Structural Heterogeneity of the Rabies Virus Virion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Strain and Cell Line
2.2. Virus Production and Purification
2.3. CryoEM Sample Preparation and Movies Acquisition
2.4. CryoET Sample Preparation and Tilt Series Acquisition
2.5. CryoET and Subtomogram Averaging (STA)
2.6. Single-Particle CryoEM Data Processing
3. Results
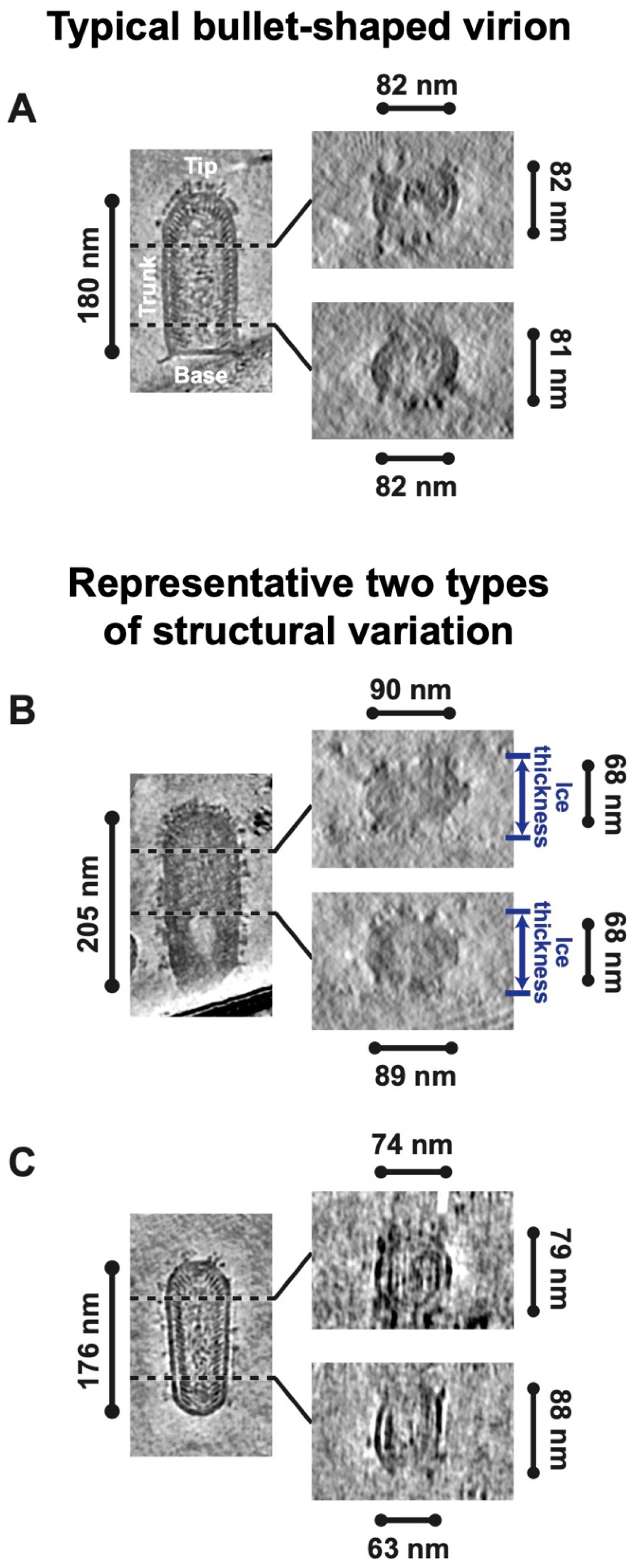
3.1. Structural Flexibility of RABV Virions
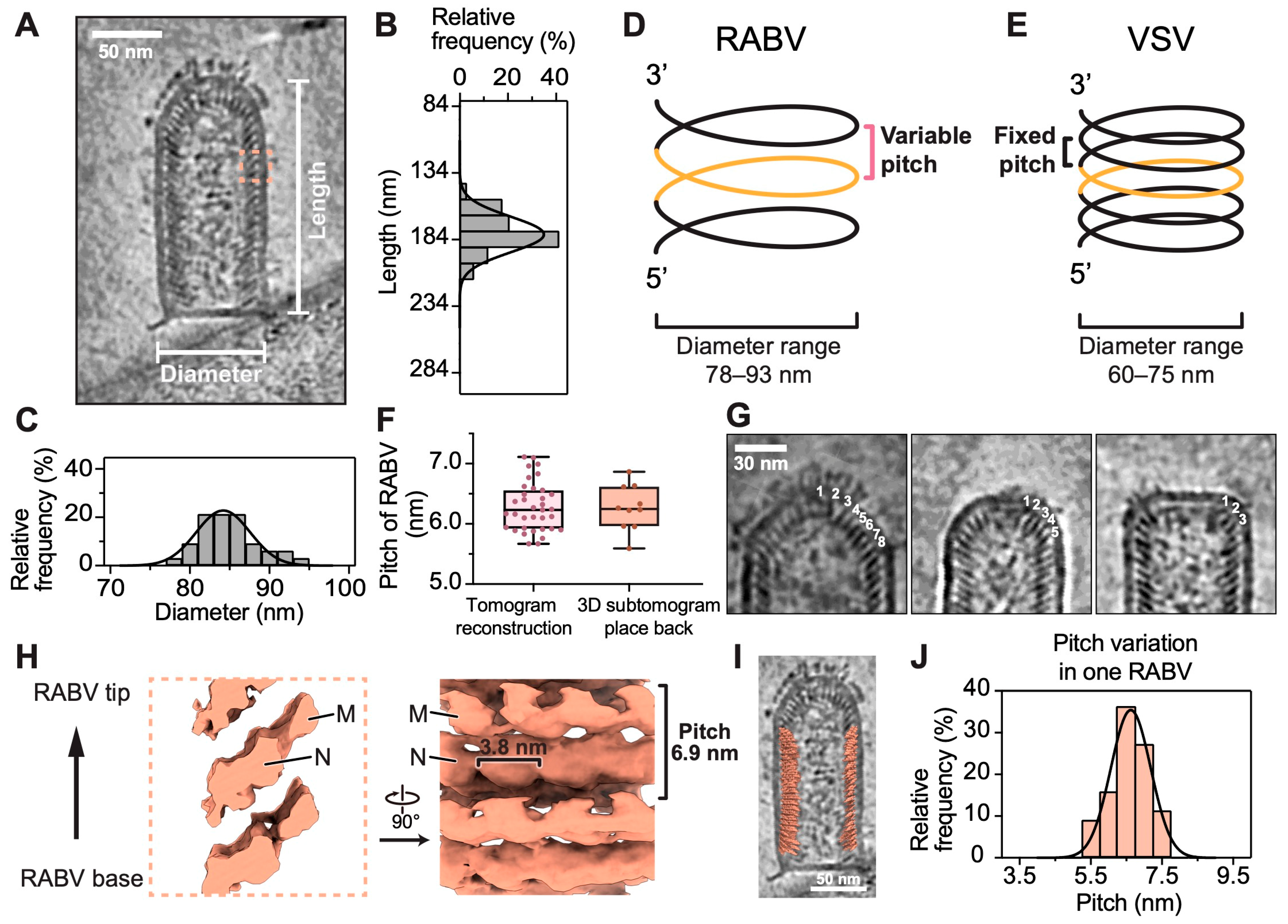
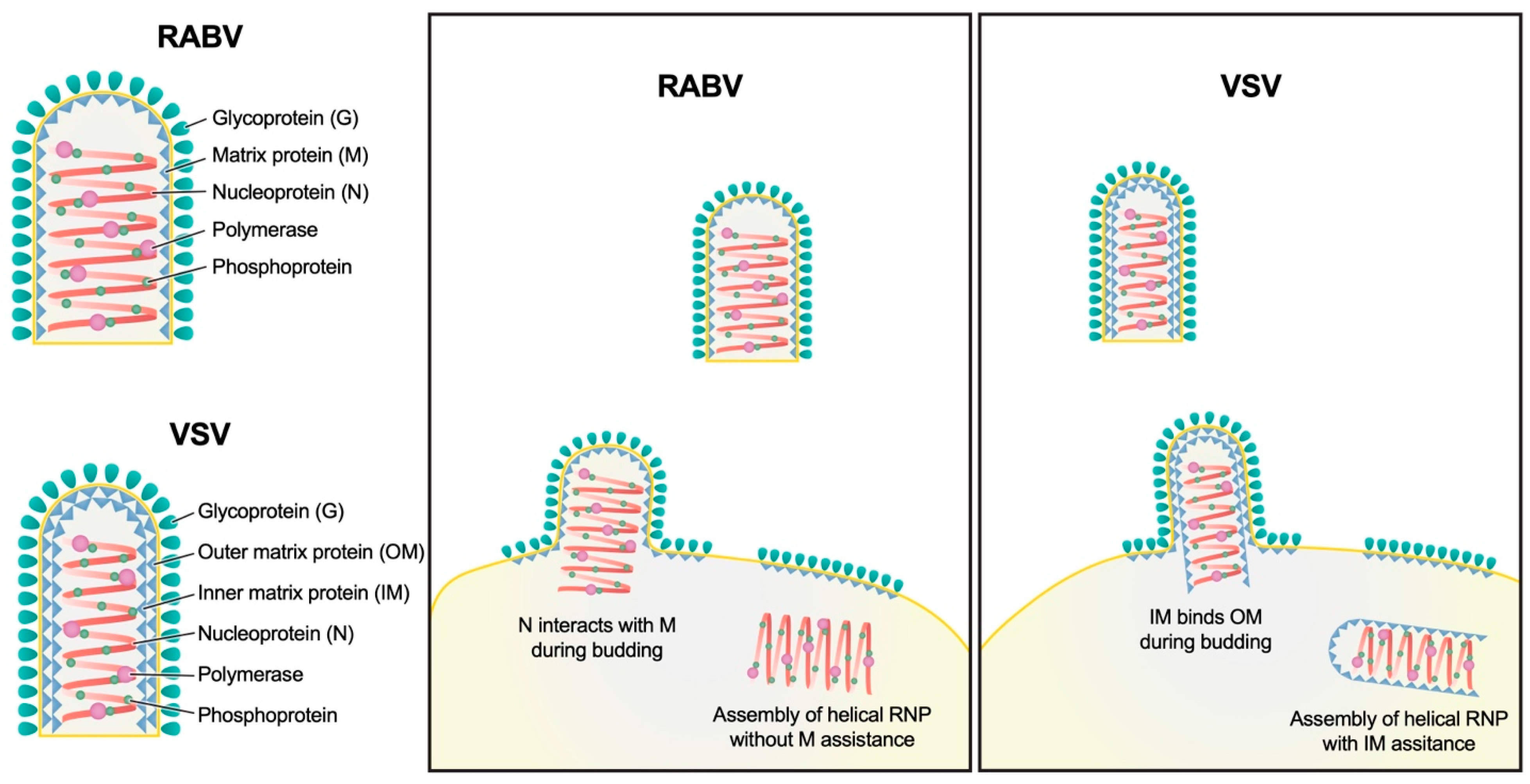
3.2. Variable Pitch and Larger Diameter of RABV Compared to VSV
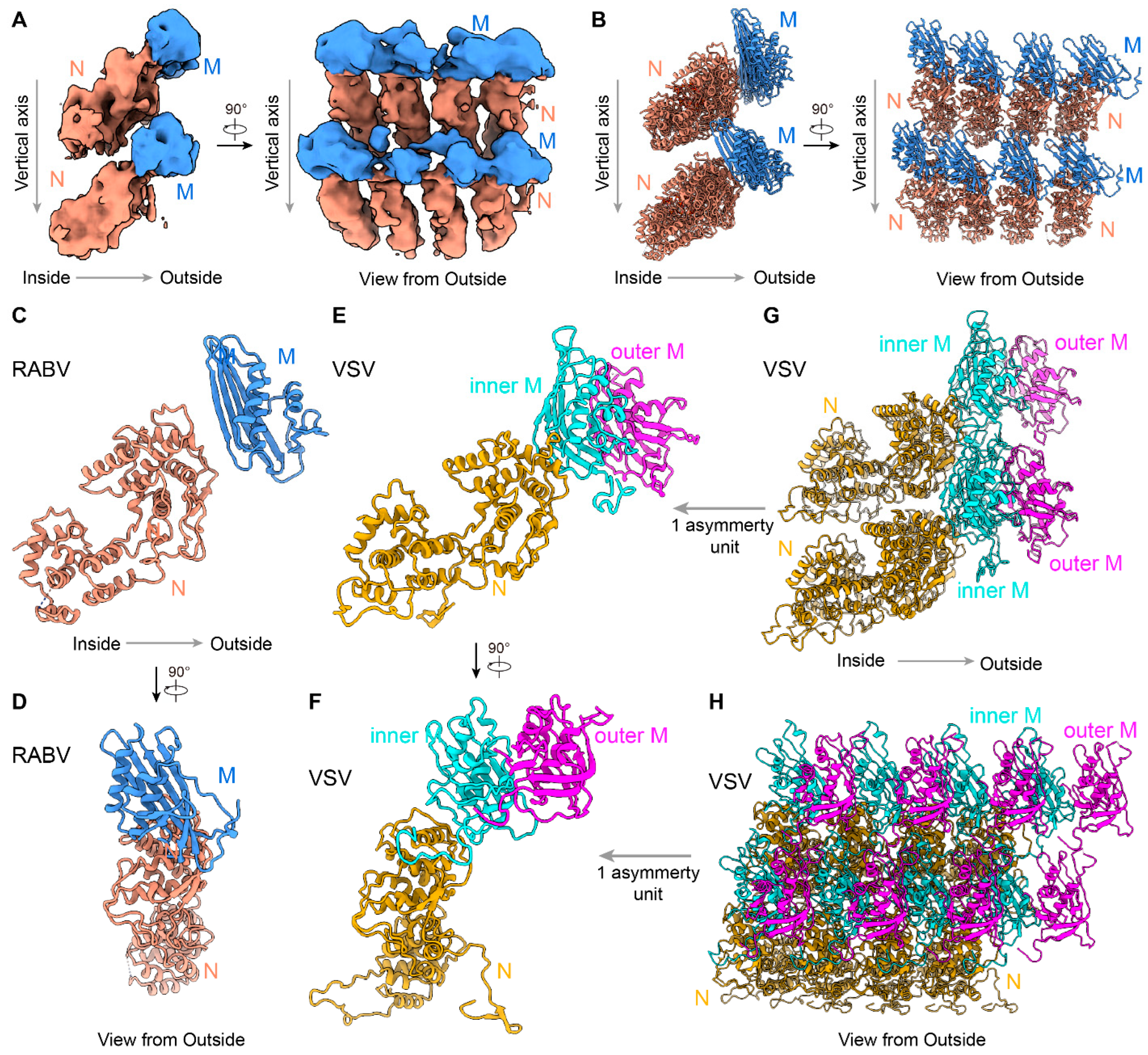
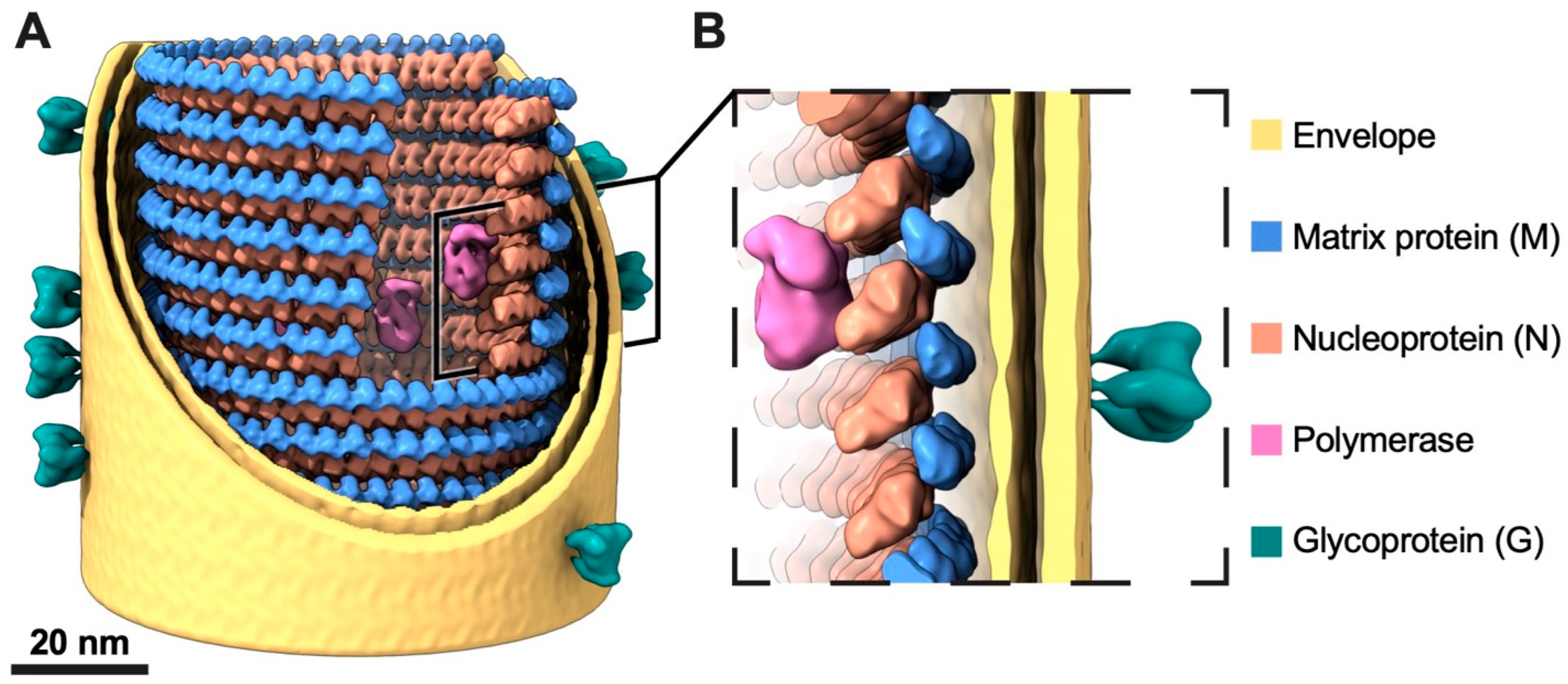
3.3. In Situ Structures of M-N by Helical Sub-Particle Reconstruction
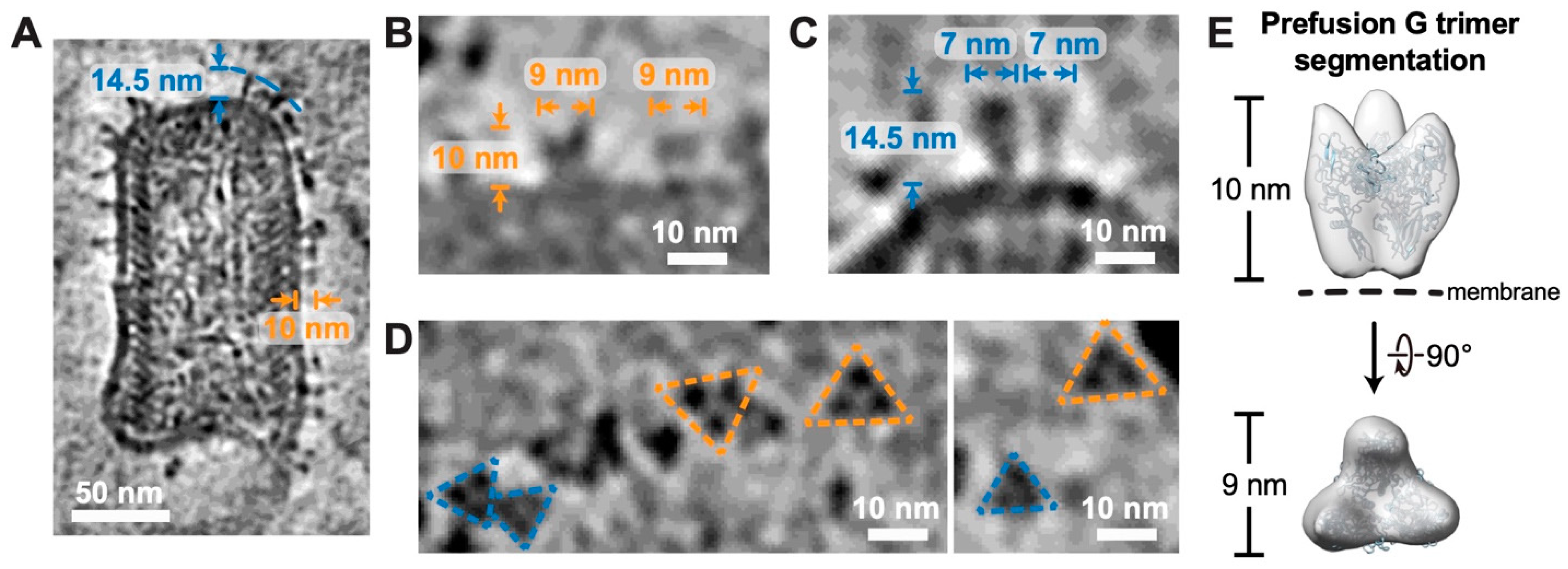
3.4. Both Prefusion and Postfusion G Trimers Captured on the Surface of RABV Virions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubos, R.J. Louis Pasteur: Freelance of Science; Little, Brown and Company: Boston, MA, USA, 1950; pp. 335–336. [Google Scholar]
