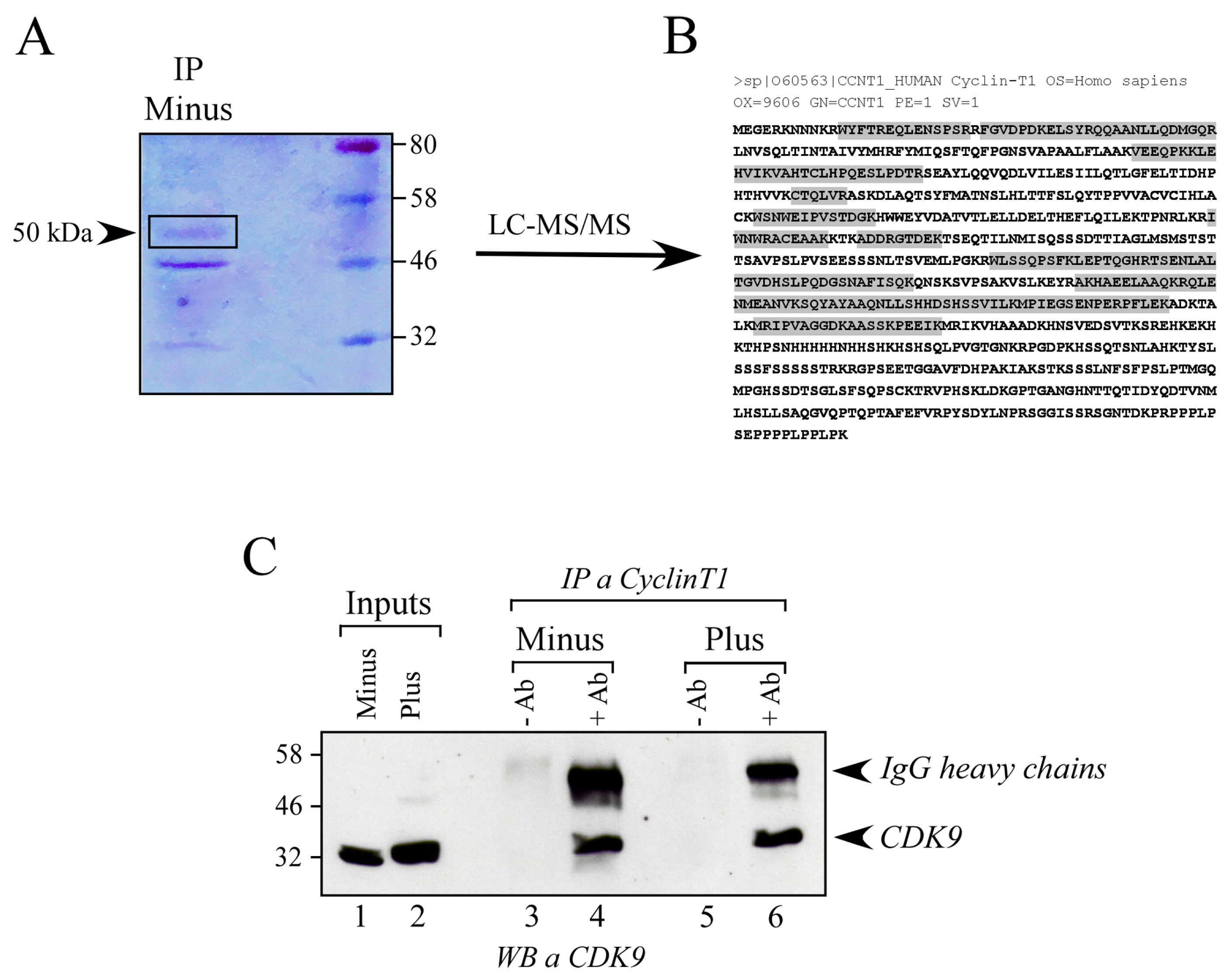
3.2. Mass Spectrometry Analysis of 50 kDa Protein Form Shows It Is Cyclin T1
3.3. The 50 kDa Cyclin T1 Protein Form Does Not Result from Alternative Splicing Nor from Proteasome-Dependent Degradation
3.4. IFNγ Treatment Reduces the Amount of 50 kDa Cyclin T1 Protein Form in U937 Minus Cells
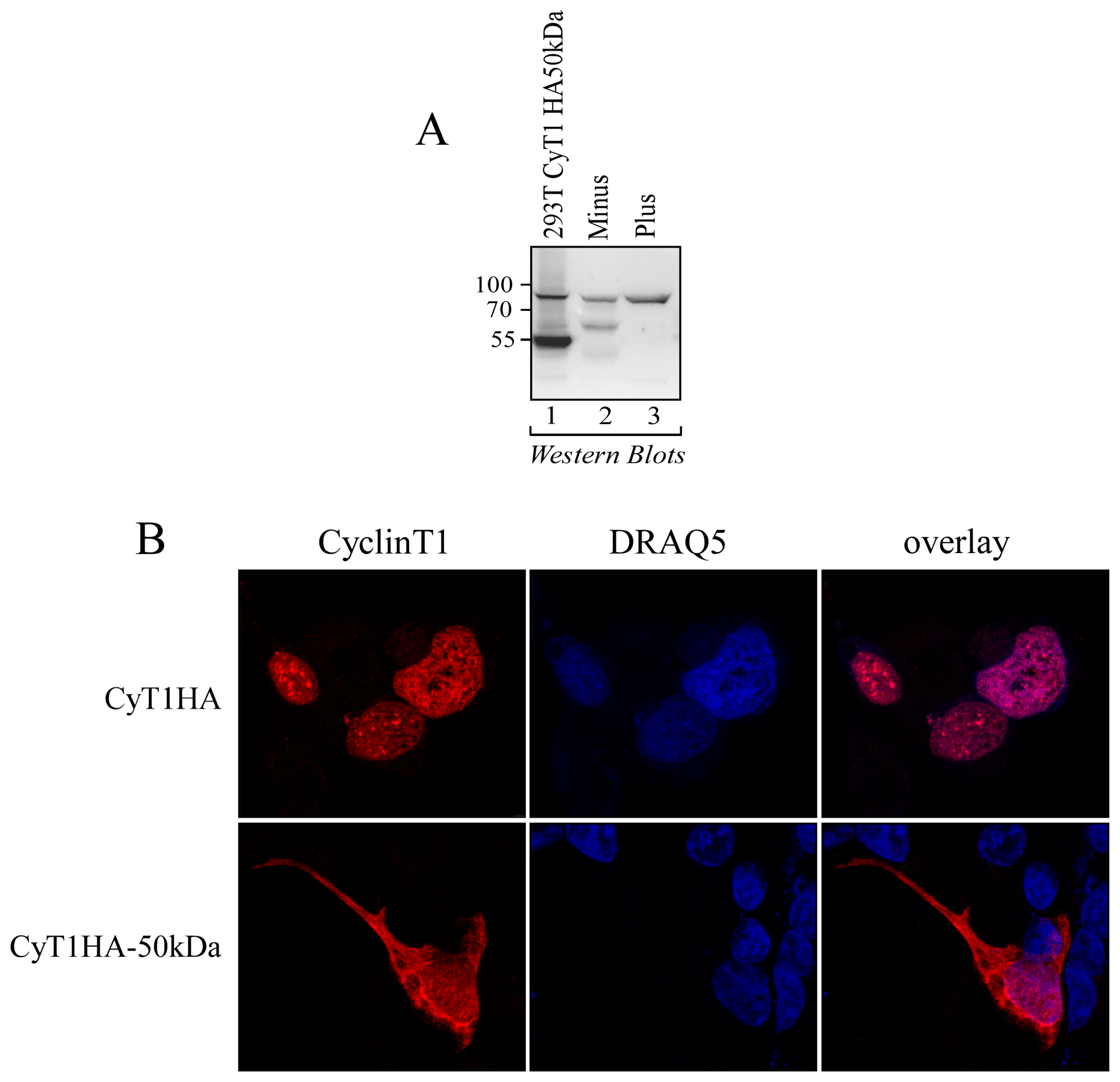
3.5. The 50 kDa Cyclin T1 Protein Form Is Localized in Both the Nucleus and the Cytoplasm
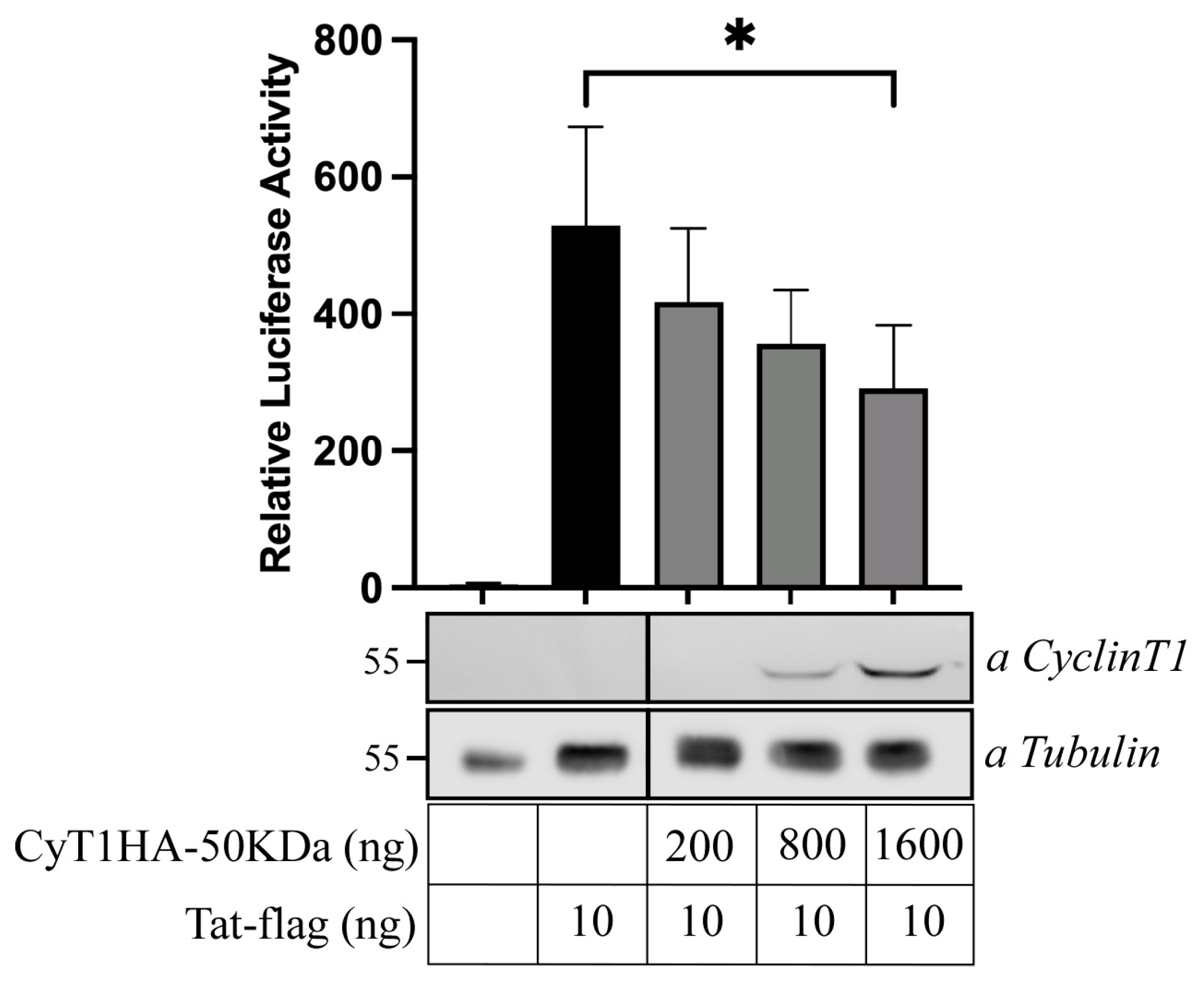
3.6. Inhibition of Tat-Mediated HIV-1 LTR Transactivation by the 50 kDa CyclinT1 Protein Form
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Price, D.H. P-TEFb, a Cyclin-Dependent Kinase Controlling Elongation by RNA Polymerase II. Mol. Cell. Biol. 2000, 20, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Garber, M.E.; Fang, S.M.; Fischer, W.H.; Jones, K.A. A Novel CDK9-Associated C-Type Cyclin Interacts Directly with HIV-1 Tat and Mediates Its High-Affinity, Loop-Specific Binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Chiang, K.; Sung, T.L.; Rice, A.P. Regulation of Cyclin T1 and HIV-1 Replication by MicroRNAs in Resting CD4+ T Lymphocytes. J. Virol. 2012, 86, 3244–3252. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yik, J.H. The Yin and Yang of P-TEFb Regulation: Implications for Human Immunodeficiency Virus Gene Expression and Global Control of Cell Growth and Differentiation. Microbiol. Mol. Biol. Rev. 2006, 70, 646–659. [Google Scholar] [CrossRef] [PubMed]
- Egloff, S. CDK9 Keeps RNA Polymerase II on Track. Cell. Mol. Life Sci. 2021, 78, 5543–5567. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wang, Y.; Shaw, C.A.; Qin, X.-F.; Rice, A.P. Induction of the HIV-1 Tat Co-Factor Cyclin T1 during Monocyte Differentiation Is Required for the Regulated Expression of a Large Portion of Cellular mRNAs. Retrovirol 2006, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Orozco, A.F.; Liu, H.; Budhiraja, S.; Siwak, E.B.; Nehete, P.N.; Sastry, K.J.; Rice, A.P.; Lewis, D.E. Regulation