Genomic Characterization of Phage ZP3 and Its Endolysin LysZP with Antimicrobial Potential against Xanthomonas oryzae pv. oryzae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Isolation, Purification, and Concentration of Phage
2.3. Optimal Multiplicity of Infection (MOI) Determination
2.4. Determination of Phage Adsorption Rate
2.5. Determination of Phage One Step Growth Curve
2.6. Transmission Electron Microscopic (TEM) Observation
2.7. Determination of the Biophysical Stability of Phage ZP3
2.8. Determination of Phage Host Range
2.9. Phage DNA Extraction, Genome Sequencing, and Bioinformatic Analysis
2.10. Recombinant Plasmids Construction
2.11. Growth Measurement
2.12. Staining of Live/Dead Cells and Flow Cytometry Monitoring
2.13. β-Galactosidase Activity Assay
2.14. Protein Expression and Antibacterial Effect Assay
3. Results
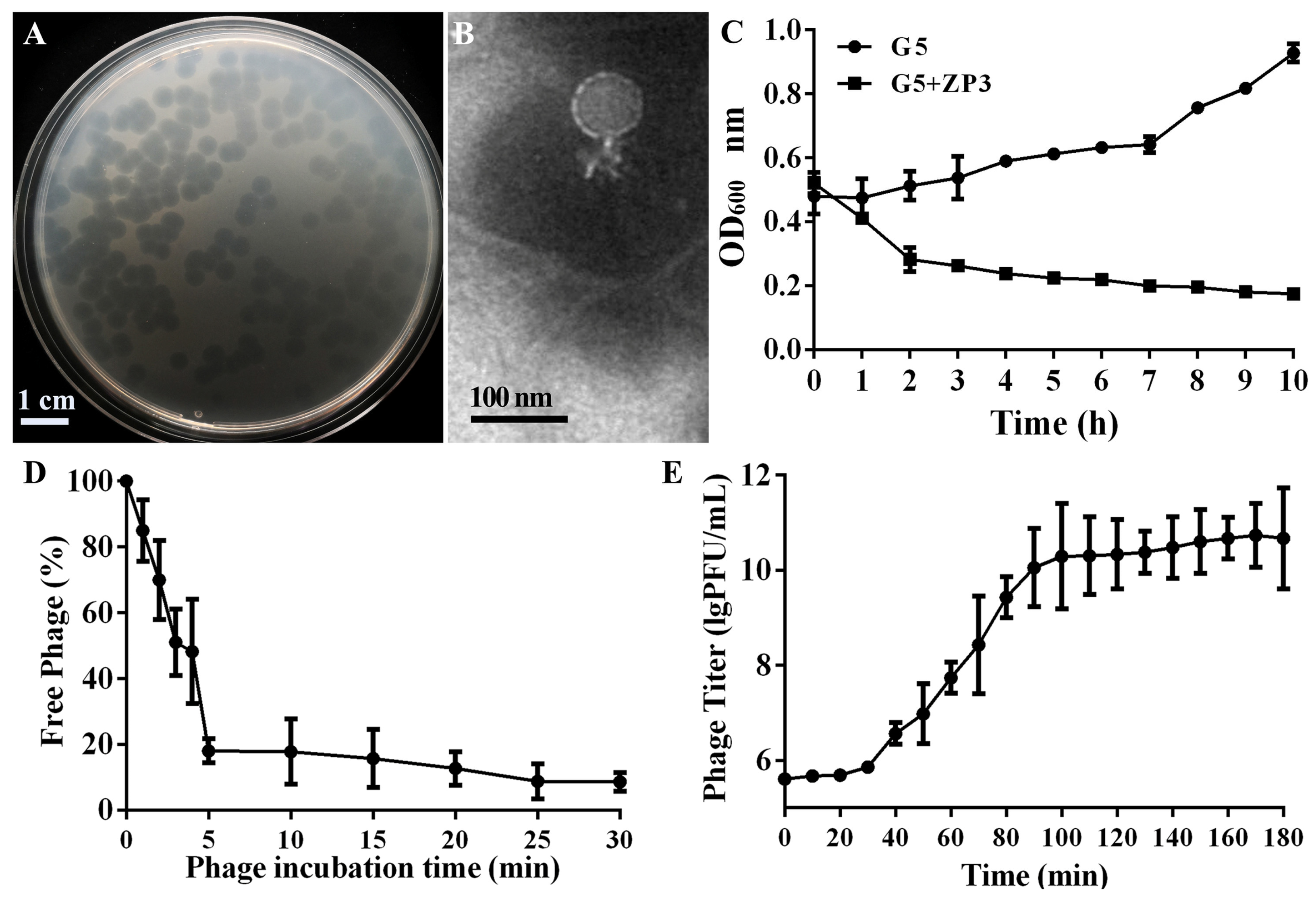
3.1. Isolation and Morphology of the Xoo Phage ZP3
3.2. Host Range of Phage ZP3
3.3. Biophysical Stability of Phage ZP3
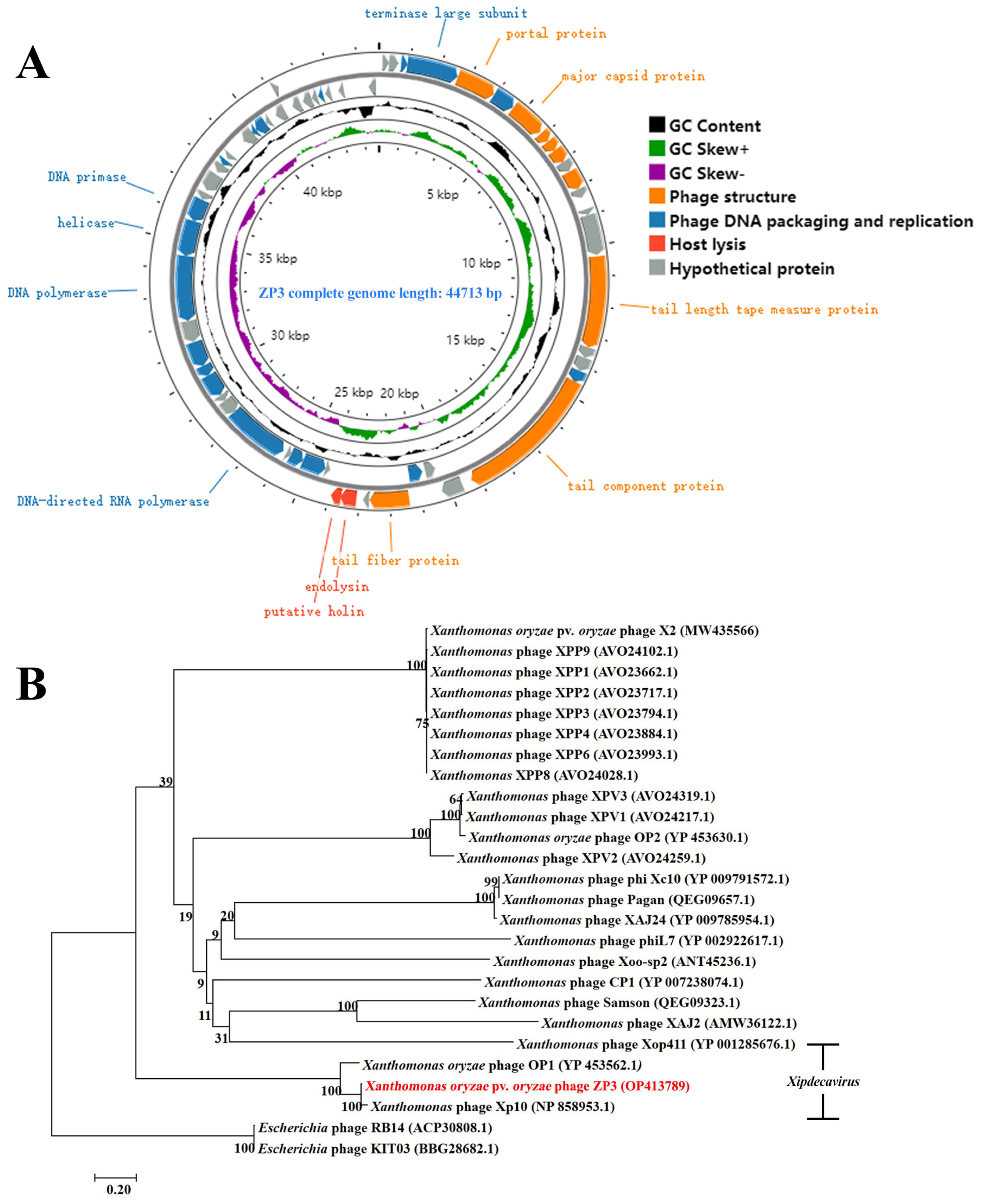
3.4. ZP3 Genome Sequencing, Annotation, and Phylogenetic Analysis
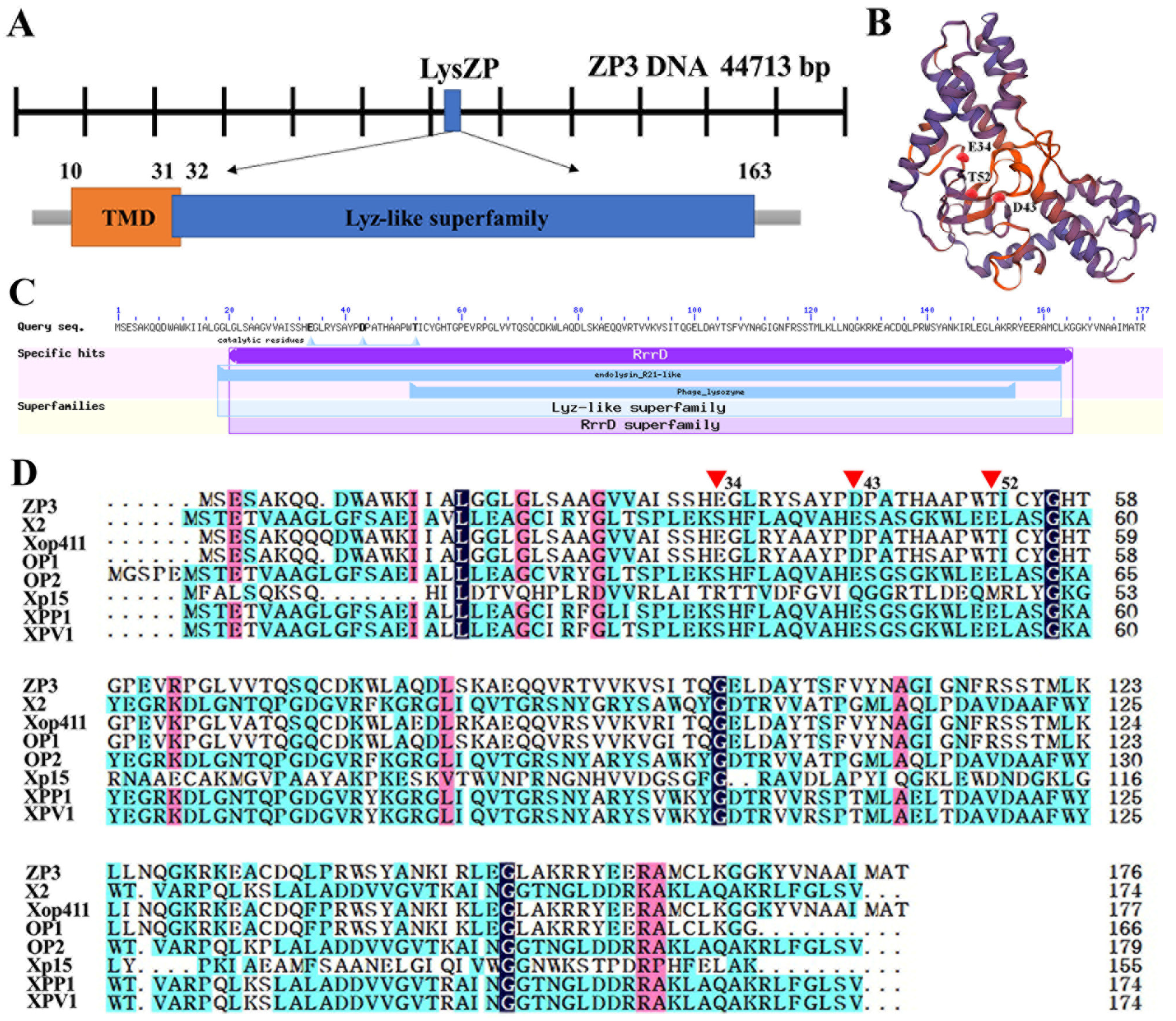
3.5. In Silico Identification of Phage ZP3 Endolysin
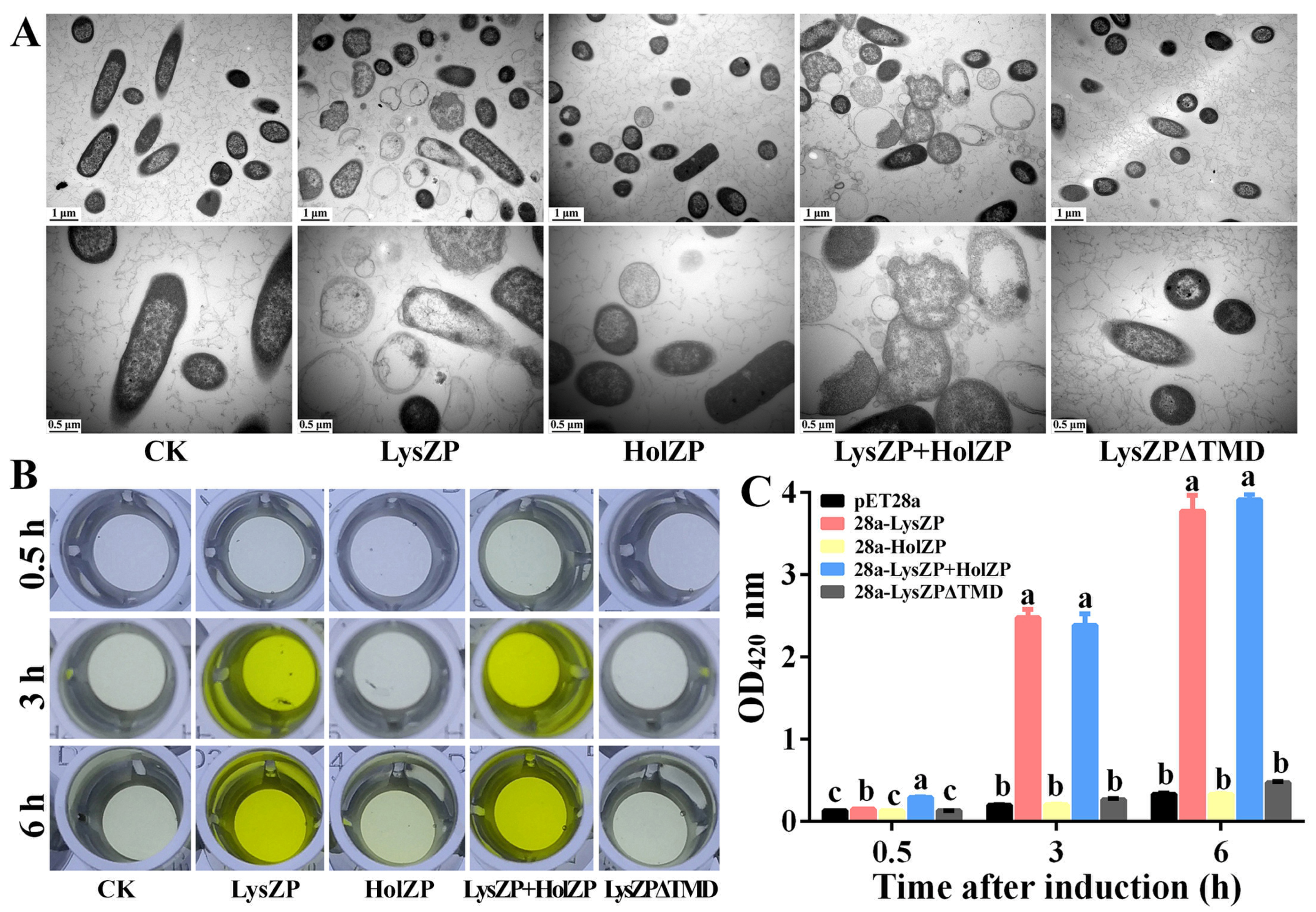
3.6. Transmembrane Domain Is the Key to Cell Lysis Induced by Endolysin
3.7. Expression of Endolysin LysZP Affects Membrane Integrity
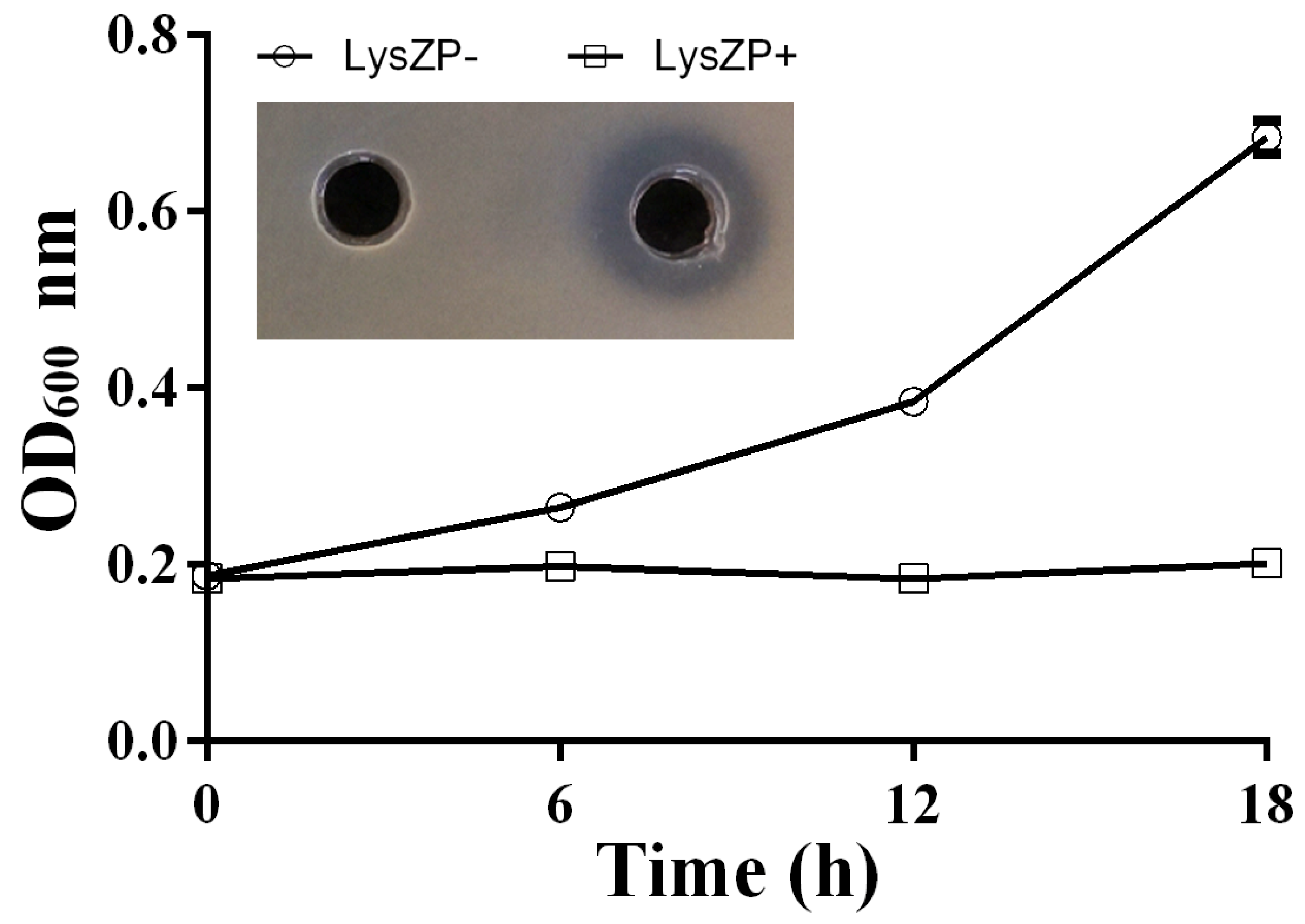