- Matsumoto, S. Electron microscopy of nerve cells infected with street rabies virus. Virology 1962, 17, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Hummeler, K.; Koprowski, H.; Wiktor, T.J. Structure and development of rabies virus in tissue culture. J. Virol. 1967, 1, 152–170. [Google Scholar] [CrossRef] [PubMed]
- Guichard, P.; Krell, T.; Chevalier, M.; Vaysse, C.; Adam, O.; Ronzon, F.; Marco, S. Three dimensional morphology of rabies virus studied by cryo-electron tomography. J. Struct. Biol. 2011, 176, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.K.; Zhang, H.; Tan, K.; Li, Z.; Liu, J.H.; Chan, P.K.; Li, S.M.; Chan, W.Y.; Au, S.W.; Joachimiak, A.; et al. Structure of the influenza virus A H5N1 nucleoprotein: Implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008, 22, 3638–3647. [Google Scholar] [CrossRef]
- Jones, S.M.; Feldmann, H.; Stroher, U.; Geisbert, J.B.; Fernando, L.; Grolla, A.; Klenk, H.D.; Sullivan, N.J.; Volchkov, V.E.; Fritz, E.A.; et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005, 11, 786–790. [Google Scholar] [CrossRef]
- Daddario-DiCaprio, K.M.; Geisbert, T.W.; Stroher, U.; Geisbert, J.B.; Grolla, A.; Fritz, E.A.; Fernando, L.; Kagan, E.; Jahrling, P.B.; Hensley, L.E.; et al. Postexposure protection against Marburg haemorrhagic fever with recombinant vesicular stomatitis virus vectors in non-human primates: An efficacy assessment. Lancet 2006, 367, 1399–1404. [Google Scholar] [CrossRef]
- Stojdl, D.F.; Lichty, B.D.; tenOever, B.R.; Paterson, J.M.; Power, A.T.; Knowles, S.; Marius, R.; Reynard, J.; Poliquin, L.; Atkins, H.; et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 2003, 4, 263–275. [Google Scholar] [CrossRef]
- Faul, E.J.; Wanjalla, C.N.; Suthar, M.S.; Gale, M.; Wirblich, C.; Schnell, M.J. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog. 2010, 6, e1001016. [Google Scholar] [CrossRef]
- Kurup, D.; Wirblich, C.; Ramage, H.; Schnell, M.J. Rabies virus-based COVID-19 vaccine CORAVAX induces high levels of neutralizing antibodies against SARS-CoV-2. npj Vaccines 2020, 5, 98. [Google Scholar] [CrossRef]
- Albertini, A.A.; Wernimont, A.K.; Muziol, T.; Ravelli, R.B.; Clapier, C.R.; Schoehn, G.; Weissenhorn, W.; Ruigrok, R.W. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 2006, 313, 360–363. [Google Scholar] [CrossRef]