of Cyclin T1 during HIV Replication and Latency Establishment in Human Memory CD4 T Cells. Virol. J. 2019, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Hafer, T.L.; Felton, A.; Delgado, Y.; Srinivasan, H.; Emerman, M. A CRISPR Screen of HIV Dependency Factors Reveals That CCNT1 Is Non-Essential in T Cells but Required for HIV-1 Reactivation from Latency. Viruses 2023, 15, 1863. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Nguyen, T.T.; Echeverria, I.; Rakesh, R.; Cary, D.C.; Paculova, H.; Sali, A.; Weiss, A.; Peterlin, B.M.; Fujinaga, K. Reversible Phosphorylation of Cyclin T1 Promotes Assembly and Stability of P-TEFb. eLife 2021, 10, e68473. [Google Scholar] [CrossRef] [PubMed]
- Gazzolo, L.; Macé, K. Regulation of HIV1 Replication in Promonocytic U937 Cells. Res. Virol. 1990, 141, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Boulerice, F.; Geleziunas, R.; Bour, S.; Li, H.L.; D’Addario, M.; Roulston, A.; Hiscott, J.; Wainberg, M.A. Differential Susceptibilities of U-937 Cell Clones to Infection by Human Immunodeficiency Virus Type 1. J. Virol. 1992, 66, 1183–1187. [Google Scholar] [CrossRef] [PubMed]
- Franzoso, G.; Biswas, P.; Poli, G.; Carlson, L.M.; Brown, K.D.; Tomita-Yamaguchi, M.; Fauci, A.S.; Siebenlist, U.K. A Family of Serine Proteases Expressed Exclusively in Myelo-Monocytic Cells Specifically Processes the Nuclear Factor-Kappa B Subunit P65 in Vitro and May Impair Human Immunodeficiency Virus Replication in These Cells. J. Exp. Med. 1994, 180, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Kajaste-Rudnitski, A.; Marelli, S.S.; Pultrone, C.; Pertel, T.; Uchil, P.D.; Mechti, N.; Mothes, W.; Poli, G.; Luban, J.; Vicenzi, E. TRIM22 Inhibits HIV-1 Transcription Independently of Its E3 Ubiquitin Ligase Activity, Tat, and NF-kappaB-Responsive Long Terminal Repeat Elements. J. Virol. 2011, 85, 5183–5196. