KSHV ORF20 Promotes Coordinated Lytic Reactivation for Increased Infectious Particle Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reverse-Transcription (RT)-Quantitative PCR (RT-qPCR)
2.2. Construction of ORF20-Null Recombinant KSHV Genome and Virus Reconstitution
2.3. BAC16-ORF20-Null Virus Reconstitution and Lytic Reactivation
2.4. Cell Culture
2.5. Western Blot Analysis and Antibodies
2.6. Purification and Quantification of Viral DNA by TaqMan Real-Time PCR
2.7. Infectious Virus Quantification
2.8. Cell Death Analysis
2.9. Recombinant Lentiviral Vectors
3. Results
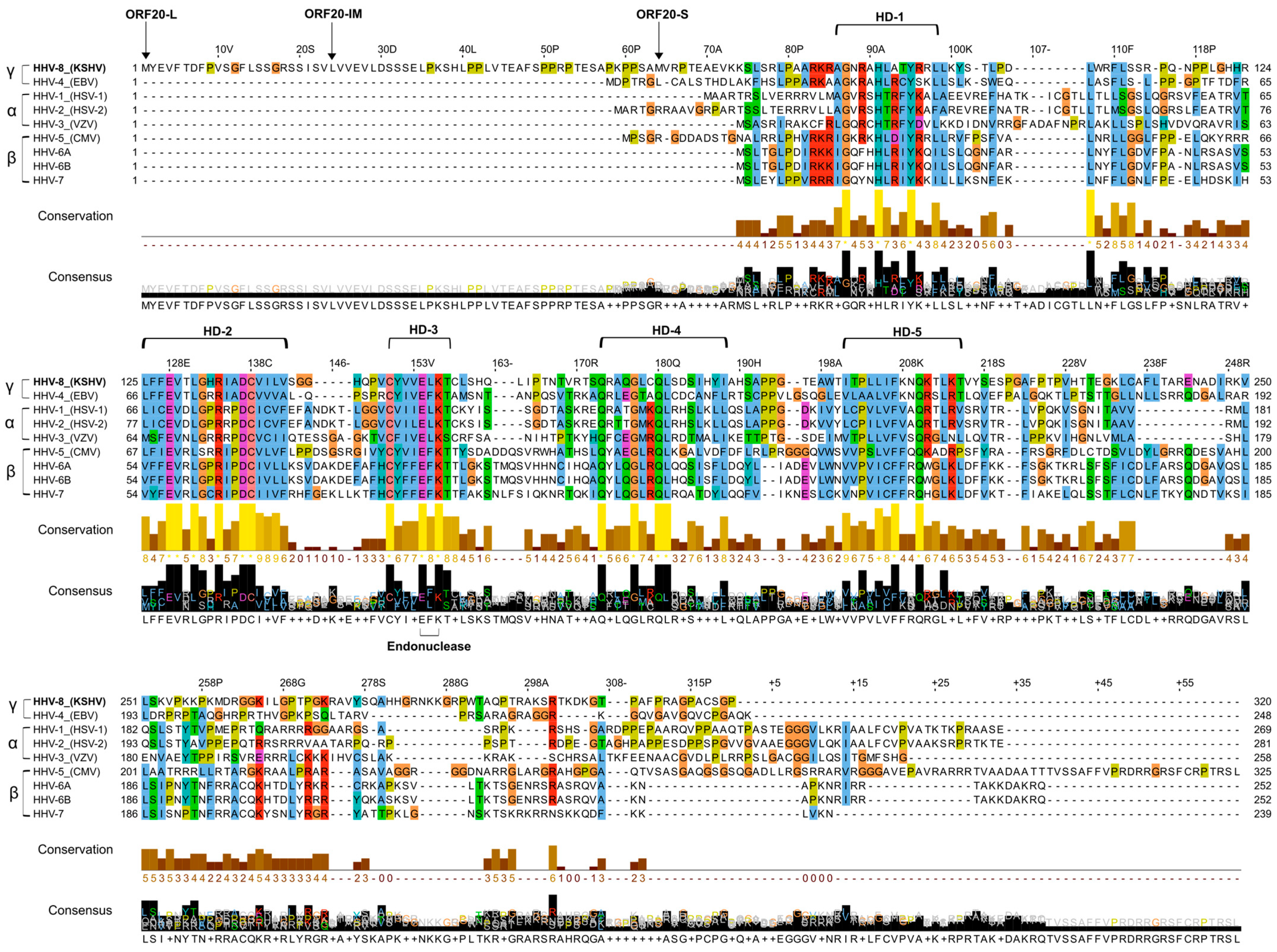
3.1. Alignment of KSHV ORF20 with UL24 Family Members Encoded by Human Herpesviruses
3.2. Kinetics of ORF20 Gene Expression
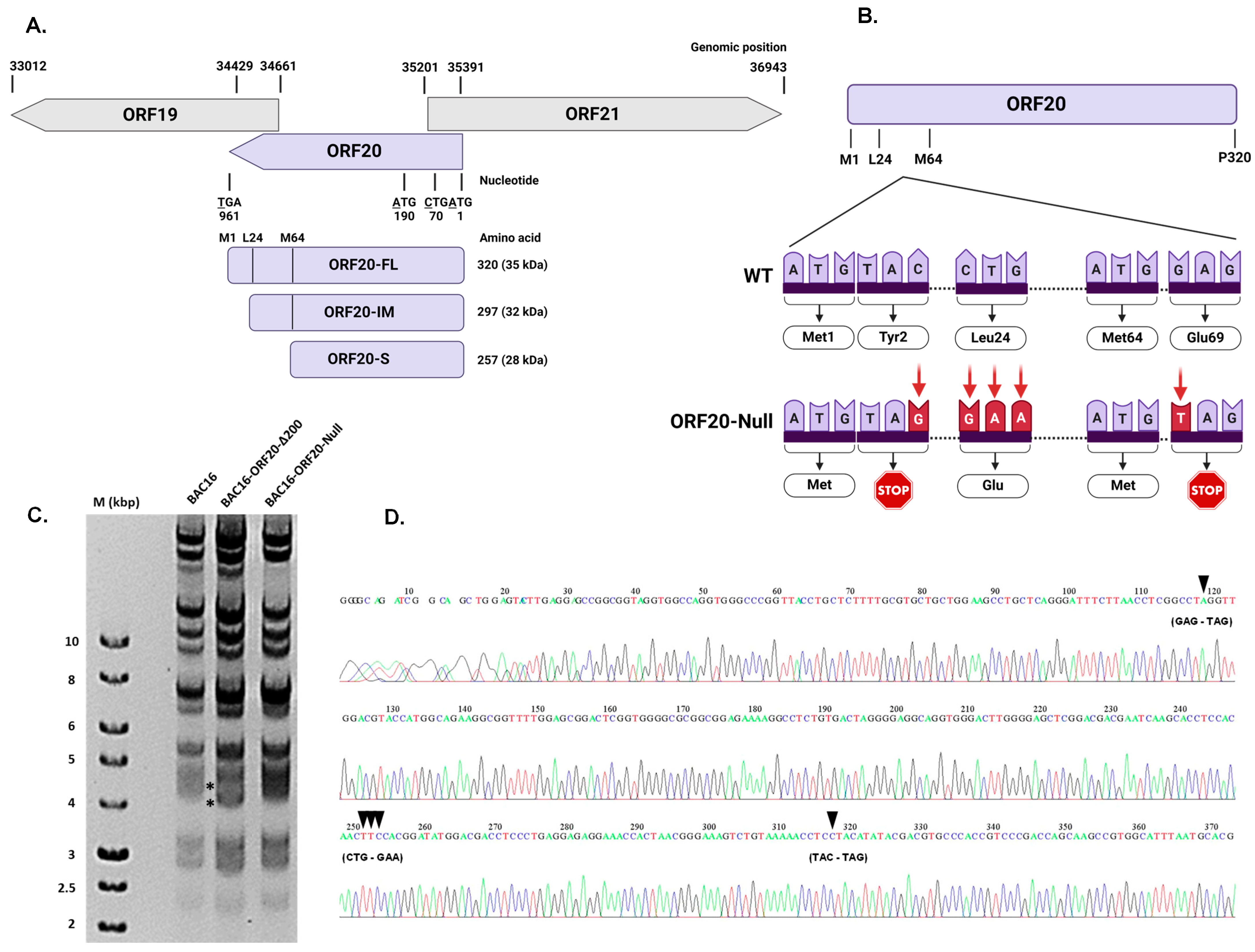
3.3. Construction of Recombinant ORF20-Null KSHV Genome
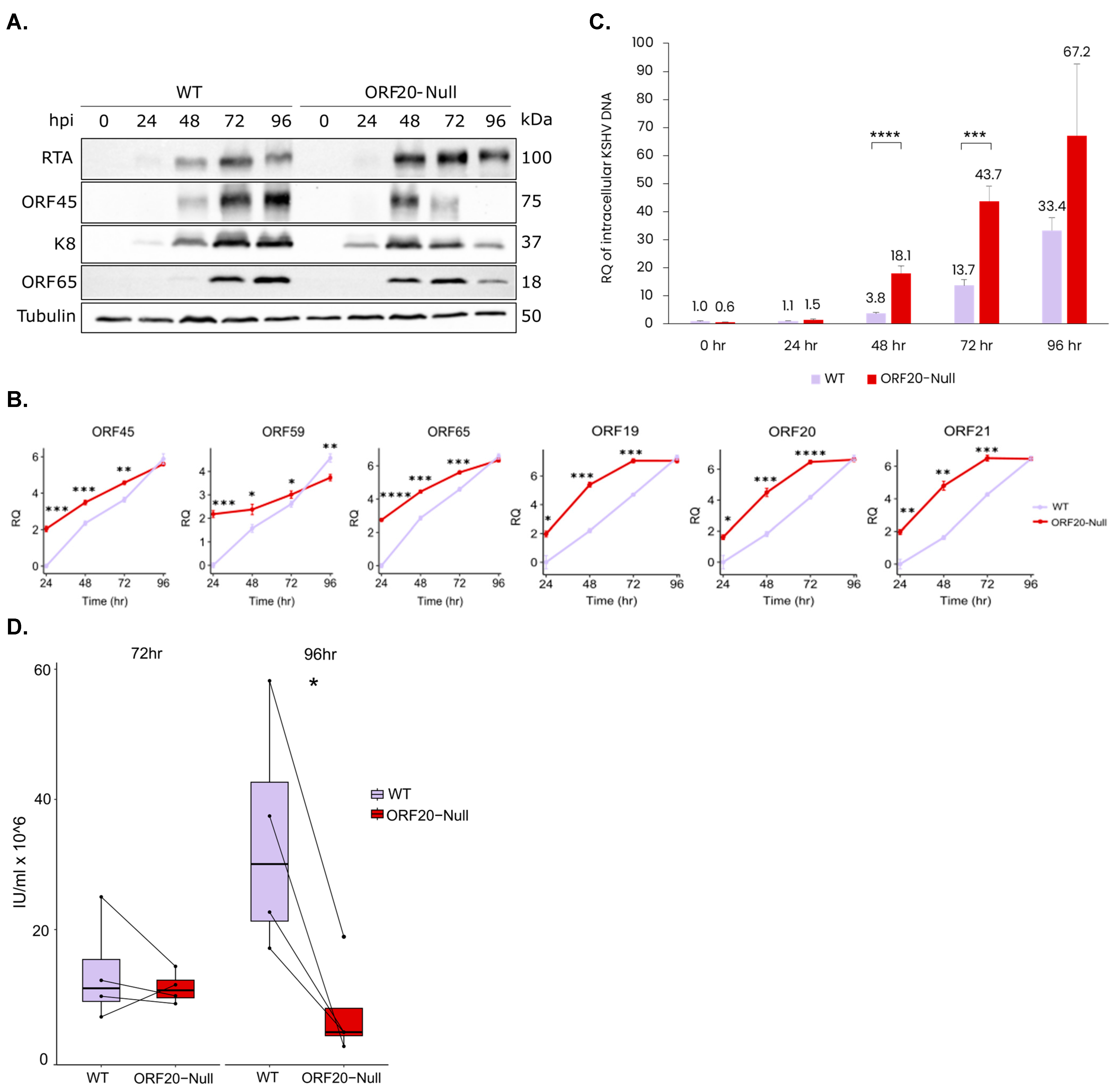
3.4. Lytic Reactivation of ORF20-Null Viruses Results in Accelerated Transcription of Viral Lytic Genes, an Earlier Accumulation of Viral Lytic Proteins, an Increase in the Quantity of Viral DNA Copies and a Decrease in Virion Production
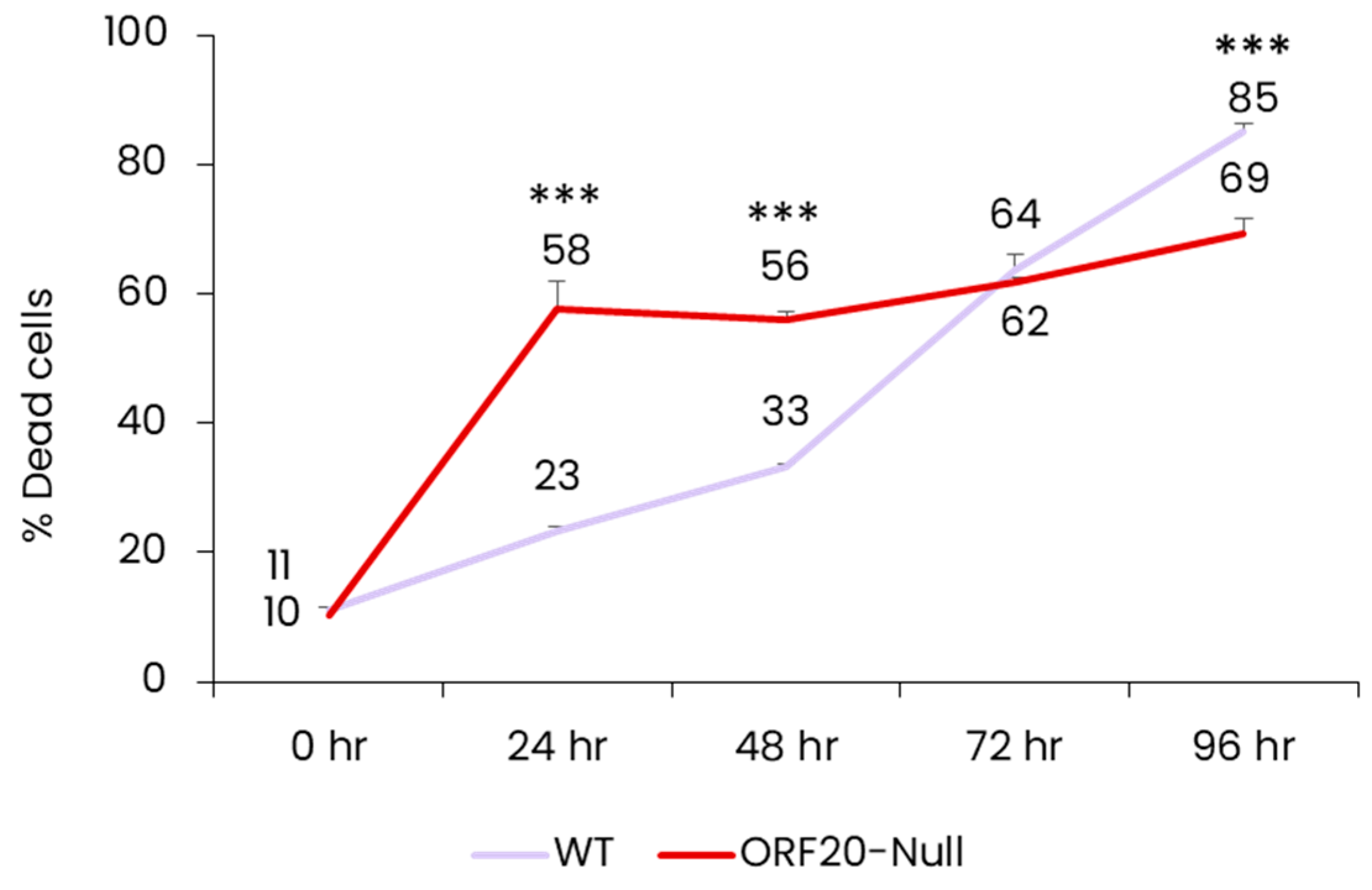
3.5. Lack of ORF20 Leads to an Increase in Cell Death at Early Time Points after Lytic Induction
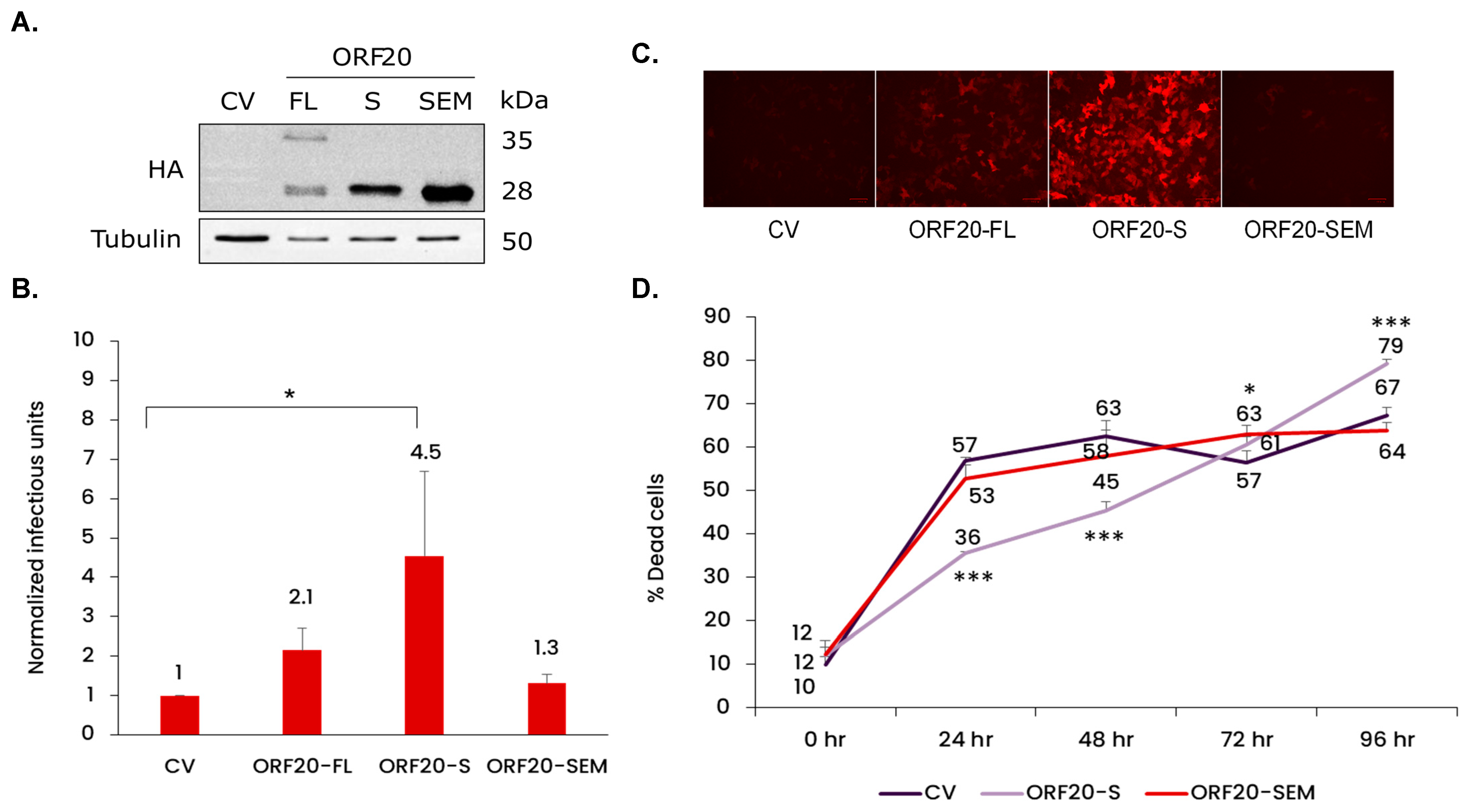
3.6. ORF20 with Its Conserved Putative Endonuclease Motif Is Required for Efficient Production of KSHV Infectious Particles
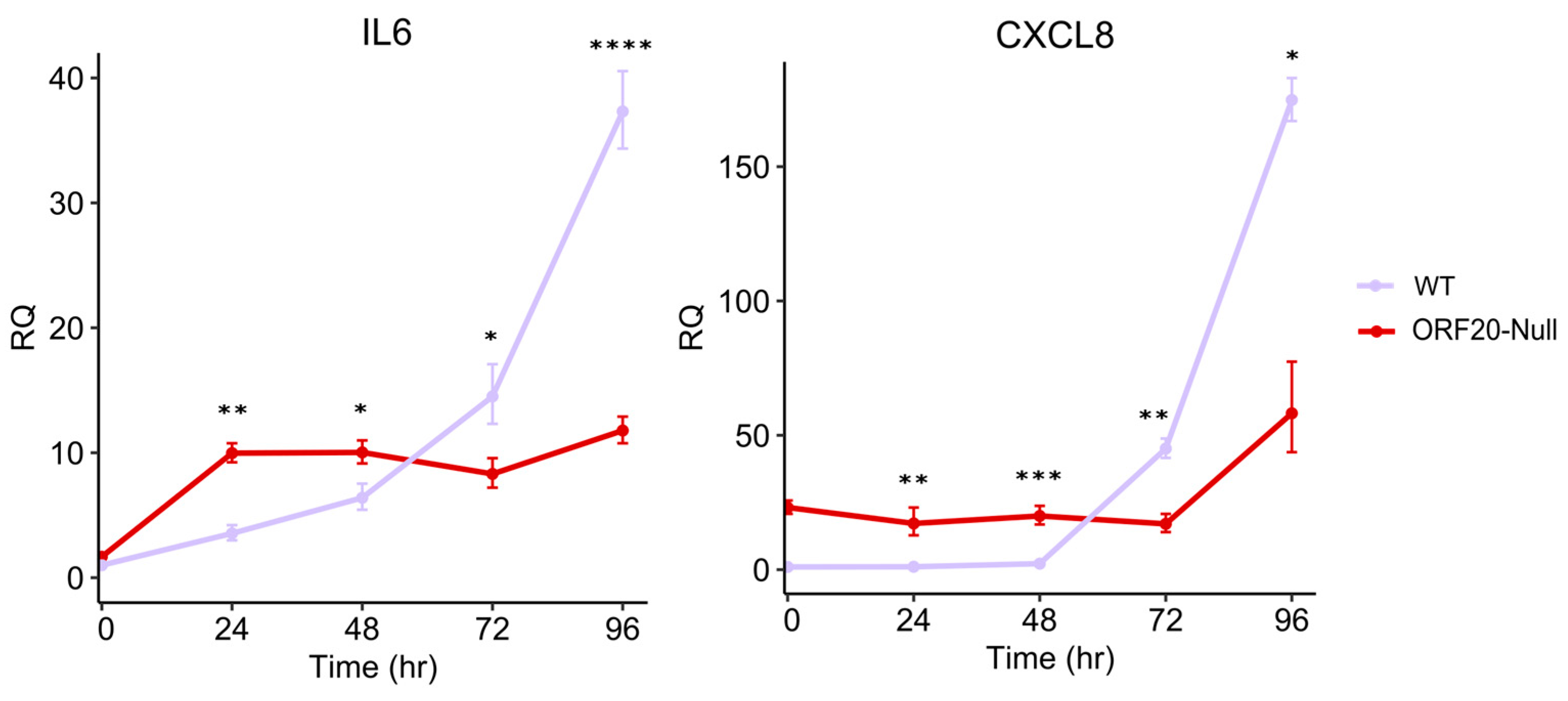
3.7. ORF20 Is Required for IL6 and CXCL8 Induction by KSHV
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sternbach, G.; Varon, J. Moritz Kaposi: Idiopathic pigmented sarcoma of the skin. J. Emerg. Med. 1995, 13, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi’s sarcoma and its associated herpesvirus. Nat. Rev. Cancer 2010, 10, 707. [Google Scholar] [CrossRef] [PubMed]
- Damania, B.; Dittmer, D.P. Today’s Kaposi sarcoma is not the same as it was 40 years ago, or is it? J. Med. Virol. 2023, 95, e28773. [Google Scholar] [CrossRef]