- Riedel, C.; Vasishtan, D.; Prazak, V.; Ghanem, A.; Conzelmann, K.K.; Rumenapf, T. Cryo EM structure of the rabies virus ribonucleoprotein complex. Sci. Rep. 2019, 9, 9639. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, J.A.; Jenni, S.; Harrison, S.C.; Whelan, S.P.J. Structure of a rabies virus polymerase complex from electron cryo-microscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lin, S.; Ye, F.; Yang, J.; Qi, J.; Chen, Z.; Lin, X.; Wang, J.; Yue, D.; Cheng, Y.; et al. Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody. Cell Host Microbe 2020, 27, 441–453.e7. [Google Scholar] [CrossRef]
- Callaway, H.M.; Zyla, D.; Larrous, F.; de Melo, G.D.; Hastie, K.M.; Avalos, R.D.; Agarwal, A.; Corti, D.; Bourhy, H.; Saphire, E.O. Structure of the rabies virus glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Sci. Adv. 2022, 8, eabp9151. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Si, Z.; Ge, P.; Tsao, J.; Luo, M.; Zhou, Z.H. Atomic model of vesicular stomatitis virus and mechanism of assembly. Nat. Commun. 2022, 13, 5980. [Google Scholar] [CrossRef]
- Si, Z.; Zhou, K.; Tsao, J.; Luo, M.; Zhou, Z.H. Locations and in situ structure of the polymerase complex inside the virion of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 2022, 119, e2111948119. [Google Scholar] [CrossRef]
- Jenni, S.; Horwitz, J.A.; Bloyet, L.M.; Whelan, S.P.J.; Harrison, S.C. Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle. Nat. Commun. 2022, 13, 4802. [Google Scholar] [CrossRef]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.F.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Wang, H.; Liao, S.; Yu, X.; Zhang, J.; Zhou, Z.H. TomoNet: A streamlined cryogenic electron tomography software pipeline with automatic particle picking on flexible lattices. Biol. Imaging 2024, 4, e7. [Google Scholar] [CrossRef]
- Liu, Y.T.; Zhang, H.; Wang, H.; Tao, C.L.; Bi, G.Q.; Zhou, Z.H. Isotropic reconstruction for electron tomography with deep learning. Nat. Commun. 2022, 13, 6482. [Google Scholar] [CrossRef] [PubMed]
