Characterization and Anti-Biofilm Activity of Lytic Enterococcus Phage vB_Efs8_KEN04 against Clinical Isolates of Multidrug-Resistant Enterococcus faecalis in Kenya
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultural Conditions
2.2. Bacteriophage Isolation, Purification, and Propagation
2.3. Phage Stability
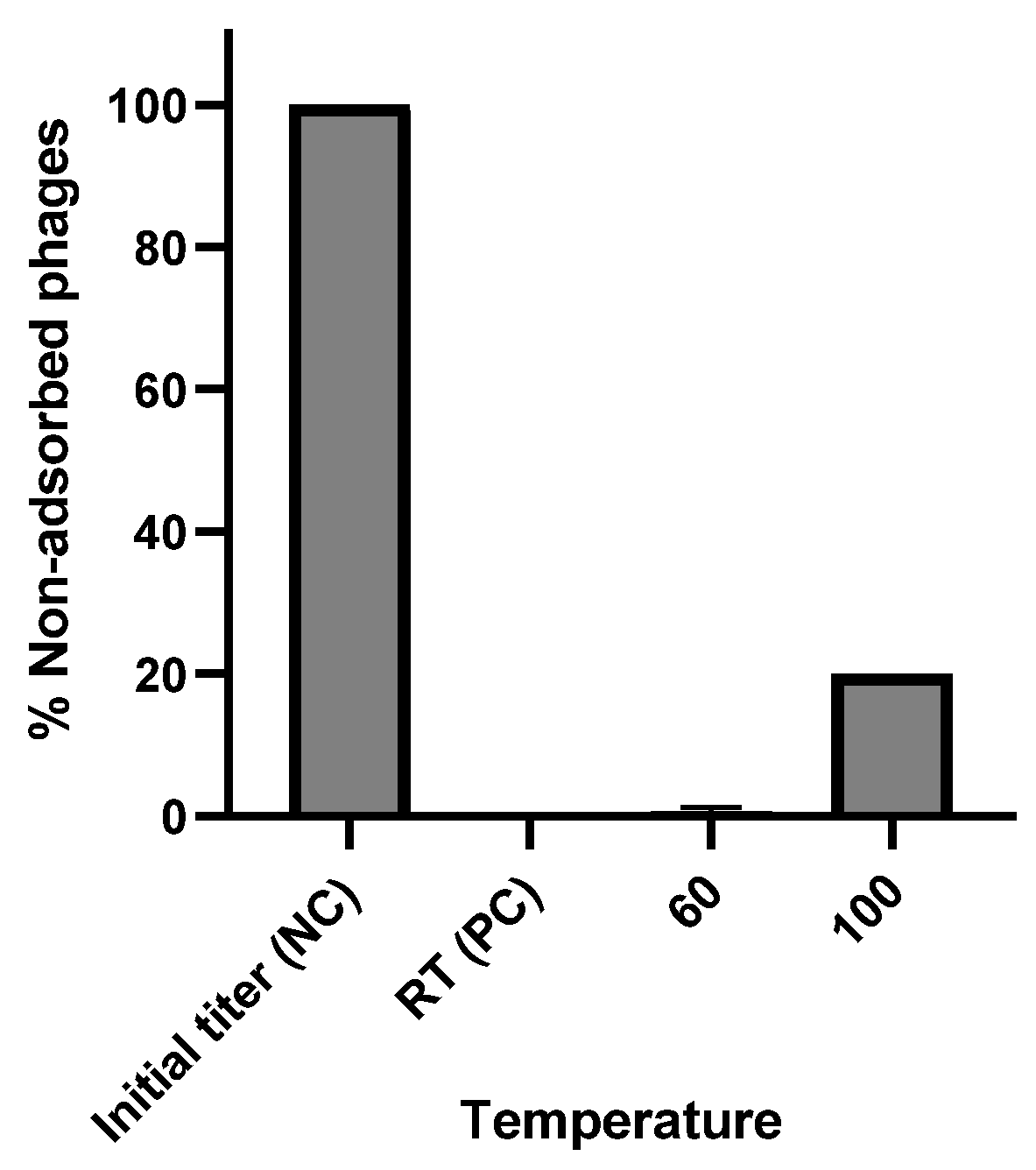
2.3.1. Thermal Stability
2.3.2. pH Stability
2.4. Host Range Analysis
2.5. Determination of Optimal Multiplicity of Infection
2.6. Lytic Properties of Phage vB_Efs8_KEN04
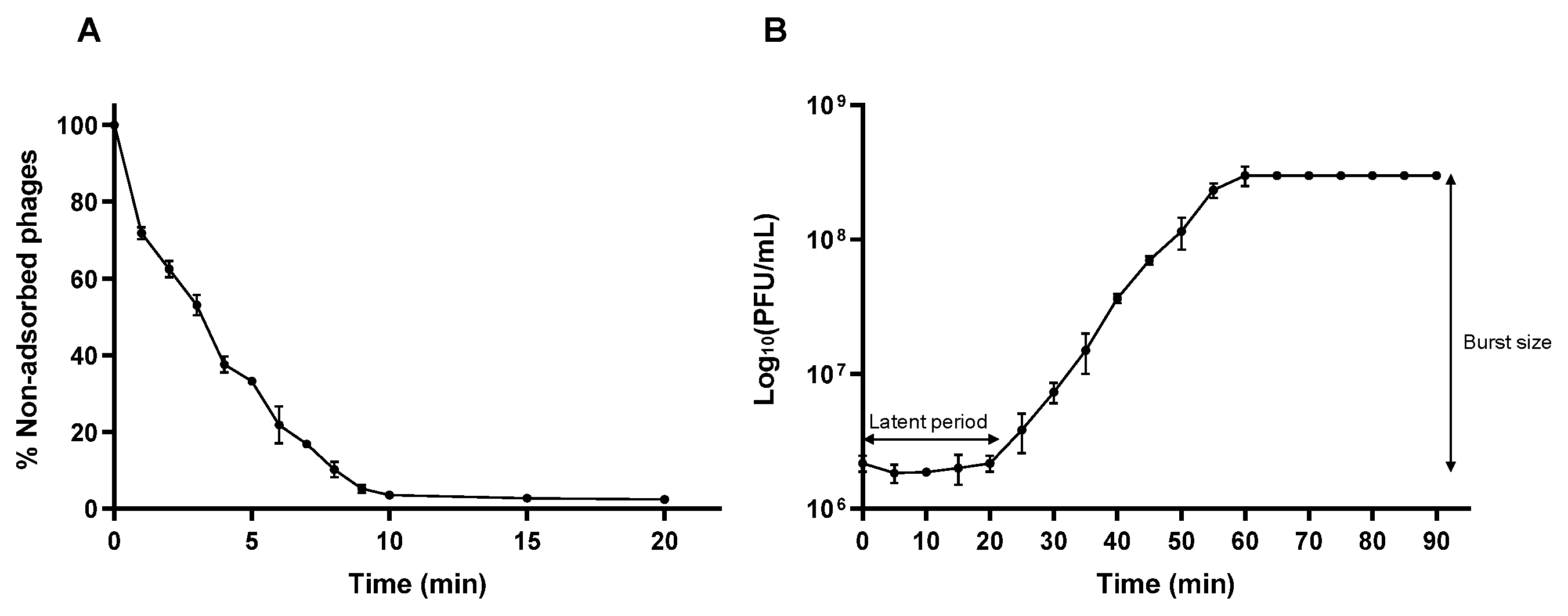
2.7. One-Step Growth Curve
2.8. Adsorption Assay
2.9. Isolation of Phage-Resistant Mutants
2.10. Determination of the Nature of Phage Receptor
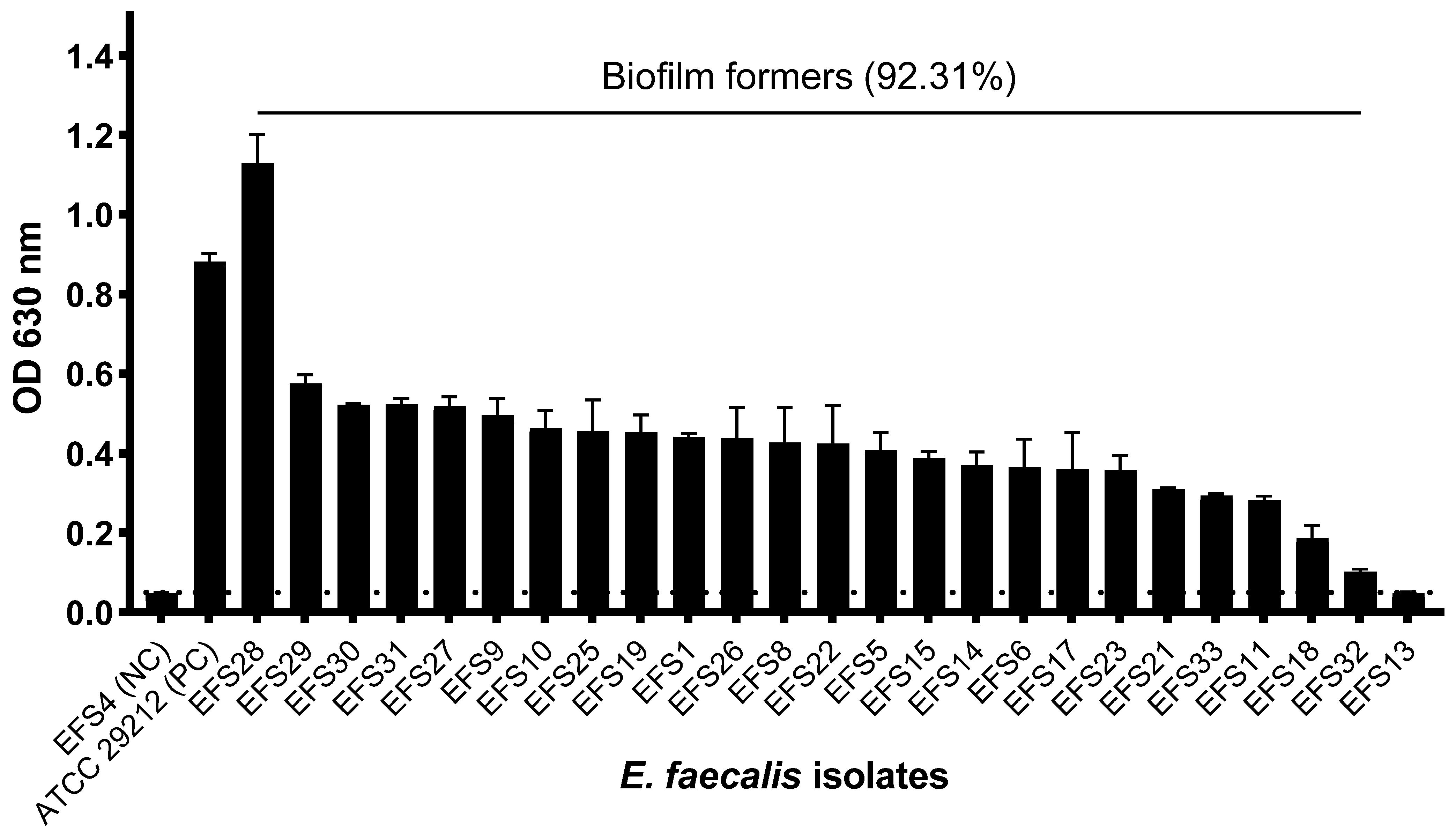
2.11. Biofilm Formation Assay
2.12. Biofilm Inhibition Assay by Phage
2.13. Biofilm Disruption Assay by Phage
2.14. Genomic DNA Extraction
2.15. Genome Sequencing and Bioinformatic Analysis of Sequencing Data
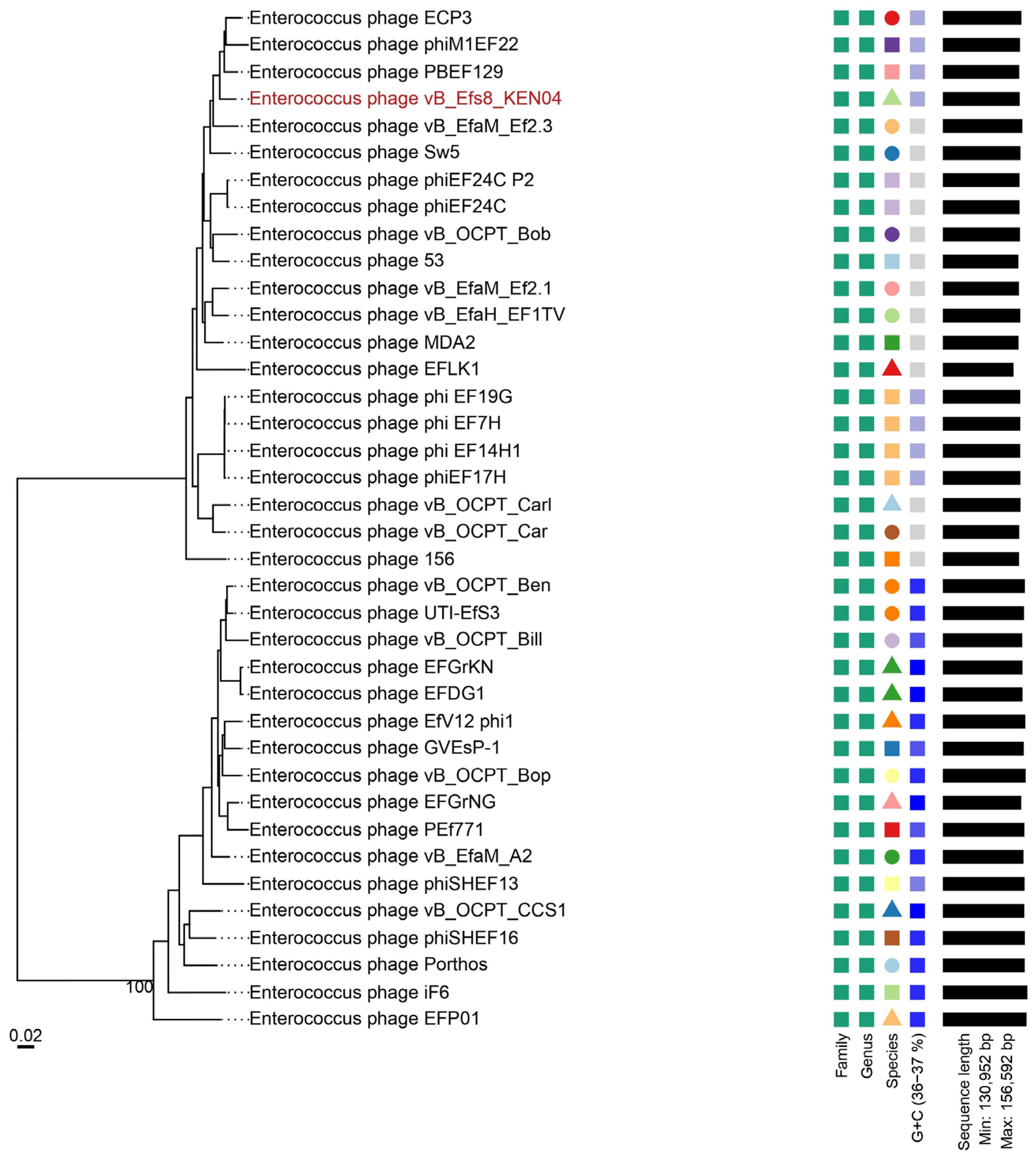
2.16. Phylogenetic Tree and Comparative Genomics of Phage Genomes
2.17. Phage Host Range Prediction Based on Phage Receptor-Binding Proteins
2.18. Statistical Analysis
3. Results
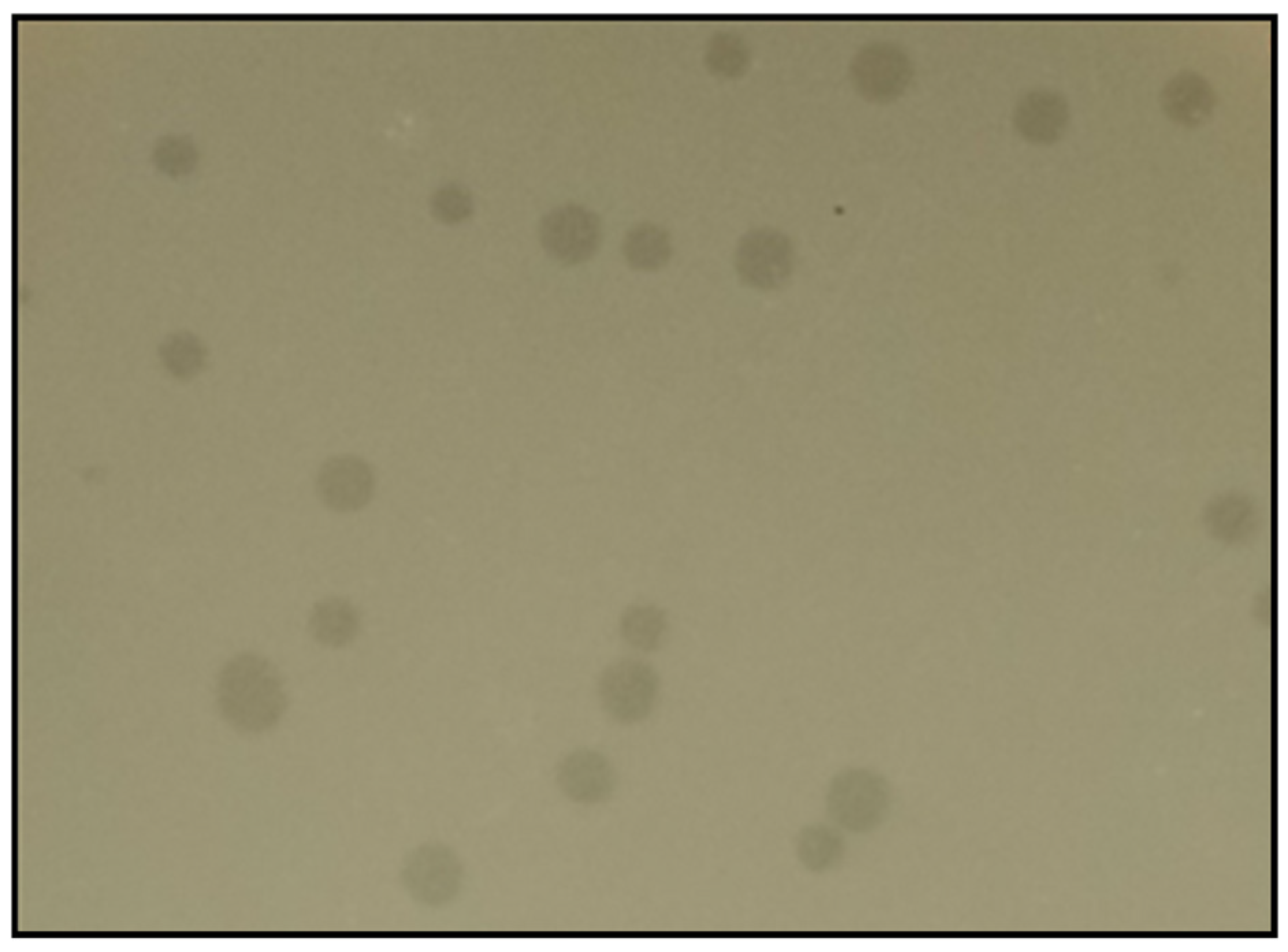
3.1. Bacteriophage Isolation and Purification
3.2. Host Range Analysis
3.3. Host Range Prediction Based on Receptor-Binding Proteins
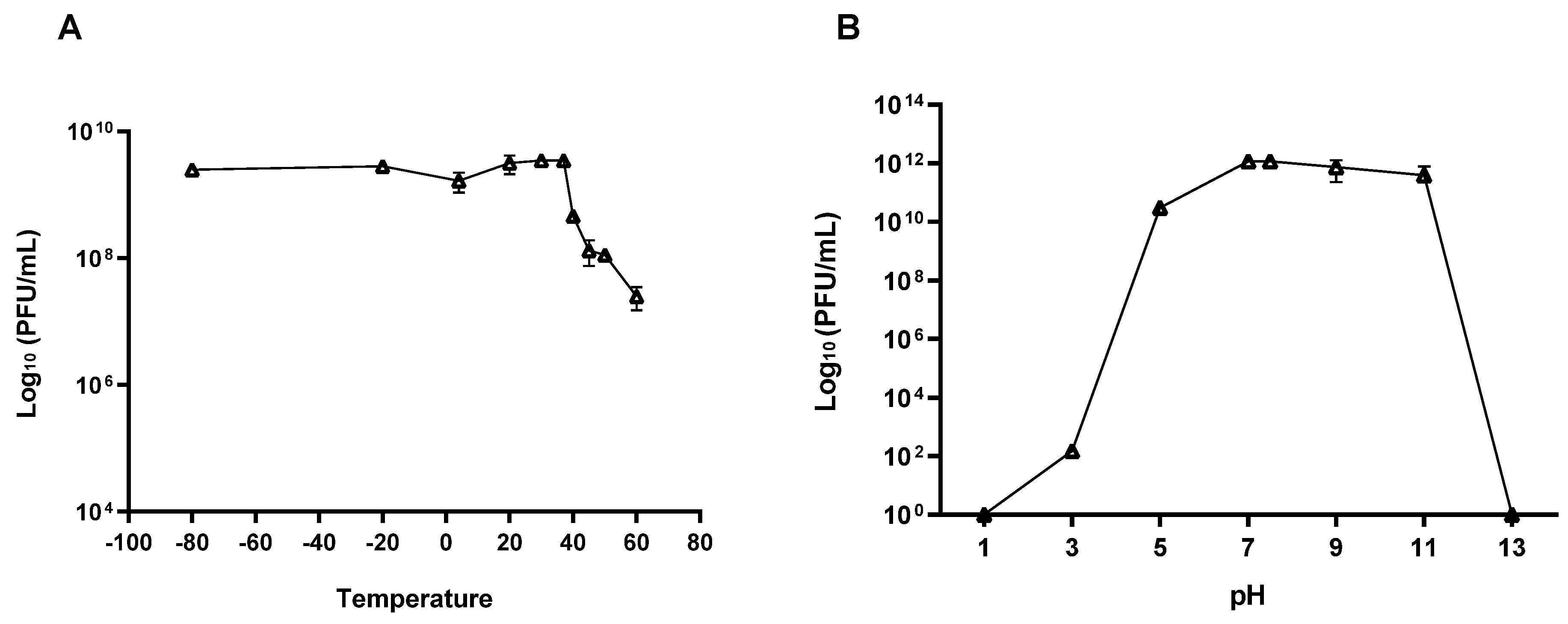
3.4. Phage Stability
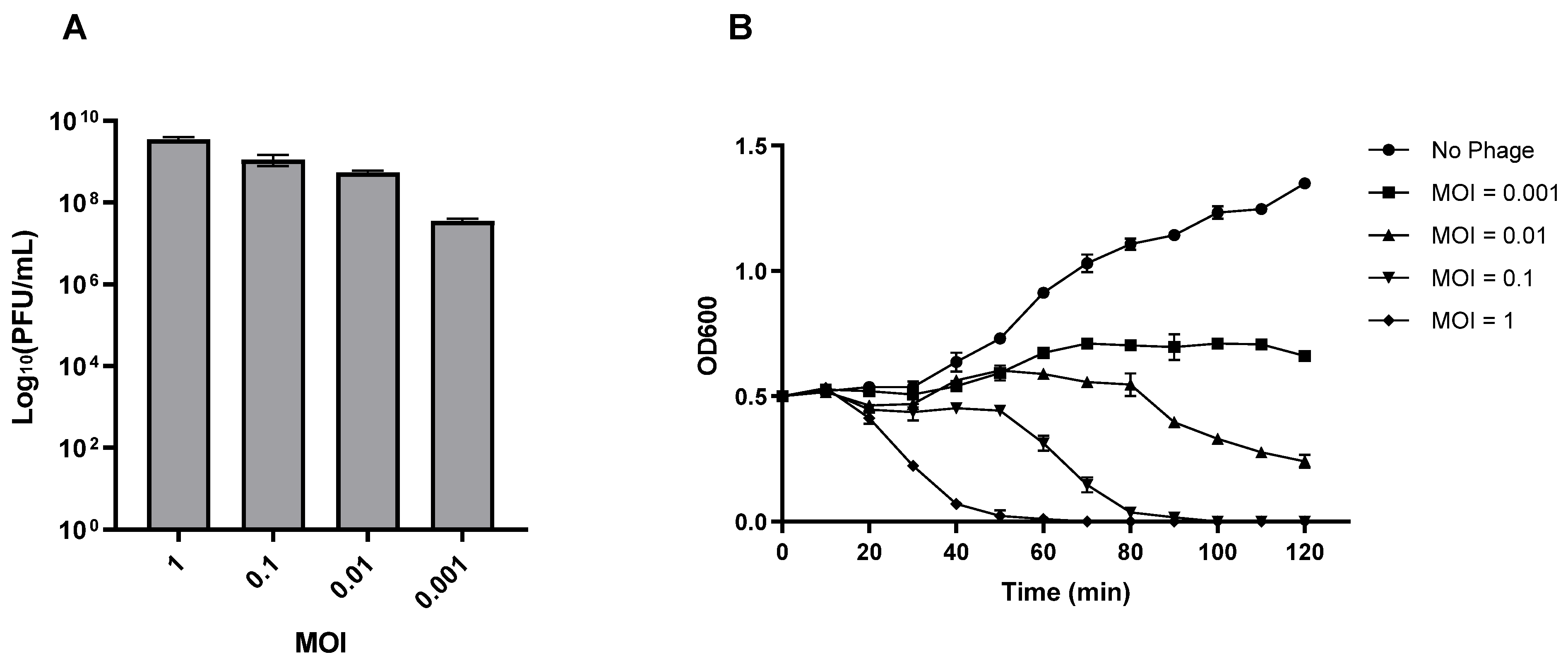
3.5. Lytic Characteristics of Phage vB_Efs8_KEN04
3.6. Adsorption Efficiency and One-Step Growth Curve
3.7. Phage Mutation Rate and Nature of Phage Receptors
3.8. Biofilm Formation of Enterococcus faecalis
3.9. Biofilm Inhibition and Disruption by Phage vB_Efs8_KEN04
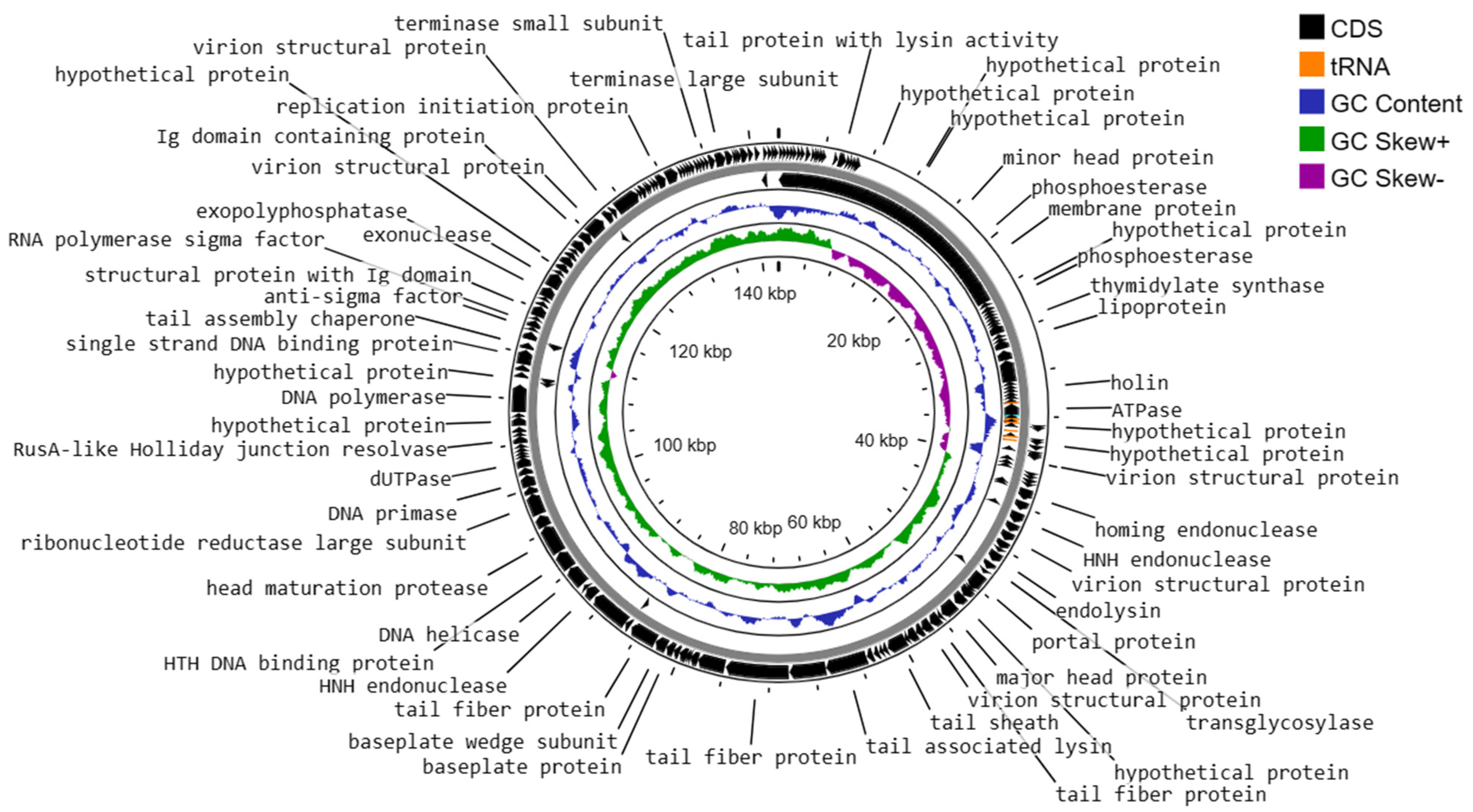
3.10. Genome Characteristics of Enterococcus faecalis Phage vB_Efs8_KEN04
- (i)
- DNA replication, transcription, translation, and nucleotide metabolism: A total of 25 CDSs were predicted to encode for DNA replication, transcription regulation, translation, and metabolism-related proteins, such as HNH homing endonuclease, DNA helicase, exonucleases, transcriptional repressor, DNA helicase, DNA primase, and a transcriptional regulator, RNA polymerase beta subunit, and thymidylate synthase.