- Oksenhendler, E.; Meignin, V. HHV-8 associated lymphoma. Curr. Opin. Oncol. 2022, 34, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Cesarman, E.; Damania, B.; Krown, S.E.; Martin, J.; Bower, M. Kaposi sarcoma. Nat. Rev. Dis. Prim. 2019, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Calabrò, M.L.; Sarid, R. Human Herpesvirus 8 and Lymphoproliferative Disorders. Mediterr. J. Hematol. Infect. Dis. 2018, 10, e2018061. [Google Scholar] [CrossRef]
- Carbone, A.; Borok, M.; Damania, B.; Gloghini, A.; Polizzotto, M.N.; Jayanthan, R.K.; Fajgenbaum, D.C.; Bower, M. Castleman disease. Nat. Rev. Dis. Prim. 2021, 7, 84. [Google Scholar] [CrossRef]
- Goncalves, P.H.; Ziegelbauer, J.; Uldrick, T.S.; Yarchoan, R. Kaposi sarcoma herpesvirus-associated cancers and related diseases. Curr. Opin. HIV AIDS 2018, 12, 47–56. [Google Scholar] [CrossRef]
- Mariggiò, G.; Koch, S.; Schulz, T.F. Kaposi sarcoma herpesvirus pathogenesis Review. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160275. [Google Scholar] [CrossRef]
- Polizzotto, M.N.; Uldrick, T.S.; Wyvill, K.M.; Aleman, K.; Marshall, V.; Wang, V.; Whitby, D.; Pittaluga, S.; Jaffe, E.S.; Millo, C.; et al. Clinical Features and Outcomes of Patients with Symptomatic Kaposi Sarcoma Herpesvirus (KSHV)—Associated In fl ammation: Prospective Characterization of KSHV In fl ammatory Cytokine Syndrome (KICS). Clin. Infect. Dis. 2016, 62, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef]
- Kook, I.; Ziegelbauer, J.; Tagawa, T.; Serqui, A. Viral non-coding RNAs: Stealth strategies in the tug-of-war between humans and herpesviruses. Semin. Cell Dev. Biol. 2021, 111, 135–147. [Google Scholar] [CrossRef]
- Purushothaman, P.; Uppal, T.; Verma, S.C. Molecular Biology of KSHV Lytic Reactivation. Viruses 2015, 7, 116–153. [Google Scholar] [CrossRef] [PubMed]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef]
- Butnaru, M.; Gaglia, M.M. Transcriptional and Post-Transcriptional Regulation of Viral Gene Expression in the Gamma-Herpesvirus Kaposi’s Sarcoma-Associated Herpesvirus. Curr. Clin. Microbiol. Rep. 2018, 5, 219–228. [Google Scholar] [CrossRef]
- Oko, L.M.; Kimball, A.K.; Kaspar, R.E.; Knox, A.N.; Coleman, C.B.; Rochford, R.; Chang, T.; Alderete, B.; van Dyk, L.F.; Clambey, E.T. Multidimensional analysis of Gammaherpesvirus RNA expression reveals unexpected heterogeneity of gene expression. PLoS Pathog. 2019, 15, e1007849. [Google Scholar] [CrossRef]
