Resistance of the CRISPR-Cas13a Gene-Editing System to Potato Spindle Tuber Viroid Infection in Tomato and Nicotiana benthamiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of pC1300-PSTVd-s
2.2. Construction of PSTVd-Targeted crRNAs
2.3. Transformation and Agrobacterium Suspension Preparation
2.4. Transient Expression of Recombinant Vectors in ‘Rutgers’ Tomato Plants
2.5. Preparation of DIG-Labeled Probe and Northern blot Hybridization
2.6. Development and Selection of Transgenic N. benthamiana Plants
2.7. PSTVd Inoculation in Transgenic N. benthamiana CCR2(+) Plants
2.8. Densitometry and Statistical Analysis
3. Results
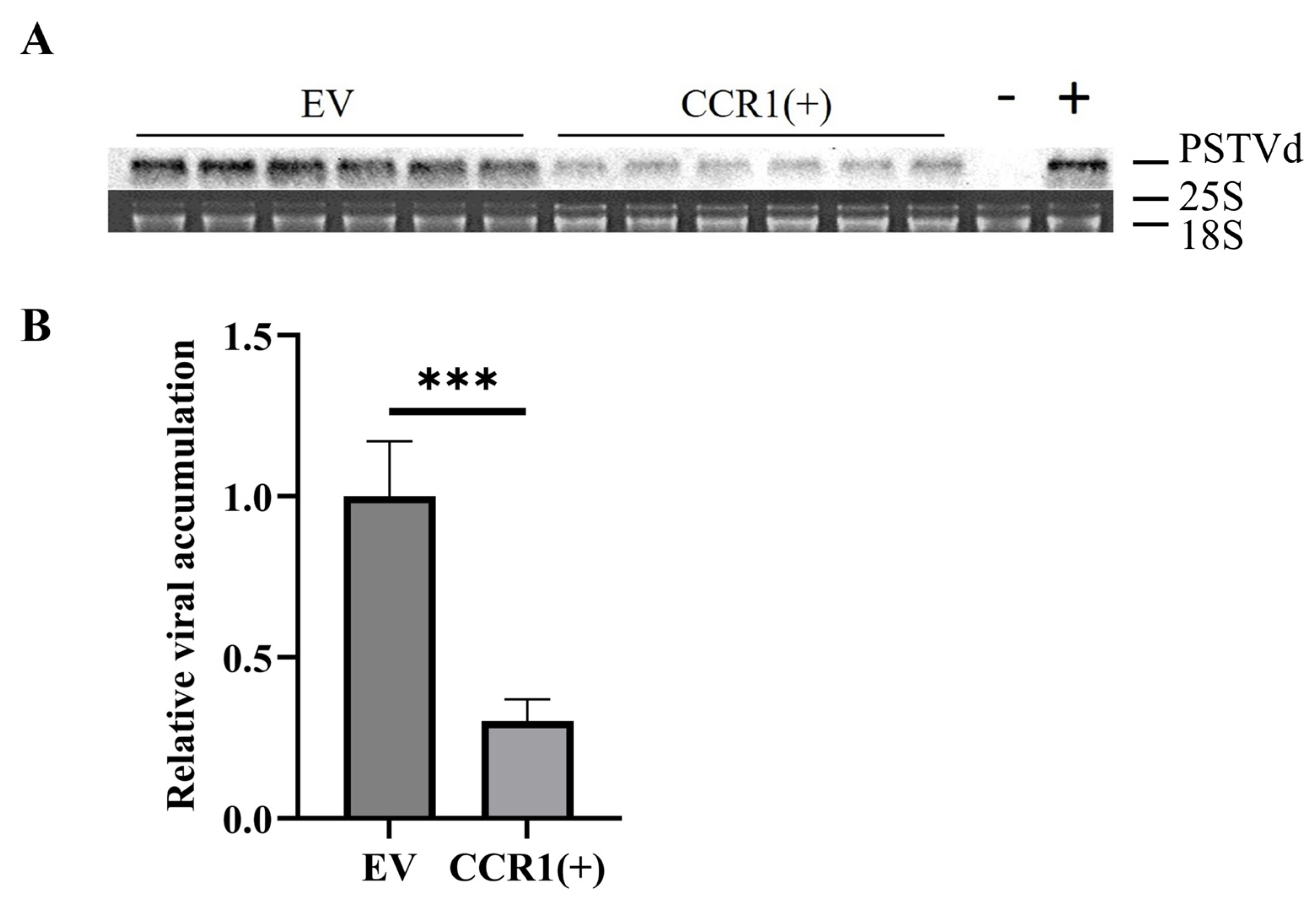
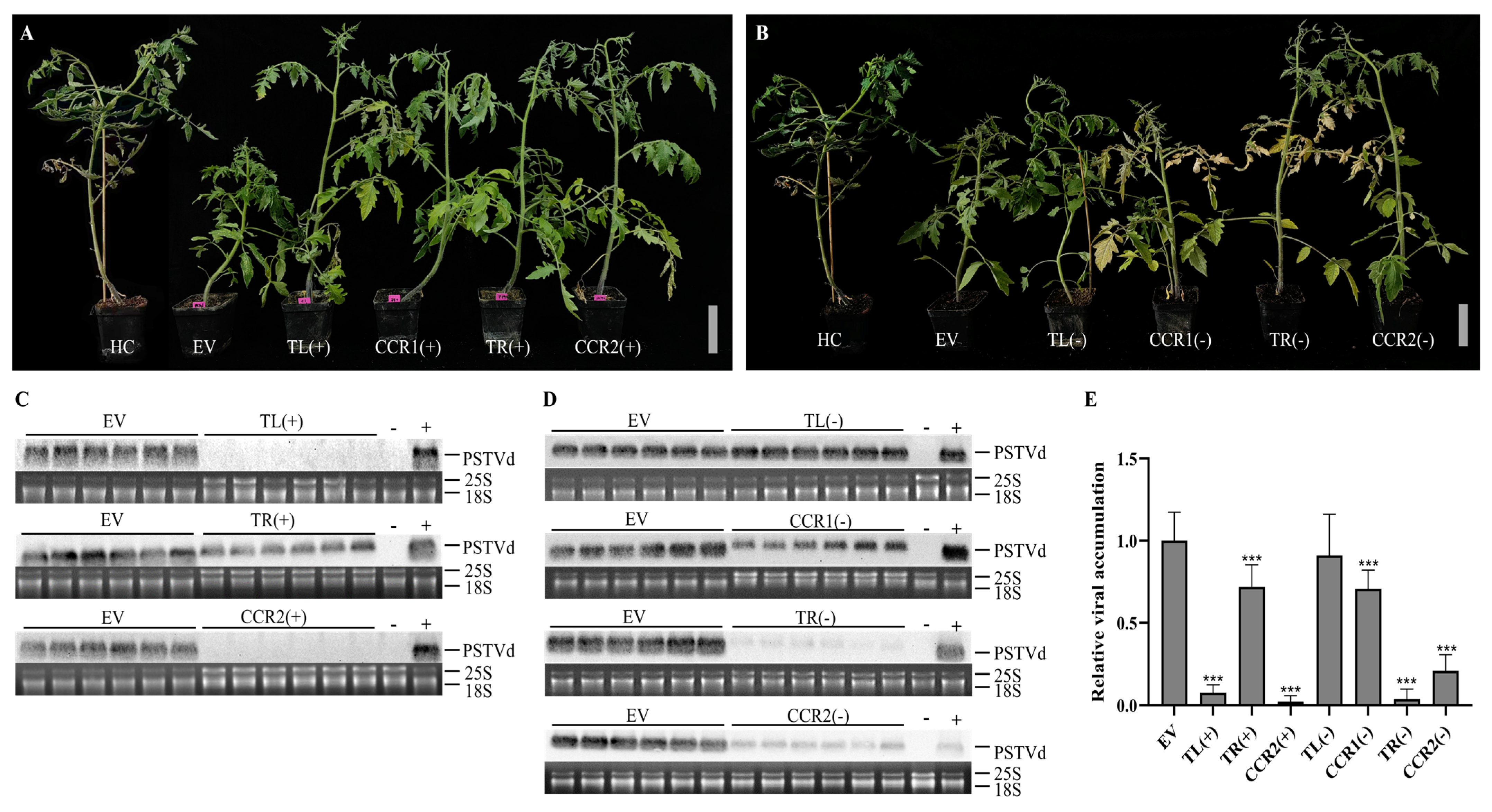
3.1. The CRISPR-Cas13a System can Efficiently Reduce Viroid Accumulation
3.2. crRNAs Targeting PSTVd Genomic RNA
3.3. Screen of crRNAs with High Efficiency Targeting PSTVd RNA
3.4. Stable Expression of the CRISPR-Cas13a System Attenuates Viroid Accumulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moriones, E.; Verdin, E. Viral diseases. In Integrated Pest and Disease Management in Greenhouse Crops. Plant Pathology in the 21st Century; Springer: Cham, Switzerland, 2020; pp. 3–31. [Google Scholar]
- Owens, R.A.; Verhoeven, J.T.J. Potato spindle tuber viroid. In Viroids and Satellites; Academic Press: London, UK, 2017; pp. 149–158. [Google Scholar] [CrossRef]
- Di Serio, F.; Chiumenti, M. Viroid taxonomy. In Fundamentals of Viroid Biology; Academic Press: London, UK, 2024; pp. 25–44. [Google Scholar] [CrossRef]
- Keese, P.; Symons, R.H. Domains in viroids: Evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 1985, 82, 4582–4586. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Randles, J.W.; Bar-Joseph, M.; Diener, T. A proposed scheme for viroid classification and nomenclature. Arch. Virol. 1998, 143, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Navarro, B.; Kovalskaya, N.; Hammond, R.W.; Di Serio, F. Engineering resistance against viroids. Curr. Opin. Virol. 2017, 26, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Góra-Sochacka, A.; Candresse, T.; Zagórski, W. Genetic variability of potato spindle tuber viroid RNA replicon. Acta Biochim. Pol. 2001, 48, 467–476. [Google Scholar] [CrossRef]