3.8. Antibacterial Effect of ZP3 Endolysin on Xoo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanya, D.R.A.; Syed-Ab-Rahman, S.F.; Jia, A.Q.; Onesime, D.; Kim, K.M.; Ahohuendo, B.C.; Rohr, J.R. A review of approaches to control bacterial leaf blight in rice. World J. Microbiol. Biotechnol. 2022, 38, 113. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.M.; Haider, M.S.; Randhir, T.O.; Randhir, R.; Ahmad, S.R. Spatial analysis of factors influencing bacterial leaf blight in rice production. Braz. J. Biol. Rev. Brasleira Biol. 2023, 83, e264249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, Y.H.; Sun, J.; Liang, W.F.; Chen, X.Y.; Wang, X.M.; Zhou, J.; Yu, C.L.; Wang, J.M.; Wu, S.L.; et al. Research Progress on Cloning and Function of Xa Genes Against Rice Bacterial Blight. Front. Plant Sci. 2022, 13, 847199. [Google Scholar] [CrossRef] [PubMed]
- Sundin, G.W.; Wang, N. Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Nakayinga, R.; Makumi, A.; Tumuhaise, V.; Tinzaara, W. Xanthomonas bacteriophages: A review of their biology and biocontrol applications in agriculture. BMC Microbiol. 2021, 21, 291. [Google Scholar] [CrossRef]
- Ranjani, P.; Gowthami, Y.; Samuel, S.G.; Palani, P. Bacteriophages: A New Weapon for the Control of Bacterial Blight Disease in Rice Caused by Xanthomonas oryzae. Microbiol. Biotechnol. Lett. 2018, 46, 346–359. [Google Scholar] [CrossRef]
- Mallmann, W.L.; Hemstreet, C. Isolation of an inhibitory substance from plants. J. Agric. Res. 1924, 28, 0599–0602. [Google Scholar]