- Landis, J.T.; Tuck, R.; Pan, Y.; Mosso, C.N.; Eason, A.B.; Moorad, R.; Marron, J.S.; Dittmer, D.P. Evidence for Multiple Subpopulations of Herpesvirus-Latently Infected Cells. mBio 2022, 13, e0347321. [Google Scholar] [CrossRef]
- Knizewski, L.; Kinch, L.; Grishin, N.V.; Rychlewski, L.; Ginalski, K. Human herpesvirus 1 UL24 gene encodes a potential PD-(D/E)XK endonuclease. J. Virol. 2006, 80, 2575–2577. [Google Scholar] [CrossRef]
- Arias, C.; Weisburd, B.; Stern-ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A Comprehensive Annotation of the Kaposi’s Sarcoma-Associated Herpesvirus Genome Using Next- Generation Sequencing Reveals Novel Genomic and Functional Features. PLoS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef]
- Nascimento, R.; Dias, J.D.; Parkhouse, R.M.E. The conserved UL24 family of human alpha, beta and gamma herpesviruses induces cell cycle arrest and inactivation of the cyclinB/cdc2 complex. Arch. Virol. 2009, 154, 1143–1149. [Google Scholar] [CrossRef]
- Nascimento, R.; Parkhouse, R.M.E. Murine gammaherpesvirus 68 ORF20 induces cell-cycle arrest in G2 by inhibiting the Cdc2-cyclin B complex. J. Gen. Virol. 2007, 88, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Siew, V.K.; Duh, C.Y.; Wang, S.K. Human cytomegalovirus UL76 induces chromosome aberrations. J. Biomed. Sci. 2009, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Davis, Z.H.; Verschueren, E.; Jang, G.M.; Kleffman, K.; Jeffrey, R.; Park, J.; Von Dollen, J.; Maher, M.C.; Johnson, T.; Jäger, S.; et al. Global mapping of herpesvirus-host protein complexes reveals a novel transcription strategy for late genes. Mol. Cell 2016, 57, 349–360. [Google Scholar] [CrossRef]
- Bussey, K.A.; Lau, U.; Schumann, S.; Gallo, A.; Osbelt, L.; Stempel, M.; Arnold, C.; Wissing, J.; Gad, H.H.; Hartmann, R.; et al. The interferon-stimulated gene product oligoadenylate synthetase-like protein enhances replication of Kaposi’s sarcoma-associated herpesvirus (KSHV) and interacts with the KSHV ORF20 protein. PLoS Pathog. 2018, 14, e1006937. [Google Scholar] [CrossRef]