- (ii)
- Structural and packaging proteins: 27 CDS were predicted to encode for tail, head, and packaging proteins such as portal proteins, head proteins, tail fiber proteins, head maturation proteases, virion structural proteins, tail proteins, tail assembly chaperones, minor and major head proteins, and terminase large and small subunits.
- (iii)
- Host lysis and adhesion proteins: Two CDS were predicted to encode holin and endolysin proteins. BLASTp analysis of the phage vB_Efs8_KEN04 genome revealed no similarities to the genes encoding integrase or excisionase. The genome of phage vB_Efs8_KEN04 lacks genes encoding toxins, virulence factors, antibiotic resistance genes, and CRISPR. These data indicate that phage vB_Efs8_KEN04 is a strictly lytic phage that can be used to treat E. faecalis infection.
- (iv)
- Sixteen CDS encode for moron, auxiliary metabolic genes, and host takeover.
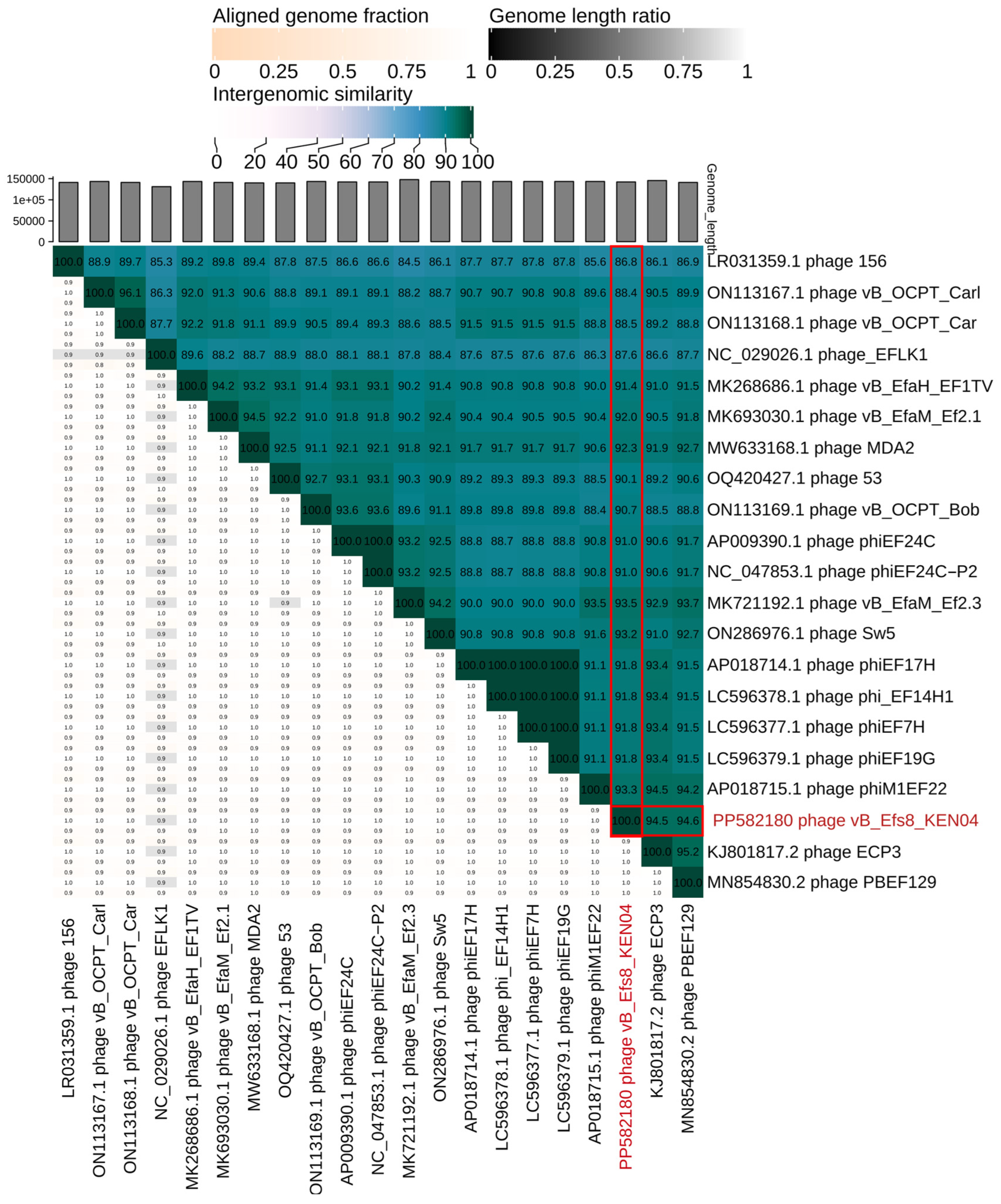
3.11. Phylogenetic Analysis
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Said, M.S.; Tirthani, E.; Lesho, E. Enterococcus Infections. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Raza, T.; Ullah, S.R.; Mehmood, K.; Andleeb, S. Vancomycin resistant Enterococci: A brief review. J. Pak. Med. Assoc. 2018, 68, 768–772. [Google Scholar]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef]
- Beganovic, M.; Luther, M.K.; Rice, L.B.; Arias, C.A.; Rybak, M.J.; LaPlante, K.L. A Review of Combination Antimicrobial Therapy for Enterococcus faecalis Bloodstream Infections and Infective Endocarditis. Clin. Infect. Dis. 2018, 67, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Rajkumari, N.; Mathur, P.; Misra, M.C. Soft Tissue and Wound Infections Due to Enterococcus spp. among Hospitalized Trauma Patients in a Developing Country. J. Glob. Infect. Dis. 2014, 6, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Shiadeh, S.M.J.; Pormohammad, A.; Hashemi, A.; Lak, P. Global prevalence of antibiotic resistance in blood-isolated Enterococcus faecalis and Enterococcus faecium: A systematic review and meta-analysis. IDR 2019, 12, 2713–2725. [Google Scholar] [CrossRef]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.