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, S.; Okamoto, T.; Peterlin, B.M. Tat Competes with CIITA for the Binding to P-TEFb and Blocks the Expression of MHC Class II Genes in HIV Infection. Immunity 2000, 12, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Accolla, R.S.; Mazza, S.; De Lerma Barbaro, A.; De Maria, A.; Tosi, G. The HLA Class II Transcriptional Activator Blocks the Function of HIV-1 Tat and Inhibits Viral Replication. Eur. J. Immunol. 2002, 32, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Turrini, F.; Ghezzi, S.; Tedeschi, A.; Poli, G.; Accolla, R.S.; Tosi, G. The MHC-II Transactivator CIITA Inhibits Tat Function and HIV-1 Replication in Human Myeloid Cells. J. Transl. Med. 2016, 14, 94. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Tosi, G.; Turrini, F.; Poli, G.; Vicenzi, E.; Accolla, R.S. Tripartite Motif-Containing Protein 22 Interacts with Class II Transactivator and Orchestrates Its Recruitment in Nuclear Bodies Containing TRIM19/PML and Cyclin T1. Front. Immunol. 2017, 8, 564. [Google Scholar] [CrossRef] [PubMed]
- Urano, E.; Miyauchi, K.; Ichikawa, R.; Futahashi, Y.; Komano, J. Regulation of Cyclin T1 Expression and Function by an Alternative Splice Variant That Skips Exon 7 and Contains a Premature Termination Codon. Gene 2012, 505, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Baumli, S.; Lolli, G.; Lowe, E.D.; Troiani, S.; Rusconi, L.; Bullock, A.N.; Debreczeni, J.E.; Knapp, S.; Johnson, L.N. The Structure of P-TEFb (CDK9/Cyclin T1), Its Complex with Flavopiridol and Regulation by Phosphorylation. EMBO J. 2008, 27, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.; Kwak, Y.T.; Nee, E.; Guo, J.; García-Martínez, L.F.; Gaynor, R.B. Cyclin T1 Domains Involved in Complex Formation with Tat and TAR RNA Are Critical for Tat-Activation. J. Mol. Biol. 1999, 288, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Steimle, V.; Siegrist, C.A.; Mottet, A.; Lisowska-Grospierre, B.; Mach, B. Regulation of MHC Class II Expression by Interferon-Gamma Mediated by the Transactivator Gene CIITA. Science 1994, 265, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Rigaud, G.; De Lerma Barbaro, A.; Nicolis, M.; Cestari, T.; Ramarli, D.; Riviera, A.P.; Accolla, R.S. Induction of CIITA and Modification of in Vivo HLA-DR Promoter Occupancy in Normal Thymic Epithelial Cells Treated with IFN-Gamma: Similarities and Distinctions with Respect to HLA-DR-Constitutive B Cells. J. Immunol. 