- Sano, T.; Nagayama, A.; Ogawa, T.; Ishida, I.; Okada, Y. Transgenic potato expressing a double-stranded RNA-specific ribonuclease is resistant to potato spindle tuber viroid. Nat. Biotechnol. 1997, 15, 1290–1294. [Google Scholar] [CrossRef] [PubMed]
- Atkins, D.; Young, M.; Uzzell, S.; Kelly, L.; Fillatti, J.; Gerlach, W.L. The expression of antisense and ribozyme genes targeting citrus exocortis viroid in transgenic plants. J. Gen. Virol. 1995, 76, 1781–1790. [Google Scholar] [CrossRef]
- Jo, K.M.; Jo, Y.; Choi, H.; Chu, H.; Lian, S.; Yoon, J.Y.; Choi, S.K.; Kim, K.H.; Cho, W.K. Development of genetically modified chrysanthemums resistant to chrysanthemum stunt viroid using sense and antisense RNAs. Sci. Hortic. 2015, 195, 17–24. [Google Scholar] [CrossRef]
- Yang, X.C.; Yie, Y.; Zhu, F.; Liu, Y.L.; Kang, L.Y.; Wang, X.F.; Tien, P. Ribozyme-mediated high resistance against potato spindle tuber viroid in transgenic potatoes. Proc. Natl. Acad. Sci. USA 1997, 94, 4861–4865. [Google Scholar] [CrossRef]
- Carbonell, A. RNAi tools for controlling viroid diseases. Virus Res. 2022, 313, 198729. [Google Scholar] [CrossRef]
- Adkar-Purushothama, C.R.; Kasai, A.; Sugawara, K.; Yamamoto, H.; Yamazaki, Y.; He, Y.H.; Takada, N.; Goto, H.; Shindo, S.; Harada, T.; et al. RNAi mediated inhibition of viroid infection in transgenic plants expressing viroid-specific small RNAs derived from various functional domains. Sci. Rep. 2015, 5, 17949. [Google Scholar] [CrossRef]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.A.; Wang, M.B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 2009, 10, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; de Alba, Á.E.M.; Flores, R.; Gago, S. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 2008, 371, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Daròs, J.A. Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol. Plant Pathol. 2017, 18, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A. Next-generation sequencing and CRISPR/Cas13 editing in viroid research and molecular diagnostics. Viruses 2019, 11, 120. [Google Scholar] [CrossRef]
- Perčulija, V.; Lin, J.; Zhang, B.; Ouyang, S. Functional Features and Current Applications of the RNA-Targeting Type VI CRISPR-Cas Systems. Adv. Sci. 2021, 8, 2004685. [Google Scholar] [CrossRef]
- Zaidi, S.S.-e.-A.; Tashkandi, M.; Mansoor, S.; Mahfouz, M.M. Engineering plant immunity: Using CRISPR/Cas9 to generate virus resistance. Front. Plant Sci. 2016, 7, 228325. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.; Kellner, M.J.; Regev, A. RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019, 76, 826–837. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1–9. [Google Scholar] [CrossRef]
- Jiao, B.L.; Hao, X.Y.; Liu, Z.M.; Liu, M.B.; Wang, J.Y.; Liu, L.; Liu, N.; Song, R.; Zhang, J.X.; Fang, Y.L.; et al. Engineering CRISPR immune systems conferring GLRaV-3 resistance in grapevine. Hortic. Res. 2022, 9, uhab023. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Pan, Z.; Wang, X.; Bian, X.; Wang, W.; Liang, Q.; Kou, M.; Ji, H.; Li, Y.; Ma, D. Targeting of SPCSV-RNase3 via CRISPR-Cas13 confers resistance against sweet potato virus disease. Mol. Plant Pathol. 2022, 23, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185. [Google Scholar] [CrossRef] [PubMed]
- Steger, G. Predicting the Structure of a Viroid: Structure, Structure Distribution, Consensus Structure, and Structure Drawing; Humana: New York, NY, USA, 2022; pp. 331–371. [Google Scholar] [CrossRef]
- Khoo, Y.W.; Li, S.; Chong, K.P. Development of rapid DNA extraction and PCR amplification methods for fungi and parasitic plants. Zemdirbyste-Agriculture 2022, 109, 283–286. [Google Scholar] [CrossRef]