- Hoffman, D.; Rodriguez, W.; Macveigh-Fierro, D.; Miles, J.; Muller, M. The KSHV ORF20 Protein Interacts with the Viral Processivity Factor ORF59 and Promotes Viral Reactivation. Microbiol. Spectr. 2021, 9, e0014521. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, L.; Pearson, A. The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J. Gen. Virol. 2008, 89, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Lymberopoulos, M.H.; Bourget, A.; Abdeljelil, N.B.; Pearson, A. Involvement of the UL24 protein in herpes simplex virus 1-induced dispersal of B23 and in nuclear egress. Virology 2011, 412, 341–348. [Google Scholar] [CrossRef]
- Sanabria-Solano, C.; Gonzalez, C.E.; Richerioux, N.; Bertrand, L.; Dridi, S.; Griffiths, A.; Langelier, Y.; Pearson, A. Regulation of viral gene expression by the herpes simplex virus 1 UL24 protein (HSV-1 UL24 inhibits accumulation of viral transcripts). Virology 2016, 495, 148–160. [Google Scholar] [CrossRef]
- Xu, H.; Su, C.; Pearson, A.; Mody, C.H.; Zheng, C. Herpes Simplex Virus 1 UL24 Abrogates the DNA Sensing Signal Pathway by Inhibiting NF-κB Activation. J. Virol. 2017, 91, e00025-17. [Google Scholar] [CrossRef]
- Wang, S.; Duh, C.; Wu, C. Human Cytomegalovirus UL76 Encodes a Novel Virion-Associated Protein That Is Able To Inhibit Viral Replication. J. Virol. 2004, 78, 9750–9762. [Google Scholar] [CrossRef] [PubMed]
- Ben Abdeljelil, N.; Rochette, P.A.; Pearson, A. The UL24 protein of herpes simplex virus 1 affects the sub-cellular distribution of viral glycoproteins involved in fusion. Virology 2013, 444, 263–273. [Google Scholar] [CrossRef]
- Isomura, H.; Stinski, M.F.; Murata, T.; Nakayama, S.; Chiba, S.; Akatsuka, Y.; Kanda, T.; Tsurumi, T. The human cytomegalovirus UL76 gene regulates the level of expression of the UL77 gene. PLoS ONE 2010, 5, e11901. [Google Scholar] [CrossRef] [PubMed]
- Westdorp, K.N.; Sand, A.; Moorman, N.J.; Terhune, S.S. Cytomegalovirus Late Protein pUL31 Alters pre-rRNA Expression and Nuclear Organization during Infection. J. Virol. 2017, 91, JVI.00593-17. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-R.; Jiang, M.J.; Wang, H.-H.; Hu, C.-H.; Hsu, M.-S.; Hsi, E.; Duh, C.-Y.; Wang, S.-K. Human Cytomegalovirus UL76 Elicits Novel Aggresome Formation via Interaction with S5a of the Ubiquitin Proteasome System. J. Virol. 2013, 87, 11562–11578. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, M.; Ye, C.; Ruan, K.; Xu, A.; Gao, F.; Tong, G.; Zheng, H. Inhibition of the DNA-Sensing pathway by pseudorabies virus UL24 protein via degradation of interferon regulatory factor 7. Vet. Microbiol. 2021, 255, 109023. [Google Scholar] [CrossRef]
