Coat Proteins of the Novel Victoriviruses FaVV1 and FaVV2 Suppress Sexual Reproduction and Virulence in the Pathogen of Fusarium Head Blight
Abstract
:1. Background
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Genome Sequence Assembly and Phylogenetic Analysis
2.3. Virus Cure and Transmission
2.4. Vector Construction and Transformation of F. asiaticum and F. graminearum
2.5. Virulence Assays
3. Results
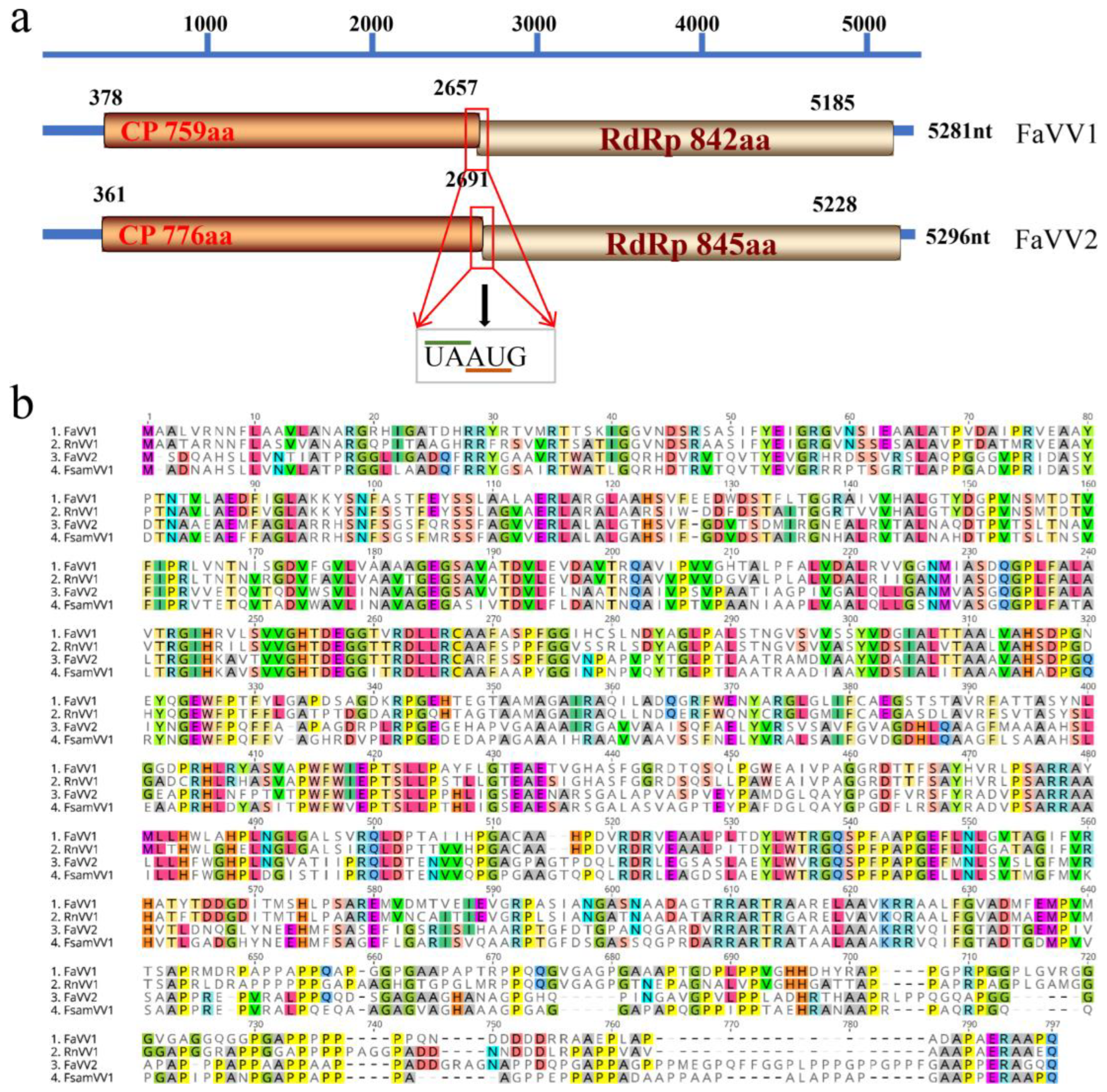
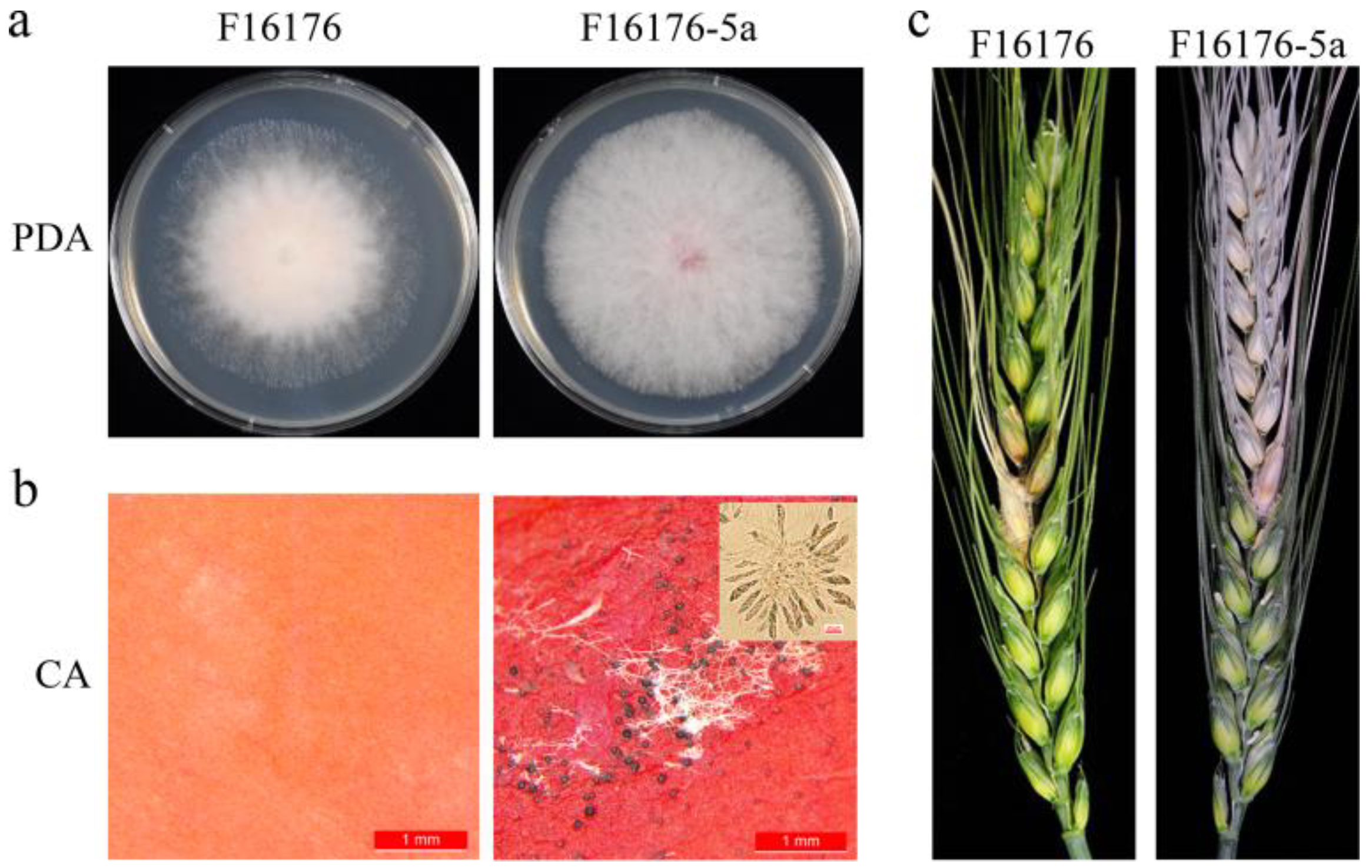
3.1. Another New Victorivirus in the F. asiaticum Strain F16176
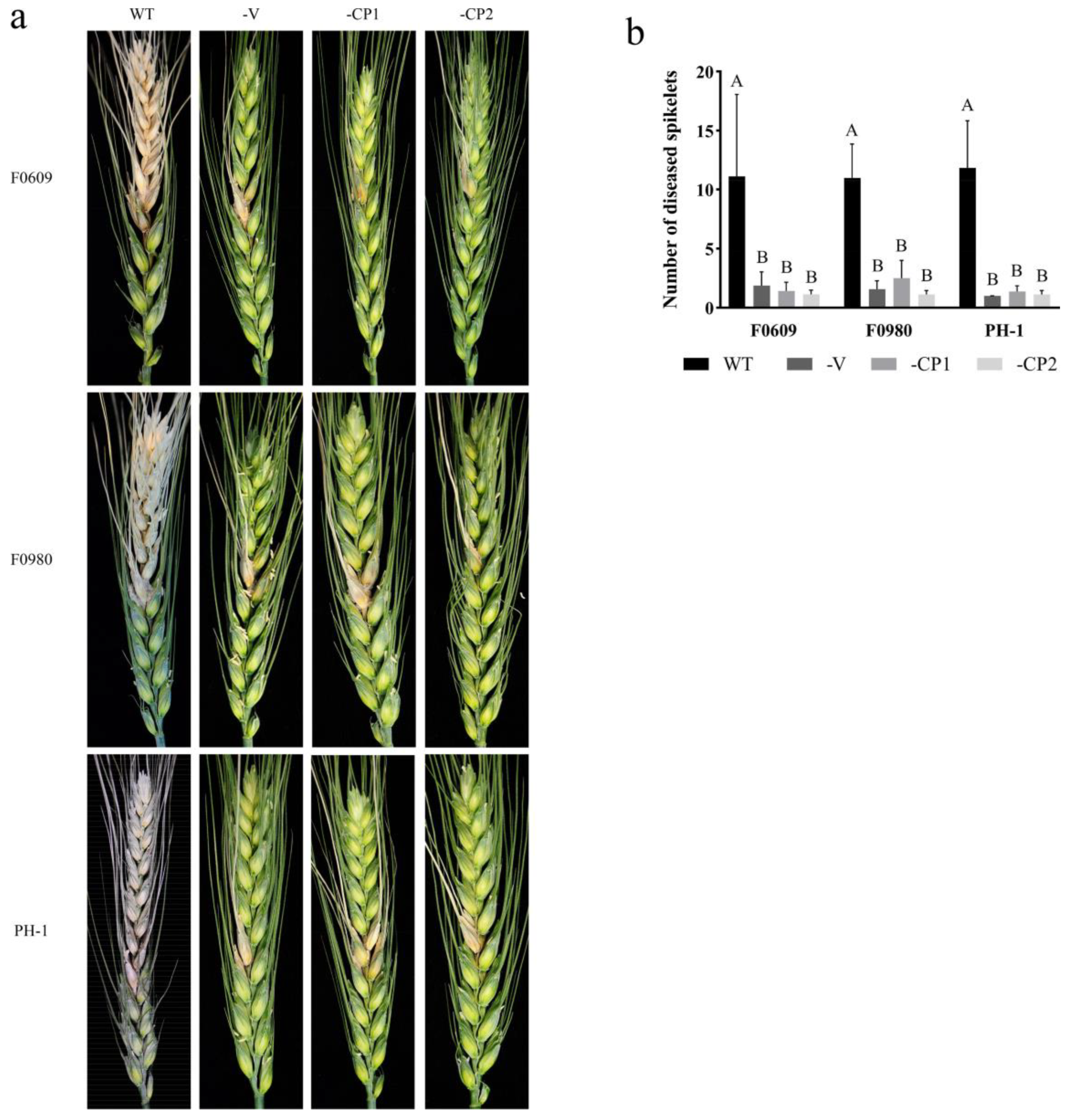
3.2. FaVV1 and FaVV2 Suppress the Sexual Reproduction and Virulence of Their Host Fusarium Strains
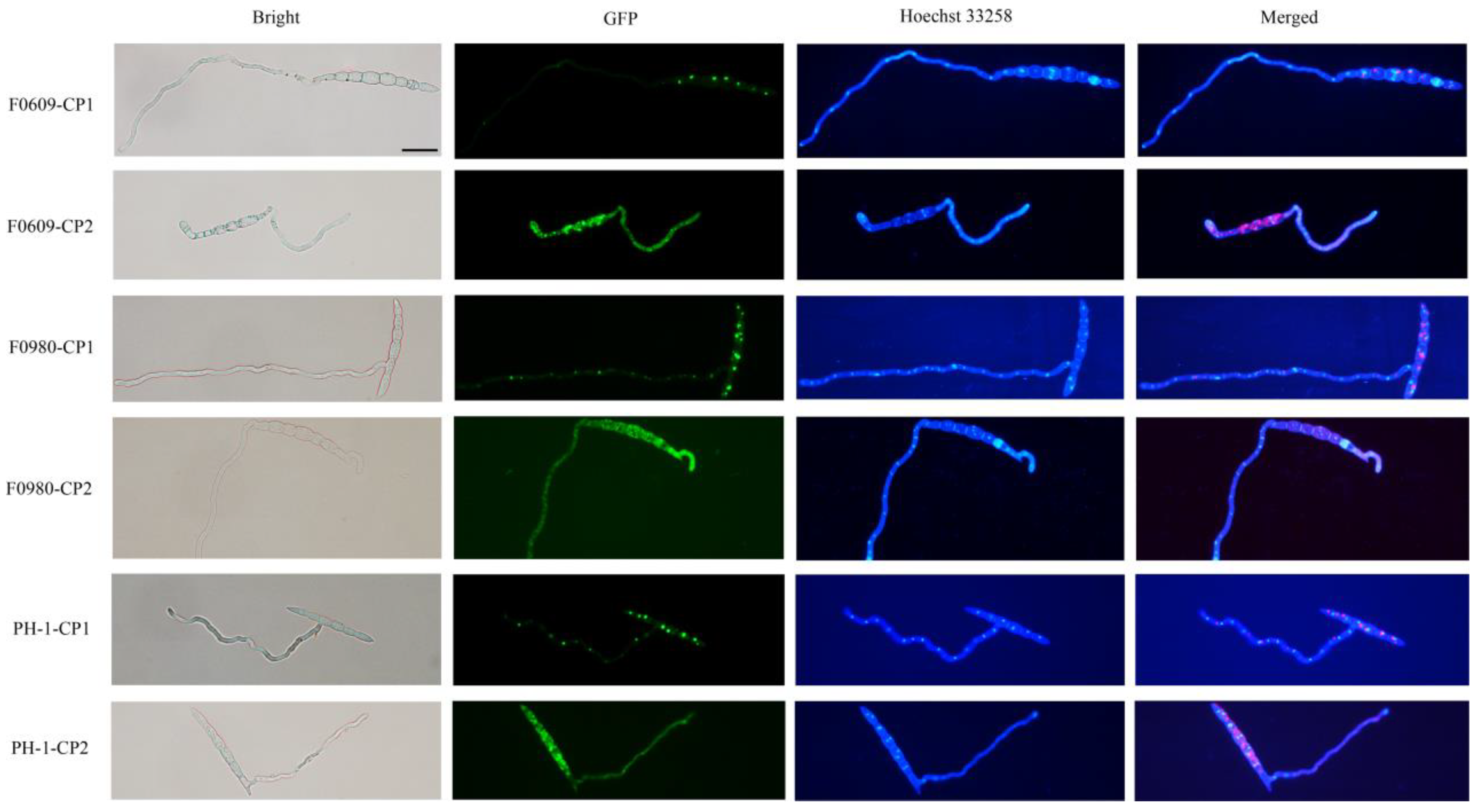
3.3. Heterologous Expression of Viral CP Proteins Elicits Virus Infection-like Symptoms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, X.; Nicholson, P. Community ecology of fungal pathogens causing wheat head blight. Annu. Rev. Phytopathol. 2009, 47, 83–103. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Islam, T.; Ma, Z. An integrated pest management program for managing fusarium head blight disease in cereals. J. Integr. Agric. 2022, 21, 3434–3444. [Google Scholar] [CrossRef]
- Moonjely, S.; Ebert, M.; Paton-Glassbrook, D.; Noel, Z.A.; Roze, L.; Shay, R.; Watkins, T.; Trail, F. Update on the state of research to manage Fusarium head blight. Fungal Genet. Biol. 2023, 169, 103829. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jiang, D. New Insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.H.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479, 356–368. [Google Scholar] [CrossRef]