- Xing, F. Identification, Detection Techniques and Pathogenicity Of Viral Pathogen Associated with Mosaic Disease of Apple Trees in China. Ph.D. Thesis, China Agricultural University, Beijing, China, 2018. [Google Scholar]
- Jiang, D.; Hou, W.; Sano, T.; Kang, N.; Qin, L.; Wu, Z.; Li, S.; Xie, L. Rapid detection and identification of viroids in the genus Coleviroid using a universal probe. J. Virol. Methods 2013, 187, 321–326. [Google Scholar] [CrossRef]
- Ma, X.; Xu, X.; Li, X.; Shang, X. Rapid and efficient production of homozygous transgenic tobacco plants with Agrobacterium tumefaciens. Acta Tabacaria Sin. 2012, 18, 66–71. [Google Scholar]
- Schrader, O.; Baumstark, T.; Riesner, D. A Mini-RNA containing the tetraloop, wobble-pair and loop E motifs of the central conserved region of potato spindle tuber viroid is processed into a minicircle. Nucleic Acids Res. 2003, 31, 988–998. [Google Scholar] [CrossRef]
- Perreault, J.-P.; Bolduc, F.; Adkar-Purushothama, C.R. Structure of viroids. In Fundamentals of Viroid Biology; Academic Press: London, UK, 2024; pp. 45–61. [Google Scholar] [CrossRef]
- Itaya, A.; Zhong, X.H.; Bundschuh, R.; Qi, Y.J.; Wang, Y.; Takeda, R.; Harris, A.R.; Molina, C.; Nelson, R.S.; Ding, B. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 2007, 81, 2980–2994. [Google Scholar] [CrossRef]
- Spiesmacher, E.; Mühlbach, H.-P.; Schnölzer, M.; Haas, B.; Sänger, H.L. Oligomeric forms of potato spindle tuber viroid (PSTV) and of its complementary RNA are present in nuclei isolated from viroid-infected potato cells. Biosci. Rep. 1983, 3, 767–774. [Google Scholar] [CrossRef]
- Branch, A.D.; Robertson, H.D. A Replication Cycle for Viroids and Other Small Infectious RNA’s. Science 1984, 223, 450–455. [Google Scholar] [CrossRef]
- Owens, R.A.; Diener, T. RNA intermediates in potato spindle tuber viroid replication. Proc. Natl. Acad. Sci. USA 1982, 79, 113–117. [Google Scholar] [CrossRef]