- Kimanius, D.; Dong, L.; Sharov, G.; Nakane, T.; Scheres, S.H.W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021, 478, 4169–4185. [Google Scholar] [CrossRef]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, P.B.; Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003, 333, 721–745. [Google Scholar] [CrossRef]
- Zivanov, J.; Nakane, T.; Forsberg, B.O.; Kimanius, D.; Hagen, W.J.; Lindahl, E.; Scheres, S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 2018, 7, e42166. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Feng, Y.; Tu, Z.; Lou, Z.; Tu, C. Proteomic Profiling of Purified Rabies Virus Particles. Virol. Sin. 2020, 35, 143–155. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Kondo, H.; Goodin, M.M.; Kurath, G.; Vasilakis, N. The family Rhabdoviridae: Mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 2017, 227, 158–170. [Google Scholar] [CrossRef]
- Albertini, A.A.; Schoehn, G.; Weissenhorn, W.; Ruigrok, R.W. Structural aspects of rabies virus replication. Cell Mol. Life Sci. 2008, 65, 282–294. [Google Scholar] [CrossRef]
- Ge, P.; Tsao, J.; Schein, S.; Green, T.J.; Luo, M.; Zhou, Z.H. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 2010, 327, 689–693. [Google Scholar] [CrossRef]
- Chouaieb, N.; Goriely, A.; Maddocks, J.H. Helices. Proc. Natl. Acad. Sci. USA 2006, 103, 9398–9403. [Google Scholar] [CrossRef] [PubMed]
- Olsen, K.; Bohr, J. The generic geometry of helices and their close-packed structures. Theor. Chem. Acc. 2010, 125, 207–215. [Google Scholar] [CrossRef]
- Gerard, F.C.A.; Bourhis, J.M.; Mas, C.; Branchard, A.; Vu, D.D.; Varhoshkova, S.; Leyrat, C.; Jamin, M. Structure and Dynamics of the Unassembled Nucleoprotein of Rabies Virus in Complex with Its Phosphoprotein Chaperone Module. Viruses 2022, 14, 2813. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Li, T.; Li, Z.; Deans, E.E.; Mittler, E.; Liu, M.; Chandran, K.; Ivanovic, T. The shape of pleomorphic virions determines resistance to cell-entry pressure. Nat. Microbiol. 2021, 6, 617–629. [Google Scholar] [CrossRef]
- Roberts, P.C.; Lamb, R.A.; Compans, R.W. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 1998, 240, 127–137. [Google Scholar] [CrossRef]
- Pollin, R.; Granzow, H.; Kollner, B.; Conzelmann, K.K.; Finke, S. Membrane and inclusion body targeting of lyssavirus matrix proteins. Cell Microbiol. 2013, 15, 200–212. [Google Scholar] [CrossRef]