- Myers, J.M.; James, T.Y. Mycoviruses. Curr. Biol. 2022, 32, R150–R155. [Google Scholar] [CrossRef]
- Wagemans, J.; Holtappels, D.; Vainio, E.; Rabiey, M.; Marzachì, C.; Herrero, S.; Ravanbakhsh, M.; Tebbe, C.C.; Ogliastro, M.; Ayllón, M.A.; et al. Going Viral: Virus-based biological control agents for plant protection. Annu. Rev. Phytopathol. 2022, 60, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Darissa, O.; Adam, G.; Schafer, W. A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Pathol. 2012, 134, 181–189. [Google Scholar] [CrossRef]
- Li, P.; Bhattacharjee, P.; Wang, S.; Zhang, L.; Ahmed, I.; Guo, L. Mycoviruses in Fusarium species: An updating review. Front. Cellular Infect. Microbiol. 2019, 9, 257. [Google Scholar] [CrossRef]
- Li, P.; Wang, S.; Zhang, L.; Qiu, D.; Zhou, X.; Guo, L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci. Adv. 2020, 6, eaay9634. [Google Scholar] [CrossRef]
- Paudel, B.; Pedersen, C.; Yen, Y.; Marzano, S.Y.L. Fusarium graminearum virus-1 strain fgv1-sd4 infection eliminates mycotoxin deoxynivalenol synthesis by Fusarium graminearum in FHB. Microorganisms 2022, 10, 1484. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Ruan, S.; Nzabanita, C.; Wang, Y.; Guo, L. A mycovirus VIGS vector confers hypovirulence to a plant pathogenic fungus to control wheat FHB. Adv. Sci. 2023, 10, e2302606. [Google Scholar] [CrossRef] [PubMed]