- Grimm, D. Small silencing RNAs: State-of-the-art. Adv. Drug Deliv. Rev. 2009, 61, 672–703. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Viral vectors applied for RNAi-based antiviral therapy. Viruses 2020, 12, 924. [Google Scholar] [CrossRef]
- Hoang, B.T.L.; Fletcher, S.J.; Brosnan, C.A.; Ghodke, A.B.; Manzie, N.; Mitter, N. RNAi as a Foliar Spray: Efficiency and Challenges to Field Applications. Int. J. Mol. Sci. 2022, 23, 6639. [Google Scholar] [CrossRef] [PubMed]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Hillary, V.E.; Ceasar, S.A. A review on the mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14 proteins utilized for genome engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef]
- Naoi, T.; Hataya, T. Tolerance even to lethal strain of potato spindle tuber viroid found in wild tomato species can be introduced by crossing. Plants 2021, 10, 575. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khoo, Y.W.; Wang, Q.; Liu, S.; Zhan, B.; Xu, T.; Lv, W.; Liu, G.; Li, S.; Zhang, Z. Resistance of the CRISPR-Cas13a Gene-Editing System to Potato Spindle Tuber Viroid Infection in Tomato and Nicotiana benthamiana. Viruses 2024, 16, 1401. https://doi.org/10.3390/v16091401
Khoo YW, Wang Q, Liu S, Zhan B, Xu T, Lv W, Liu G, Li S, Zhang Z. Resistance of the CRISPR-Cas13a Gene-Editing System to Potato Spindle Tuber Viroid Infection in Tomato and Nicotiana benthamiana. Viruses. 2024; 16(9):1401. https://doi.org/10.3390/v16091401
Chicago/Turabian StyleKhoo, Ying Wei, Qingsong Wang, Shangwu Liu, Binhui Zhan, Tengfei Xu, Wenxia Lv, Guangjing Liu, Shifang Li, and Zhixiang Zhang. 2024. "Resistance of the CRISPR-Cas13a Gene-Editing System to Potato Spindle Tuber Viroid Infection in Tomato and Nicotiana benthamiana" Viruses 16, no. 9: 1401. https://doi.org/10.3390/v16091401
APA StyleKhoo, Y. W., Wang, Q., Liu, S., Zhan, B., Xu, T., Lv, W., Liu, G., Li, S., & Zhang, Z. (2024). Resistance of the CRISPR-Cas13a Gene-Editing System to Potato Spindle Tuber Viroid Infection in Tomato and Nicotiana benthamiana. Viruses, 16(9), 1401. https://doi.org/10.3390/v16091401