-P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.L.; Nilson, J.L.; Barnes, A.M.T.; Dunny, G.M. Restructuring of Enterococcus faecalis biofilm architecture in response to antibiotic-induced stress. npj Biofilms Microbiomes 2017, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, M.S.; Worthington, R.J.; Melander, C. Biologically inspired strategies for combating bacterial biofilms. Curr. Opin. Pharmacol. 2013, 13, 699–706. [Google Scholar] [CrossRef]
- Chaudhary, N.; Maurya, R.; Singh, D.; Mohan, B.; Taneja, N. Genome Analysis and Antibiofilm Activity of Phage 590B against Multidrug-Resistant and Extensively Drug-Resistant Uropathogenic Escherichia coli Isolates, India. Pathogens 2022, 11, 1448. [Google Scholar] [CrossRef]
- Shrestha, L.; Fan, H.-M.; Tao, H.-R.; Huang, J.-D. Recent Strategies to Combat Biofilms Using Antimicrobial Agents and Therapeutic Approaches. Pathogens 2022, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W.; Smith, M.C.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197. [Google Scholar] [CrossRef]
- Ferriol-González, C.; Domingo-Calap, P. Phages for Biofilm Removal. Antibiotics 2020, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Bacteriophages and their Enzymes in Biofilm Control. Curr. Pharm. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front. Pharmacol. 2019, 10, 513. [Google Scholar] [CrossRef]
- Song, M.; Wu, D.; Hu, Y.; Luo, H.; Li, G. Characterization of an Enterococcus faecalis Bacteriophage vB_EfaM_LG1 and Its Synergistic Effect with Antibiotic. Front. Cell. Infect. Microbiol. 2021, 11, 698807. [Google Scholar] [CrossRef] [PubMed]