- Bormann, J.; Heinze, C.; Blum, C.; Mentges, M.; Brockmann, A.; Alder, A.; Landt, S.K.; Josephson, B.; Indenbirken, D.; Spohn, M.; et al. Expression of a structural protein of the mycovirus FgV-ch9 negatively affects the transcript level of a novel symptom alleviation factor and causes virus infection-like symptoms in Fusarium graminearum. J. Virol. 2018, 92, e00326-18. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Park, J.Y.; Heo, J.I.; Kim, K.H. The ORF2 protein of Fusarium graminearum virus 1 suppresses the transcription of FgDICER2 and FgAGO1 to limit host antiviral defences. Mol. Plant Pathol. 2020, 21, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Van der Lee, T.; Waalwijk, C.; Chen, W.; Xu, J.; Xu, J.; Zhang, Y.; Feng, J. Population analysis of the Fusarium graminearum Species Complex from wheat in China show a shift to more aggressive isolates. PLoS ONE 2012, 7, e31722. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Sun, H.Y.; Li, W.; Xia, Y.L.; Deng, Y.Y.; Zhang, A.X.; Chen, H.G. Fitness of three chemotypes of Fusarium graminearum species complex in major winter wheat-producing areas of China. PLoS ONE 2017, 12, e0174040. [Google Scholar] [CrossRef]
- Li, W.; Xia, Y.; Zhang, H.; Zhang, X.; Chen, H. A victorivirus from Fusarium asiaticum, the pathogen of fusarium head blight in China. Arch. Virol. 2019, 164, 313–316. [Google Scholar] [CrossRef]