1996, 156, 4254–4258. [Google Scholar] [CrossRef] [PubMed]
- Bovolenta, C.; Lorini, A.L.; Mantelli, B.; Camorali, L.; Novelli, F.; Biswas, P.; Poli, G. A Selective Defect of IFN-Gamma- but Not of IFN-Alpha-Induced JAK/STAT Pathway in a Subset of U937 Clones Prevents the Antiretroviral Effect of IFN-Gamma against HIV-1. J. Immunol. 1999, 162, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, C.H.; Mancini, M.A. The Cdk9 and Cyclin T Subunits of TAK/P-TEFb Localize to Splicing Factor-Rich Nuclear Speckle Regions. J. Cell. Sci. 2001, 114, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Moriuchi, H.; Moriuchi, M.; Arthos, J.; Hoxie, J.; Fauci, A.S. Promonocytic U937 Subclones Expressing CD4 and CXCR4 Are Resistant to Infection with and Cell-to-Cell Fusion by T-Cell-Tropic Human Immunodeficiency Virus Type 1. J. Virol. 1997, 71, 9664–9671. [Google Scholar] [CrossRef] [PubMed]
- Accolla, R.S.; Jotterand-Bellomo, M.; Scarpellino, L.; Maffei, A.; Carra, G.; Guardiola, J. aIr-1, a Newly Found Locus on Mouse Chromosome 16 Encoding a Trans-Acting Activator Factor for MHC Class II Gene Expression. J. Exp. Med. 1986, 164, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Steimle, V.; Otten, L.A.; Zufferey, M.; Mach, B. Complementation Cloning of an MHC Class II Transactivator Mutated in Hereditary MHC Class II Deficiency (or Bare Lymphocyte Syndrome). Cell 1993, 75, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Wu, X.; Zhou, J.; He, M.; He, J.J.; Guo, D. Inhibition of HIV-1 Transcription and Replication by a Newly Identified Cyclin T1 Splice Variant. J. Biol. Chem. 2013, 288, 14297–14309. [Google Scholar] [CrossRef]