- Di Lallo, G.; Falconi, M.; Iacovelli, F.; Frezza, D.; D’Addabbo, P. Analysis of Four New Enterococcus faecalis Phages and Modeling of a Hyaluronidase Catalytic Domain from Saphexavirus. Phage 2021, 2, 131–141. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, J.; Lin, Y.; Yuan, L.; El-Telbany, M.; Maung, A.; Abdelaziz, M.; Masuda, Y.; Honjoh, K.; Miyamoto, T. Isolation, Characterization of Enterococcus phages and their application in control of E. faecalis in milk. J. Appl. Microbiol. 2023, 134, lxad250. [Google Scholar] [CrossRef]
- Zhang, W.; Mi, Z.; Yin, X.; Fan, H.; An, X.; Zhang, Z.; Chen, J.; Tong, Y. Characterization of Enterococcus faecalis Phage IME-EF1 and Its Endolysin. PLoS ONE 2013, 8, e80435. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, F.; Hallajzadeh, M.; Sholeh, M.; Talebi, M.; Pirhajati Mahabadi, V.; Amirmozafari, N. Anti-biofilm Activity of a Lytic Phage Against Vancomycin-Resistant Enterococcus faecalis. Iran. J. Pathol. 2022, 17, 285–293. [Google Scholar] [CrossRef] [PubMed]
- El-Telbany, M.; Lin, C.-Y.; Abdelaziz, M.; Maung, A.; El-Shibiny, A.; Noor Mohammadi, T.; Zayda, M.; Wang, C.; Lwin, S.; Zhao, J.; et al. Potential application of phage vB_EfKS5 to control Enterococcus faecalis and its biofilm in food. AMB Express 2023, 13, 130. [Google Scholar] [CrossRef] [PubMed]
- El-Atrees, D.M.; El-Kased, R.F.; Abbas, A.M.; Yassien, M.A. Characterization and anti-biofilm activity of bacteriophages against urinary tract Enterococcus faecalis isolates. Sci. Rep. 2022, 12, 13048. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.-A.; Beyth, N.; Hazan, R. Targeting Enterococcus faecalis Biofilms with Phage Therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fouts, D.E.; DePew, J.; Stevens, R.H. Genetic modifications to temperate Enterococcus faecalis phage Ef11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology 2013, 159, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, J.M.; Buttaro, B.; Zhang, H.; Liss, N.; Sassone, L.; Stevens, R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch. Oral Biol. 2016, 71, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Prasad, Y. Efficacy of polyvalent bacteriophage P-27/HP to control multidrug resistant Staphylococcus aureus associated with human infections. Curr. Microbiol. 2011, 62, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Carlton, R.M. Phage therapy: Past history and future prospects. Arch. Immunol. Ther. Exp. 1999, 47, 267–274. [Google Scholar]