- Huang, H.; Hua, X.; Pang, X.; Zhang, Z.; Ren, J.; Cheng, J.; Fu, Y.; Xiao, X.; Lin, Y.; Chen, T.; et al. Discovery and characterization of putative glycoprotein-encoding mycoviruses in the Bunyavirales. J. Virol. 2023, 97, e01381-22. [Google Scholar] [CrossRef]
- Chiba, S.; Lin, Y.H.; Kondo, H.; Kanematsu, S.; Suzuki, N. A novel victorivirus from a phytopathogenic fungus, Rosellinia necatrix, is infectious as particles and targeted by RNA silencing. J. Virol. 2013, 87, 6727–6738. [Google Scholar] [CrossRef]
- Xie, J.; Havens, W.M.; Lin, Y.H.; Suzuki, N.; Ghabrial, S.A. The victorivirus Helminthosporium victoriae virus 190S is the primary cause of disease/hypovirulence in its natural host and a heterologous host. Virus Res. 2016, 213, 238–245. [Google Scholar] [CrossRef]
- Cavinder, B.; Sikhakolli, U.; Fellows, K.M.; Trail, F. Sexual development and ascospore discharge in Fusarium graminearum. J. Vis. Exp. 2012, 29, 3895. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic, an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Yu, J.; Son, M.; Lee, Y.W.; Kim, K.H. Transmission of Fusarium boothii mycovirus via protoplast fusion causes hypovirulence in other phytopathogenic fungi. PLoS ONE 2011, 6, e21629. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, G.; Xu, J.R. Efficient approaches for generating GFP gusion and epitope-tagging constructs in filamentous fungi. Methods Mol. Biol. 2011, 722, 199–212. [Google Scholar]