- Mebatsion, T.; Weiland, F.; Conzelmann, K.K. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 1999, 73, 242–250. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, X.; Zhou, K.; Alvarez-Cabrera, A.L.; Si, Z.; Wang, H.; He, Y.; Li, C.; Zhou, Z.H. Structural Heterogeneity of the Rabies Virus Virion. Viruses 2024, 16, 1447. https://doi.org/10.3390/v16091447
Cai X, Zhou K, Alvarez-Cabrera AL, Si Z, Wang H, He Y, Li C, Zhou ZH. Structural Heterogeneity of the Rabies Virus Virion. Viruses. 2024; 16(9):1447. https://doi.org/10.3390/v16091447
Chicago/Turabian StyleCai, Xiaoying, Kang Zhou, Ana Lucia Alvarez-Cabrera, Zhu Si, Hui Wang, Yao He, Cally Li, and Z. Hong Zhou. 2024. "Structural Heterogeneity of the Rabies Virus Virion" Viruses 16, no. 9: 1447. https://doi.org/10.3390/v16091447
APA StyleCai, X., Zhou, K., Alvarez-Cabrera, A. L., Si, Z., Wang, H., He, Y., Li, C., & Zhou, Z. H. (2024). Structural Heterogeneity of the Rabies Virus Virion. Viruses, 16(9), 1447. https://doi.org/10.3390/v16091447