- Voelker, R. FDA Approves Bacteriophage Trial. JAMA 2019, 321, 638. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, P.D.M. The Perfect Predator: A Scientist’s Race to Save Her Husband, by Drs. Steffanie Strathdee and Thomas Patterson. Ann. Glob. Health 2020, 86, 97. [Google Scholar] [CrossRef]
- Gupta, P.; Singh, H.S.; Shukla, V.K.; Nath, G.; Bhartiya, S.K. Bacteriophage Therapy of Chronic Nonhealing Wound: Clinical Study. Int. J. Low. Extrem. Wounds 2019, 18, 171–175. [Google Scholar] [CrossRef]
- Ling, H.; Lou, X.; Luo, Q.; He, Z.; Sun, M.; Sun, J. Recent advances in bacteriophage-based therapeutics: Insight into the post-antibiotic era. Acta Pharm. Sin. B 2022, 12, 4348–4364. [Google Scholar] [CrossRef] [PubMed]
- Blasco, L.; López-Hernández, I.; Rodríguez-Fernández, M.; Pérez-Florido, J.; Casimiro-Soriguer, C.S.; Djebara, S.; Merabishvili, M.; Pirnay, J.-P.; Rodríguez-Baño, J.; Tomás, M.; et al. Case report: Analysis of phage therapy failure in a patient with a Pseudomonas aeruginosa prosthetic vascular graft infection. Front. Med. 2023, 10, 1199657. [Google Scholar] [CrossRef] [PubMed]
- Song’oro, E.; Nyerere, A.; Magoma, G.; Gunturu, R. Occurrence of Highly Resistant Microorganisms in Ruai Wastewater Treatment Plant and Dandora Dumpsite in Nairobi County, Kenya. Adv. Microbiol. 2019, 9, 479–494. [Google Scholar] [CrossRef]
- D’Souza, R.; Filippov, A.A.; Sergueev, K.V.; He, Y.; Ward, A.M.; Goglin, K.; Vashee, S.; Nikolich, M.P.; Fouts, D.E. Complete Genome Sequence of Broad-Host-Range Staphylococcus aureus Myophage ESa1. Microbiol. Resour. Announc. 2020, 9, e00730-20. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.J.; Kropinski, A.M. Bacteriophages: Methods and Protocols; Humana: Totowa, NJ, USA, 2009; Volume 1—Isolation, Characterization, and Interactions; Available online: https://link.springer.com/book/10.1007/978-1-60327-164-6 (accessed on 3 March 2024).