- Bourett, T.M.; Sweigard, J.A.; Czymmek, K.J.; Carroll, A.; Howard, R.J. Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet. Biol. 2002, 37, 211–220. [Google Scholar] [CrossRef]
- Royer, J.C.; Moyer, D.L.; Reiwitch, S.G.; Madden, M.S.; Jensen, E.B.; Brown, S.H.; Onker, C.C.; Johnston, J.A.; Golightly, E.J.; Yoder, W.T.; et al. Fusarium graminearum A 3/5 as a novel host for heterologous protein production. Bio Technol. 1995, 13, 1479–1483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Sun, H.Y.; Shen, C.M.; Li, W.; Yu, H.S.; Chen, H.G. Survery of Fusarium spp. causing wheat crown rot in major winter wheat growing regions of China. Plant Dis. 2015, 99, 1610–1615. [Google Scholar] [CrossRef]
- Mizutani, Y.; Uesaka, K.; Ota, A.; Calassanzio, M.; Ratti, C.; Suzuki, T.; Fujimori, F.; Chiba, S. De novo sequencing of novel mycoviruses from Fusarium sambucinum: An attempt on direct RNA sequencing of viral dsRNAs. Front. Microbiol. 2021, 12, 641484. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Mäkinen, K. Coat proteins, host factors and plant viral replication. Curr. Opin. Virol. 2012, 2, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Lõhmus, A.; Hafrén, A.; Mäkinen, K. Coat protein regulation by CK2, CPIP, HSP70, and CHIP is required for potato virus A replication and coat protein accumulation. J. Virol. 2017, 91, e01316-16. [Google Scholar] [CrossRef]


| Strain | Viruses or Viral CP Content | Background Strain | Antibiotic Resistance |
|---|---|---|---|
| F16176 | FaVV1, FaVV2 | F16176 | |