- Chen, X.; Kong, N.; Xu, J.; Wang, J.; Zhang, M.; Ruan, K.; Li, L.; Zhang, Y.; Zheng, H.; Tong, W.; et al. Pseudorabies virus UL24 antagonizes OASL-mediated antiviral effect. Virus Res. 2021, 295, 198276. [Google Scholar] [CrossRef]
- Wang, T.Y.; Yang, Y.L.; Feng, C.; Sun, M.X.; Peng, J.M.; Tian, Z.J.; Tang, Y.D.; Cai, X.H. Pseudorabies virus UL24 abrogates tumor necrosis factor alpha-induced NF-κB activation by degrading p65. Viruses 2020, 12, 51. [Google Scholar] [CrossRef]
- Chen, X.; Sun, D.; Dong, S.; Zhai, H.; Kong, N.; Zheng, H.; Tong, W.; Li, G.; Shan, T.; Tong, G. Host Interferon-Stimulated Gene 20 Inhibits Pseudorabies Virus Proliferation. Virol. Sin. 2021, 36, 1027–1035. [Google Scholar] [CrossRef]
- Chen, X.; Shan, T.; Sun, D.; Zhai, H.; Dong, S.; Kong, N.; Zheng, H.; Tong, W.; Tong, G. Host Zinc-finger CCHC-type containing protein 3 inhibits pseudorabies virus proliferation by regulating type I interferon signaling. Gene 2022, 827, 146480. [Google Scholar] [CrossRef]
- Gelgor, A.; Kalt, I.; Bergson, S.; Brulois, K.F.; Jung, J.U.; Sarid, R. Viral Bcl-2 Encoded by the Kaposi’s Sarcoma-Associated Herpesvirus Is Vital for Virus Reactivation. J. Virol. 2015, 89, 5298–5307. [Google Scholar] [CrossRef]
- Brulois, K.F.; Chang, H.; Lee, A.S.-Y.; Ensser, A.; Wong, L.-Y.; Toth, Z.; Lee, S.H.; Lee, H.-R.; Myoung, J.; Ganem, D.; et al. Construction and Manipulation of a New Kaposi’s Sarcoma-Associated Herpesvirus Bacterial Artificial Chromosome Clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef] [PubMed]
- Gam Ze Letova, C.; Kalt, I.; Shamay, M.; Sarid, R. Latently kshv-infected cells promote further establishment of latency upon superinfection with kshv. Int. J. Mol. Sci. 2021, 22, 11994. [Google Scholar] [CrossRef]