- Chang, Y.; Shin, H.; Lee, J.-H.; Park, C.J.; Paik, S.-Y.; Ryu, S. Isolation and Genome Characterization of the Virulent Staphylococcus aureus Bacteriophage SA97. Viruses 2015, 7, 5225–5242. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.K.; Nilsson, A.S. Correction: Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0127606. [Google Scholar] [CrossRef] [PubMed]
- Mutai, I.J.; Juma, A.A.; Inyimili, M.I.; Nyachieo, A.; Nyamache, A.K. Efficacy of diversely isolated lytic phages against multi-drug resistant Enterobacter cloacae isolates in Kenya. Afr. J. Lab. Med. 2022, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Mohan, B.; Kaur, H.; Modgil, V.; Kant, V.; Bhatia, A.; Taneja, N. Vibrio Phage VMJ710 Can Prevent and Treat Disease Caused by Pathogenic MDR V. cholerae O1 in an Infant Mouse Model. Antibiotics 2023, 12, 1046. [Google Scholar] [CrossRef] [PubMed]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Brabban, A.D.; Diez-Gonzalez, F. Isolation and characterization of lytic bacteriophages against enterohaemorrhagic Escherichia coli. J. Appl. Microbiol. 2011, 110, 1323–1331. [Google Scholar] [CrossRef]
- Mencke, J.L.; He, Y.; Filippov, A.A.; Nikolich, M.P.; Belew, A.T.; Fouts, D.E.; McGann, P.T.; Swierczewski, B.E.; Getnet, D.; Ellison, D.W.; et al. Identification and Characterization of vB_PreP_EPr2, a Lytic Bacteriophage of Pan-Drug Resistant Providencia rettgeri. Viruses 2022, 14, 708. [Google Scholar] [CrossRef]
- Lee, D.; Im, J.; Na, H.; Ryu, S.; Yun, C.-H.; Han, S.H. The Novel Enterococcus Phage vB_EfaS_HEf13 Has Broad Lytic Activity Against Clinical Isolates of Enterococcus faecalis. Front. Microbiol. 2019, 10, 2877. [Google Scholar] [CrossRef] [PubMed]
- Sváb, D.; Falgenhauer, L.; Rohde, M.; Szabó, J.; Chakraborty, T.; Tóth, I. Identification and Characterization of T5-Like Bacteriophages Representing Two Novel Subgroups from Food Products. Front. Microbiol. 2018, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Yong, D. Two Novel Bacteriophages Improve Survival in Galleria mellonella Infection and Mouse Acute Pneumonia Models Infected with Extensively Drug-Resistant Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2019, 85, e02900-18. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zeng, Y.; Wang, M.; Bao, R.; Chen, Y.; Li, X.; Pan, J.; Zhu, T.; Hu, B.; Tan, D. Characterization of Phage Resistance and Their Impacts on Bacterial Fitness in Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e02072-22. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.A.; Alorabi, J.A.; Al-Otaibi, L.M.; Ali, S.S.; Elsilk, S.E. Antibiotic Resistance and Biofilm Formation in Enterococcus spp. Isolated from Urinary Tract Infections. Pathogens 2022, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Caixeta Magalhães Tibúrcio, A.A.; Paiva, A.D.; Pedrosa, A.L.; Rodrigues, W.F.; Bernardes da Silva, R.; Oliveira, A.G. Effect of sub-inhibitory concentrations of antibiotics on biofilm formation and expression of virulence genes in penicillin-resistant, ampicillin-susceptible Enterococcus faecalis. Heliyon 2022, 8, e11154. [Google Scholar] [CrossRef] [PubMed]
- Tiria, F.; Odoyo, E.; Georges, M.; Nyerere, A.; Musila, L. Molecular Detection of Key Virulence-associated Genes and Phenotypic Analysis of Virulence Traits of Klebsiella pneumoniae Clinical Isolates from Kenya. J. Pure Appl. Microbiol. 2023, 17, 2194. [Google Scholar] [CrossRef]
- Donelli, G.; Vuotto, C.; Cardines, R.; Mastrantonio, P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol. Med. Microbiol. 2012, 65, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Mirzaie, A.; Ranjbar, R. Antibiotic resistance, virulence-associated genes analysis and molecular typing of Klebsiella pneumoniae strains recovered from clinical samples. AMB Express 2021, 11, 122. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 February 2024).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: A fast scalable bacteriophage annotation tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Rajandas, H.; Parimannan, S.; Manickam, R.; Marimuthu, K.; Petersen, B.; Clokie, M.R.J.; Millard, A.; Sicheritz-Pontén, T. PhageLeads: Rapid Assessment of Phage Therapeutic Suitability Using an Ensemble Machine Learning Approach. Viruses 2022, 14, 342. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In Gene Prediction. Methods in Molecular Biology; Kollmar, M., Ed.; Humana: New York, NY, USA, 2019; Volume 1962, pp. 1–14. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antimicrobial and Virulence Genes. Github. 2017. Available online: https://github.com/tseemann/abricate (accessed on 27 November 2023).
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Henz, S.R.; Huson, D.H.; Auch, A.F.; Nieselt-Struwe, K.; Schuster, S.C. Whole-genome prokaryotic phylogeny. Bioinformatics 2005, 21, 2329–2335. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Boeckaerts, D.; Stock, M.; Criel, B.; Gerstmans, H.; De Baets, B.; Briers, Y. Predicting bacteriophage hosts based on sequences of annotated receptor-binding proteins. Sci. Rep. 2021, 11, 1467. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Prokhorov, N.S.; Loessner, M.J.; Leiman, P.G. Reprogramming bacteriophage host range: Design principles and strategies for engineering receptor binding proteins. Curr. Opin. Biotechnol. 2021, 68, 272–281. [Google Scholar] [CrossRef]
- Degroux, S.; Effantin, G.; Linares, R.; Schoehn, G.; Breyton, C. Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger. J. Virol. 2023, 97, e0158422. [Google Scholar] [CrossRef] [PubMed]
- El Haddad, L.; Angelidakis, G.; Clark, J.R.; Mendoza, J.F.; Terwilliger, A.L.; Chaftari, C.P.; Duna, M.; Yusuf, S.T.; Harb, C.P.; Stibich, M.; et al. Genomic and Functional Characterization of Vancomycin-Resistant Enterococci-Specific Bacteriophages in the Galleria mellonella Wax Moth Larvae Model. Pharmaceutics 2022, 14, 1591. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Matsui, H.; Murakami, H.; Kato, S.-I.; Watanabe, N.; Nasukawa, T.; Mizukami, K.; Ogata, M.; Sakaguchi, M.; Matsuzaki, S.; et al. Potential Application of Bacteriophages in Enrichment Culture for Improved Prenatal Streptococcus agalactiae Screening. Viruses 2018, 10, 552. [Google Scholar] [CrossRef]
- Kang, H.-Y.; Kim, S.; Kim, J. Isolation and characterization of an Enterococcus faecalis bacteriophage. Korean J. Microbiol. 2015, 51, 194–198. [Google Scholar] [CrossRef]
- Wandro, S.; Ghatbale, P.; Attai, H.; Hendrickson, C.; Samillano, C.; Suh, J.; Dunham, S.J.B.; Pride, D.T.; Whiteson, K. Phage Cocktails Constrain the Growth of Enterococcus. mSystems 2022, 7, e00019-22. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Rashel, M.; Maeda, Y.; Takemura, I.; Sugihara, S.; Akechi, K.; Muraoka, A.; Wakiguchi, H.; Matsuzaki, S. Isolation and characterization of a novel Enterococcus faecalis bacteriophage φEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 2008, 278, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Moryl, M.; Palatyńska-Ulatowska, A.; Maszewska, A.; Grzejdziak, I.; Dias de Oliveira, S.; Pradebon, M.C.; Steier, L.; Różalski, A.; Poli de Figueiredo, J.A. Benefits and Challenges of the Use of Two Novel vB_Efa29212_2e and vB_Efa29212_3e Bacteriophages in Biocontrol of the Root Canal Enterococcus faecalis Infections. J. Clin. Med. 2022, 11, 6494. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Coppenhagen-Glazer, S.; Shlezinger, M.; Kott-Gutkowski, M.; Adini, O.; Beyth, N.; Hazan, R. Complete Genome Sequence of Enterococcus Bacteriophage EFLK1. Genome Announc. 2015, 3, e01308-15. [Google Scholar] [CrossRef]