| F16176-5a | F16176 | ||
| F0609 | F0609 | ||
| F0609R | F0609 | hyg | |
| F0980 | F0980 | ||
| F0980R | F0980 | hyg | |
| PH-1 | PH-1 | ||
| PH-1R | PH-1 | hyg | |
| F0609-V | FaVV1, FaVV2 | F0609R | hyg |
| F0980-V | FaVV1, FaVV2 | F0980R | hyg |
| PH-1-V | FaVV1, FaVV2 | PH-1R | hyg |
| F0609-CP1 | CP1 | F0609 | hyg |
| F0609-CP2 | CP2 | F0609 | hyg |
| F0980-CP1 | CP1 | F0980 | hyg |
| F0980-CP2 | CP2 | F0980 | hyg |
| PH-1-CP1 | CP1 | PH-1 | hyg |
| PH-1-CP2 | CP2 | PH-1 | hyg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, S.; Yang, X.; Xia, L.; Zhang, X.; Sun, H.; Deng, Y.; Shu, Y.; Zhang, A.; Chen, H.; Li, W. Coat Proteins of the Novel Victoriviruses FaVV1 and FaVV2 Suppress Sexual Reproduction and Virulence in the Pathogen of Fusarium Head Blight. Viruses 2024, 16, 1424. https://doi.org/10.3390/v16091424
Cao S, Yang X, Xia L, Zhang X, Sun H, Deng Y, Shu Y, Zhang A, Chen H, Li W. Coat Proteins of the Novel Victoriviruses FaVV1 and FaVV2 Suppress Sexual Reproduction and Virulence in the Pathogen of Fusarium Head Blight. Viruses. 2024; 16(9):1424. https://doi.org/10.3390/v16091424
Chicago/Turabian StyleCao, Shulin, Xiaoyue Yang, Lele Xia, Xing Zhang, Haiyan Sun, Yuanyu Deng, Yan Shu, Aixiang Zhang, Huaigu Chen, and Wei Li. 2024. "Coat Proteins of the Novel Victoriviruses FaVV1 and FaVV2 Suppress Sexual Reproduction and Virulence in the Pathogen of Fusarium Head Blight" Viruses 16, no. 9: 1424. https://doi.org/10.3390/v16091424