- Tischer, B.K.; Von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 2006, 40, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Dünn-Kittenplon, D.; Kalt, I.; Lellouche, J.P.; Sarid, R. The KSHV portal protein ORF43 is essential for the production of infectious viral particles. Virology 2019, 529, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Bergson, S.; Kalt, I.; Itzhak, I.; Brulois, K.F.; Jung, J.U.; Sarid, R. Fluorescent Tagging and Cellular Distribution of the Kaposi’s Sarcoma-Associated Herpesvirus ORF45 Tegument Protein. J. Virol. 2014, 88, 12839–12852. [Google Scholar] [CrossRef]
- Bergson, S.; Itzhak, I.; Wasserman, T.; Gelgor, A.; Kalt, I.; Sarid, R. The Kaposi’s-sarcoma-associated herpesvirus orf35 gene product is required for ef fi cient lytic virus reactivation. Virology 2016, 499, 91–98. [Google Scholar] [CrossRef]
- Lukac, D.M.; Garibyan, L.; Kirshner, J.R.; Palmeri, D.; Ganem, D.O.N. DNA Binding by Kaposi’s Sarcoma-Associated Herpesvirus Lytic Switch Protein Is Necessary for Transcriptional Activation of Two Viral Delayed Early Promoters. J. Virol. 2001, 75, 6786–6799. [Google Scholar] [CrossRef]
- Myoung, J.; Ganem, D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 2011, 174, 12–21. [Google Scholar] [CrossRef]
- Stürzl, M.; Gaus, D.; Dirks, W.G.; Ganem, D.; Jochmann, R. Kaposi’s sarcoma-derived cell line SLK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. Int. J. Cancer 2013, 1958, 1954–1958. [Google Scholar] [CrossRef]
- Zhu, F.X.; Chong, J.M.; Wu, L.; Yuan, Y. Virion Proteins of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2005, 79, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Tang, Q.; Maul, G.G.; Yuan, Y.; Al, W.E.T.; Irol, J.V. Kaposi’s Sarcoma-Associated Herpesvirus ori-Lyt -Dependent DNA Replication: Involvement of Host Cellular Factors. J. Virol. 2008, 82, 2867–2882. [Google Scholar] [CrossRef]
- Ueda, K.; Ishikawa, K.; Nishimura, K.; Sakakibara, S.; Do, E.; Yamanishi, K. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) replication and transcription factor activates the K9 (vIRF) gene through two distinct cis elements by a non-DNA-binding mechanism. J. Virol. 2002, 76, 12044–12054. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Zhou, F.; Bedolla, R.G.; Jones, T.; Lei, X.; Kang, T. Reactive Oxygen Species Hydrogen Peroxide Mediates Kaposi’s Sarcoma-Associated Herpesvirus Reactivation from Latency. PLoS Pathog. 2011, 7, e1002054. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, J.G.; Martin, S.L.; Coen, D.M. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J. Virol. 1989, 63, 1839–1843. [Google Scholar] [CrossRef]