- Khalifa, L.; Gelman, D.; Shlezinger, M.; Dessal, A.L.; Coppenhagen-Glazer, S.; Beyth, N.; Hazan, R. Defeating Antibiotic- and Phage-Resistant Enterococcus faecalis Using a Phage Cocktail in Vitro and in a Clot Model. Front. Microbiol. 2018, 9, 326. [Google Scholar] [CrossRef]
- Lu, M.J.; Stierhof, Y.D.; Henning, U. Location and unusual membrane topology of the immunity protein of the Escherichia coli phage T4. J. Virol. 1993, 67, 4905–4913. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Egido, J.E.; Costa, A.R.; Aparicio-Maldonado, C.; Haas, P.-J.; Brouns, S.J.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 2022, 46, fuab048. [Google Scholar] [CrossRef]
- Jiang, W.; Han, G.; Zhou, C.; Zhang, Y.; Via, B.K. Thermal Degradation Kinetics of Polysaccharides in Fibrous Materials during Hot Water Treatment: A Preliminary Study. BioResources 2014, 9, 4205–4211. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, Y.; Vincent, M.; Sun, Y.; Yu, H.; Wang, J.; Bao, Q.; Kong, H.; Hu, S. Complete genome sequence of bacteriophage T5. Virology 2005, 332, 45–65. [Google Scholar] [CrossRef]
- El Haddad, L.; Harb, C.P.; Gebara, M.A.; Stibich, M.A.; Chemaly, R.F. A Systematic and Critical Review of Bacteriophage Therapy Against Multidrug-resistant ESKAPE Organisms in Humans. Clin. Infect. Dis. 2019, 69, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, F.; Hallajzadeh, M.; Sholeh, M.; Talebi, M.; Mahabadi, V.P.; Amirmozafari, N. Biological characteristics and anti-biofilm activity of a lytic phage against vancomycin-resistant Enterococcus faecium. Iran. J. Microbiol. 2021, 13, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. Expression of animal virus genomes. Bacteriol. Rev. 1971, 35, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ma, Y.; Wang, Y.; Yang, H.; Shen, W.; Chen, X. Transcription regulation mechanisms of bacteriophages: Recent advances and future prospects. Bioengineered 2014, 5, 300–304. [Google Scholar] [CrossRef]
- van den Berg, D.F.; van der Steen, B.A.; Costa, A.R.; Brouns, S.J.J. Phage tRNAs evade tRNA-targeting host defenses through anticodon loop mutations. eLife 2023, 12, e85183. [Google Scholar] [CrossRef] [PubMed]
- Abed, S.; Sholeh, M.; Asforooshani, M.K.; Shafiei, M.; Shahraki, A.H.; Nasr, S. Insights into the novel Enterococcus faecalis phage: A comprehensive genome analysis. PLoS ONE 2024, 19, e0301292. [Google Scholar] [CrossRef]
- Topka-Bielecka, G.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Jurczak-Kurek, A.; Górniak, M.; Dydecka, A.; Necel, A.; Węgrzyn, G.; Węgrzyn, A. Characterization of the Bacteriophage vB_EfaS-271 Infecting Enterococcus faecalis. Int. J. Mol. Sci. 2020, 21, 6345. [Google Scholar] [CrossRef]
- Oh, H.K.; Hwang, Y.J.; Hong, H.W.; Myung, H. Comparison of Enterococcus faecalis Biofilm Removal Efficiency among Bacteriophage PBEF129, Its Endolysin, and Cefotaxime. Viruses 2021, 13, 426. [Google Scholar] [CrossRef]
- Zaburlin, D.; Quiberoni, A.; Mercanti, D. Changes in Environmental Conditions Modify Infection Kinetics of Dairy Phages. Food Environ. Virol. 2017, 9, 270–276. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.R.; Ferreira, R.; Costa, A.R.; Oliveira, H.; Azeredo, J. Efficacy and safety assessment of two enterococci phages in an in vitro biofilm wound model. Sci. Rep. 2019, 9, 6643. [Google Scholar] [CrossRef] [PubMed]
- Duerkop, B.A.; Palmer, K.L.; Horsburgh, M.J. Enterococcal Bacteriophages and Genome Defense. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed]
- Drexler, K.; Riede, I.; Montag, D.; Eschbach, M.-L.; Henning, U. Receptor specificity of the Escherichia coli T-even type phage Ox2: Mutational alterations in host range mutants. J. Mol. Biol. 1989, 207, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Guo, W.; Lu, J.; Pan, W.; Song, F.; Wang, P. Enterococcus faecalis Bacteriophage vB_EfaS_efap05-1 Targets the Surface Polysaccharide and ComEA Protein as the Receptors. Front. Microbiol. 2022, 13, 866382. [Google Scholar] [CrossRef] [PubMed]
- Duerkop, B.A.; Huo, W.; Bhardwaj, P.; Palmer, K.L.; Hooper, L.V. Molecular Basis for Lytic Bacteriophage Resistance in Enterococci. mBio 2016, 7, e01304-16. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Li, Y.; Lu, Y.; Xiong, K.; Zhong, Q.; Wang, J. Bacteriophage-Resistant Mutant of Enterococcus faecalis Is Impaired in Biofilm Formation. Front. Microbiol. 2022, 13, 913023. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Leptihn, S. Defense and anti-defense mechanisms of bacteria and bacteriophages. J. Zhejiang Univ. Sci. B 2024, 25, 181. [Google Scholar] [CrossRef]
- Millman, A.; Melamed, S.; Leavitt, A.; Doron, S.; Bernheim, A.; Hör, J.; Garb, J.; Bechon, N.; Brandis, A.; Lopatina, A.; et al. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 2022, 30, 1556–1569.e5. [Google Scholar] [CrossRef]
- Georjon, H.; Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef]
- Molineux, I.J.; Panja, D. Popping the cork: Mechanisms of phage genome ejection. Nat. Rev. Microbiol. 2013, 11, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.-W.; Tremblay, D.; Moineau, S. Long-term bacteriophage preservation. WFCC Newsl. 2004, 38, 35–40. [Google Scholar]
- Sanz, M.; Beighton, D.; Curtis, M.A.; Cury, J.A.; Dige, I.; Dommisch, H.; Ellwood, R.; Giacaman, R.A.; Herrera, D.; Herzberg, M.C.; et al. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J. Clin. Periodontol. 2017, 44, S5–S11. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Bamford, N.C.; MacPhee, C.E.; Stanley-Wall, N.R. Microbial Primer: An introduction to biofilms—What they are, why they form and their impact on built and natural environments. Microbiology 2023, 169, 001338. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. Biofilms and antimicrobial resistance. Clin. Orthop. Relat. Res. 2005, 437, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S.; Burgess, J.G. Extracellular DNA in oral microbial biofilms. Microbes Infect. 2015, 17, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Winans, J.B.; Wucher, B.R.; Nadell, C.D. Multispecies biofilm architecture determines bacterial exposure to phages. PLoS Biol. 2022, 20, e3001913. [Google Scholar] [CrossRef]
- Schmelcher, M.; Loessner, M.J. Bacteriophage endolysins: Applications for food safety. Curr. Opin. Biotechnol. 2016, 37, 76–87. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef]
- Kaplan, J.B.; Mlynek, K.D.; Hettiarachchi, H.; Alamneh, Y.A.; Biggemann, L.; Zurawski, D.V.; Black, C.C.; Bane, C.E.; Kim, R.K.; Granick, M.S. Extracellular polymeric substance (EPS)-degrading enzymes reduce staphylococcal surface attachment and biocide resistance on pig skin in vivo. PLoS ONE 2018, 13, e0205526. [Google Scholar] [CrossRef] [PubMed]