- Zhou, J.; Gao, G.; Hou, P.; Li, C.-M.; Guo, D. Regulation of the Alternative Splicing and Function of Cyclin T1 by the Serine-Arginine-Rich Protein ASF/SF2. J. Cell. Biochem. 2017, 118, 4020–4032. [Google Scholar] [CrossRef] [PubMed]
- Frolova, A.S.; Chepikova, O.E.; Deviataikina, A.S.; Solonkina, A.D.; Zamyatnin, A.A. New Perspectives on the Role of Nuclear Proteases in Cell Death Pathways. Biology 2023, 12, 797. [Google Scholar] [CrossRef] [PubMed]
- Frolova, A.S.; Petushkova, A.I.; Makarov, V.A.; Soond, S.M.; Zamyatnin, A.A. Unravelling the Network of Nuclear Matrix Metalloproteinases for Targeted Drug Design. Biology 2020, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual Proteome-Scale Networks Reveal Cell-Specific Remodeling of the Human Interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef] [PubMed]
- Fujinaga, K.; Taube, R.; Wimmer, J.; Cujec, T.P.; Peterlin, B.M. Interactions between Human Cyclin T, Tat, and the Transactivation Response Element (TAR) Are Disrupted by a Cysteine to Tyrosine Substitution Found in Mouse Cyclin T. Proc. Natl. Acad. Sci. USA 1999, 96, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Garber, M.E.; Wei, P.; KewalRamani, V.N.; Mayall, T.P.; Herrmann, C.H.; Rice, A.P.; Littman, D.R.; Jones, K.A. The Interaction between HIV-1 Tat and Human Cyclin T1 Requires Zinc and a Critical Cysteine Residue That Is Not Conserved in the Murine CycT1 Protein. Genes. Dev. 1998, 12, 3512–3527. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, K.; Hibi, Y.; Imai, K.; Victoriano, A.F.B.; Kurimoto, E.; Kato, K.; Okamoto, T. Functional Characterization of Human Cyclin T1 N-Terminal Region for Human Immunodeficiency Virus-1 Tat Transcriptional Activation. J. Mol. Biol. 2011, 410, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Asamitsu, K.; Victoriano, A.F.B.; Cueno, M.E.; Fujinaga, K.; Okamoto, T. Cyclin T1 Stabilizes Expression Levels of HIV-1 Tat in Cells. FEBS J. 2009, 276, 7124–7133. [Google Scholar] [CrossRef] [PubMed]