- Bertrand, L.; Leiva-Torres, G.A.; Hyjazie, H.; Pearson, A. Conserved Residues in the UL24 Protein of Herpes Simplex Virus 1 Are Important for Dispersal of the Nucleolar Protein Nucleolin. J. Virol. 2010, 84, 109–118. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Dridi, S.; Richerioux, N.; Gonzalez, E.; Vanharen, M.; Sanabria-solano, C. A Mutation in the UL24 Gene Abolishes Expression of the Newly Identified UL24.5 Protein of Herpes Simplex Virus 1 and Leads to an Increase in Pathogenicity in Mice. J. Virol. 2018, 92, e00671-18. [Google Scholar] [CrossRef]
- Chang, P.; Wang, S.; Chen, L.; Hung, C.; Huang, H. ORF50-dependent and ORF50-independent activation of the ORF45 gene of Kaposi’s sarcoma-associated herpesvirus. Virology 2013, 442, 38–50. [Google Scholar] [CrossRef]
- Costa, H.; Nascimento, R.; Sinclair, J.; Parkhouse, R.M.E. Human Cytomegalovirus Gene UL76 Induces IL-8 Expression through Activation of the DNA Damage Response. PLoS Pathog. 2013, 9, e1003609. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, B.; Dutia, B.M.; Roberts, K.L.; Garcia-Ramirez, J.J.; Dickinson, P.; Stewart, J.P.; Ghazal, P.; Roy, D.J.; Nash, A.A. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 2003, 84, 99–109. [Google Scholar] [CrossRef]
- Tombácz, D.; Tóth, J.S.; Petrovszki, P.; Boldogkoi, Z. Whole-genome analysis of pseudorabies virus gene expression by real-time quantitative RT-PCR assay. BMC Genom. 2009, 10, 491. [Google Scholar] [CrossRef] [PubMed]
- Gabaev, I.; Williamson, J.C.; Crozier, T.W.M.; Schulz, T.F.; Lehner, P.J. Quantitative Proteomics Analysis of Lytic KSHV Infection in Human Endothelial Cells Reveals Targets of Viral Immune Modulation. Cell Rep. 2020, 33, 108249. [Google Scholar] [CrossRef] [PubMed]
- Leiva-Torres, G.A.; Rochette, P.A.; Pearson, A. Differential importance of highly conserved residues in UL24 for herpes simplex virus 1 replication in vivo and reactivation. J. Gen. Virol. 2010, 91, 1109–1116. [Google Scholar] [CrossRef]