- Srey, S.; Jahid, I.K.; Ha, S.-D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Taati Moghadam, M.; Farahani, I.; Jazireian, P.; Shariati, A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 45. [Google Scholar] [CrossRef]
- Gu, J.; Liu, X.; Li, Y.; Han, W.; Lei, L.; Yang, Y.; Zhao, H.; Gao, Y.; Song, J.; Lu, R.; et al. A method for generation phage cocktail with great therapeutic potential. PLoS ONE 2012, 7, e31698. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.; Koley, H.; Ghosh, A.; Palit, A.; Sarkar, B. Efficacy of cocktail phage therapy in treating Vibrio cholerae infection in rabbit model. Microbes Infect. 2013, 15, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Latz, S.; Krüttgen, A.; Häfner, H.; Buhl, E.M.; Ritter, K.; Horz, H.-P. Differential Effect of Newly Isolated Phages Belonging to PB1-Like, phiKZ-Like and LUZ24-Like Viruses against Multi-Drug Resistant Pseudomonas aeruginosa under Varying Growth Conditions. Viruses 2017, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.-J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef]
- Chang, C.; Yu, X.; Guo, W.; Guo, C.; Guo, X.; Li, Q.; Zhu, Y. Bacteriophage-Mediated Control of Biofilm: A Promising New Dawn for the Future. Front. Microbiol. 2022, 13, 825828. [Google Scholar] [CrossRef]


| No. | Bacterial Isolates | Sequence Types (ST) | Origin | Spot Assay | Efficiency of Plating (EOP) |
|---|---|---|---|---|---|
| 1 | E. faecalis EFS8 * | 1904 | Urinary tract infection | ++ | 1 |
| 2 | E. faecalis EFS1 | 6 | Skin and soft tissue infection | ++ | 0.15 |
| 3 | E. faecalis EFS4 | 947 | Skin and soft tissue infection | + | <0.001 |
| 4 | E. faecalis EFS5 | 6 | Skin and soft tissue infection | ++ | 0.076 |
| 5 | E. faecalis EFS6 | 6 | Skin and soft tissue infection | ++ | 0.05 |
| 6 | E. faecalis EFS9 | 6 | Urinary tract infection | ++ | 0.01 |
| 7 | E. faecalis EFS10 | 6 | Urinary tract infection | ++ | 1.1 |
| 8 | E. faecalis EFS11 | 368 | Urinary tract infection | ++ | 0.0004 |
| 9 | E. faecalis EFS13 | 59 | Skin and soft tissue infection | ++ | 0.5 |
| 10 | E. faecalis EFS14 | 6 | Skin and soft tissue infection | ++ | 1.7 |
| 11 | E. faecalis EFS15 | 16 | Urinary tract infection | + | <0.001 |
| 12 | E. faecalis EFS17 | 6 | Skin and soft tissue infection | + | <0.001 |
| 13 | E. faecalis EFS18 | 368 | Urinary tract infection | ++ | 0.0011 |
| 14 | E. faecalis EFS19 | 1907 | Urinary tract infection | ++ | 0.12 |
| 15 | E. faecalis EFS21 | 44 | Skin and soft tissue infection | + | <0.001 |
| 16 | E. faecalis EFS22 | 1908 | Skin and soft tissue infection | + | <0.001 |
| 17 | E. faecalis EFS23 | 6 | Urinary tract infection | ++ | 3 |
| 18 | E. faecalis EFS25 | 6 | Surgical site infection | ++ | 1.5 |
| 19 | E. faecalis EFS26 | 6 | Skin and soft tissue infection | ++ | 1.2 |
| 20 | E. faecalis EFS27 | 1903 | Urinary tract infection | ++ | 0.14 |
| 21 | E. faecalis EFS28 | 28 | Skin and soft tissue infection | ++ | 0.6 |
| 22 | E. faecalis EFS29 | 6 | Blood infection | ++ | 0.8 |
| 23 | E. faecalis EFS30 | 28 | Skin and soft tissue infection | ++ | 0.8 |
| 24 | E. faecalis EFS31 | 6 | Urinary tract infection | ++ | 1.2 |
| 25 | E. faecalis EFS32 | 1903 | Urinary tract infection | ++ | 0.9 |
| 26 | E. faecalis EFS33 | 1903 | Skin and soft tissue infection | ++ | 6 |
| 27 | E. faecium EFM5 | 80 | Urinary tract infection | + | <0.001 |
| 28 | E. faecium EFM1 | Skin and soft tissue infection | − | N/A | |
| 29 | E. faecium EFM2 | 80 | Skin and soft tissue infection | − | N/A |
| 30 | E. faecium EFM3 | Skin and soft tissue infection | − | N/A | |
| 31 | E. faecium EFM4 | 80 | Skin and soft tissue infection | − | N/A |
| 32 | E. faecium EFM6 | 612 | Skin and soft tissue infection | − | N/A |
| 33 | E. faecium EFM7 | 761 | Skin and soft tissue infection | − | N/A |
| 34 | E. faecium EFM8 | 80 | Urinary tract infection | − | N/A |
| 35 | E. faecium EFM9 | 80 | Skin and soft tissue infection | − | N/A |
| 36 | E. faecium EFM10 | 2672 | Urinary tract infection | − | N/A |
| 37 | E. faecium EFM11 | 761 | Surgical site infection | − | N/A |
| Protein Name | Phage Name | % Identity | Protein Accession Number | Protein Length | Phage Hosts |
|---|---|---|---|---|---|
| Tail fiber protein * | Enterococcus phage vB_Efs8_KEN04 | 100% | WZP34890.1 | 1832 | E. faecalis EFS8 |
| Putative tail fiber protein | Enterococcus phage MDA2 | 99.89% | QVW28137.1 | 1825 | E. faecium VREfm (VRE001, VRE004, VRE008, VRE1147, VRE1181) [73] |
| Putative tail fiber | Enterococcus phage phiM1EF22 | 99.95% | BBE37304.1 | 1822 | E. faecalis KUEF22 [74] |
| Tail fiber protein | Enterococcus phage ECP3 | 99.89% | YP_009147083.1 | 1822 | E. faecalis 10K28 [75] |
| Putative tail fiber | Enterococcus phage phiEF17H | 99.89% | BBE37101.1 | 1822 | E. faecalis EF17 [74] |
| Tail fiber protein | Enterococcus phage vB_OCPT_Carl | 98.68% | UQT00063.1 | 1825 | E. faecalis strains (DP11, EF07, EF116PII, EF11, EF09PII, Ent6, V587, Yi6-1) [76] |
| Tail fiber protein | Enterococcus phage EF24C | 98.47% | YP_001504140.1 | 1825 | E. faecalis EF24 [77] |
| Tail fiber protein | Enterococcus phage vB_OCPT_Bob | 98.30% | UQT00475.1 | 1825 | E. faecalis strains (B3286, DP6, EF06, DP11, EF07, EF116PII, EF11, EF09PII, Ent6, V587, Yi6-1) [76] |
| Tail fiber protein | Enterococcus phage vB_OCPT_Car | 97.10% | UQT00278.1 | 1825 | E. faecalis strains (DP11, EF07, EF116PII, EF11, EF09PII, Ent6, V587, Yi6-1) [76] |
| Putative tail fiber | Enterococcus phage Sw5 | 96.38% | USL84310.1 | 1825 | E. faecalis OG1RF |
| Tail fiber protein | Enterococcus phage vB_Efa29212_3e | 96.66% | UYB00790.1 | 1825 | E. faecalis ATCC 29212TM [78] |
| Tail fiber protein | Enterococcus phage EFLK1 | 95.99% | YP_009219864.2 | 1822 | E. faecalis V583 and E. faecalis V583 phage-resistant mutant (EFDG1r) [79,80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soro, O.; Kigen, C.; Nyerere, A.; Gachoya, M.; Georges, M.; Odoyo, E.; Musila, L. Characterization and Anti-Biofilm Activity of Lytic Enterococcus Phage vB_Efs8_KEN04 against Clinical Isolates of Multidrug-Resistant Enterococcus faecalis in Kenya. Viruses 2024, 16, 1275. https://doi.org/10.3390/v16081275
Soro O, Kigen C, Nyerere A, Gachoya M, Georges M, Odoyo E, Musila L. Characterization and Anti-Biofilm Activity of Lytic Enterococcus Phage vB_Efs8_KEN04 against Clinical Isolates of Multidrug-Resistant Enterococcus faecalis in Kenya. Viruses. 2024; 16(8):1275. https://doi.org/10.3390/v16081275
Chicago/Turabian StyleSoro, Oumarou, Collins Kigen, Andrew Nyerere, Moses Gachoya, Martin Georges, Erick Odoyo, and Lillian Musila. 2024. "Characterization and Anti-Biofilm Activity of Lytic Enterococcus Phage vB_Efs8_KEN04 against Clinical Isolates of Multidrug-Resistant Enterococcus faecalis in Kenya" Viruses 16, no. 8: 1275. https://doi.org/10.3390/v16081275
APA StyleSoro, O., Kigen, C., Nyerere, A., Gachoya, M., Georges, M., Odoyo, E., & Musila, L. (2024). Characterization and Anti-Biofilm Activity of Lytic Enterococcus Phage vB_Efs8_KEN04 against Clinical Isolates of Multidrug-Resistant Enterococcus faecalis in Kenya. Viruses, 16(8), 1275. https://doi.org/10.3390/v16081275