- Pagani, I.; Demela, P.; Ghezzi, S.; Vicenzi, E.; Pizzato, M.; Poli, G. Host Restriction Factors Modulating HIV Latency and Replication in Macrophages. Int. J. Mol. Sci. 2022, 23, 3021. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Accolla, R.S. Tripartite Motif 22 and Class II Transactivator Restriction Factors: Unveiling Their Concerted Action against Retroviruses. Front. Immunol. 2017, 8, 1362. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.; Aimola, G.; Forlani, G.; Turrini, F.; Accolla, R.S.; Vicenzi, E.; Poli, G. Reversible Human Immunodeficiency Virus Type-1 Latency in Primary Human Monocyte-Derived Macrophages Induced by Sustained M1 Polarization. Sci. Rep. 2018, 8, 14249. [Google Scholar] [CrossRef] [PubMed]



| Primer ID | Sequence |
|---|---|
| T1-FW | cac cgc cac cat gga ggg agg agg aag aac aac aac |
| T1-9RW | gct aaa ttc tca cta gtc cga tga ccc |
| T1-9FW | ggg tca tcg gac tag tga gaa ttt agc |
| T1-RW | ccc ggg cct cga gct ctt agg aag ggg tgg aag tgg tgg |
| T1-6FW | caa gca agg act tag cac aga c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alberio, T.; Shallak, M.; Shaik, A.K.B.; Accolla, R.S.; Forlani, G. A Truncated Isoform of Cyclin T1 Could Contribute to the Non-Permissive HIV-1 Phenotype of U937 Promonocytic Cells. Viruses 2024, 16, 1176. https://doi.org/10.3390/v16081176
Alberio T, Shallak M, Shaik AKB, Accolla RS, Forlani G. A Truncated Isoform of Cyclin T1 Could Contribute to the Non-Permissive HIV-1 Phenotype of U937 Promonocytic Cells. Viruses. 2024; 16(8):1176. https://doi.org/10.3390/v16081176
Chicago/Turabian StyleAlberio, Tiziana, Mariam Shallak, Amruth Kaleem Basha Shaik, Roberto Sergio Accolla, and Greta Forlani. 2024. "A Truncated Isoform of Cyclin T1 Could Contribute to the Non-Permissive HIV-1 Phenotype of U937 Promonocytic Cells" Viruses 16, no. 8: 1176. https://doi.org/10.3390/v16081176
APA StyleAlberio, T., Shallak, M., Shaik, A. K. B., Accolla, R. S., & Forlani, G. (2024). A Truncated Isoform of Cyclin T1 Could Contribute to the Non-Permissive HIV-1 Phenotype of U937 Promonocytic Cells. Viruses, 16(8), 1176. https://doi.org/10.3390/v16081176