- Jacobson, J.G.; Chen, S.H.; Cook, W.J.; Kramer, M.F.; Coen, D.M. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 1998, 242, 161–169. [Google Scholar] [CrossRef]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 2003, 100, 14223–14228. [Google Scholar] [CrossRef]
- Yu, D.; Silva, M.C.; Shenk, T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 12396–12401. [Google Scholar] [CrossRef]
- Ito, H.; Sommer, M.H.; Zerboni, L.; Baiker, A.; Sato, B.; Liang, R.; Hay, J.; Ruyechan, W.; Arvin, A.M. Role of the varicella-zoster virus gene product encoded by open reading frame 35 in viral replication in vitro and in differentiated human skin and T cells in vivo. J. Virol. 2005, 79, 4819–4827. [Google Scholar] [CrossRef]
- Craigen, J.L.; Yong, K.L.; Jordan, N.J.; Maccormac, L.P.; Westwick, J.; Akbar, A.N.; Grundy, J.E. Human cytomegalovirus infection up-regulates interleukin-8 gene expression and stimulates neutrophil transendothelial migration. Immunology 1997, 92, 138–145. [Google Scholar] [CrossRef]
- Murayama, T.; Kuno, K.; Jisaki, F.; Obuchi, M.; Sakamuro, D.; Furukawa, T.; Mukaida, N.; Matsushima, K. Enhancement human cytomegalovirus replication in a human lung fibroblast cell line by interleukin-8. J. Virol. 1994, 68, 7582–7585. [Google Scholar] [CrossRef] [PubMed]
- The, T.H.; van den Berg, A.P.; Harmsen, M.C.; van der Bij, W.; van Son, W.J. The cytomegalovirus antigenemia assay: A plea for standardization. Scand. J. Infect. Dis. Suppl. 1995, 99, 25–29. [Google Scholar] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orbaum-Harel, O.; Sloutskin, A.; Kalt, I.; Sarid, R. KSHV ORF20 Promotes Coordinated Lytic Reactivation for Increased Infectious Particle Production. Viruses 2024, 16, 1418. https://doi.org/10.3390/v16091418
Orbaum-Harel O, Sloutskin A, Kalt I, Sarid R. KSHV ORF20 Promotes Coordinated Lytic Reactivation for Increased Infectious Particle Production. Viruses. 2024; 16(9):1418. https://doi.org/10.3390/v16091418
Chicago/Turabian StyleOrbaum-Harel, Odelia, Anna Sloutskin, Inna Kalt, and Ronit Sarid. 2024. "KSHV ORF20 Promotes Coordinated Lytic Reactivation for Increased Infectious Particle Production" Viruses 16, no. 9: 1418. https://doi.org/10.3390/v16091418