- Retamales, J.; Nunez, P.; Alvarado, R.; Campan, E.D.M.; Otto, T.; Segovia, C.; Vasquez, I.; Santander, J. Characterization of Xanthomonas arboricola pv. juglandis bacteriophages against bacterial walnut blight and field evaluation. Viruses 2022, 14, 1380. [Google Scholar] [CrossRef]
- Ahmad, A.A.; Ogawa, M.; Kawasaki, T.; Fujie, M.; Yamada, T. Characterization of bacteriophages Cp1 and Cp2, the strain-typing agents for Xanthomonas axonopodis pv. citri. Appl. Environ. Microbiol. 2014, 80, 77–85. [Google Scholar] [CrossRef]
- Ogunyemi, S.O.; Chen, J.; Zhang, M.C.; Wang, L.; Masum, M.M.I.; Yan, C.Q.; An, Q.L.; Li, B.; Chen, J.P. Identification and characterization of five new OP2-related Myoviridae bacteriophages infecting different strains of Xanthomonas oryzae pv. oryzae. J. Plant Pathol. 2019, 101, 263–273. [Google Scholar] [CrossRef]
- Nazir, A.; Dong, Z.X.; Liu, J.; Tahir, R.A.; Ashraf, N.; Qing, H.; Peng, D.H.; Tong, Y.G. Isolation, characterization, and genome sequence analysis of a novel lytic phage, Xoo-sp15 infecting Xanthomonas oryzae pv. oryzae. Curr. Microbiol. 2021, 78, 3192–3200. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.N.; Hu, R.M.; Chow, T.Y.; Lin, J.W.; Chen, H.Y.; Tseng, Y.H.; Weng, S.F. Comparison of genomes of three Xanthomonas oryzae bacteriophages. BMC Genom. 2007, 8, 442. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.P.; et al. Engineered Endolysin-Based “Artilysins” To Combat Multidrug-Resistant Gram-Negative Pathogens. Mbio 2014, 5, e01379-14. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, Ł.; Łepek, K.; Stasiłojć, M.; Kosznik-Kwaśnicka, K.; Zdrojewska, K.; Maciąg-Dorszyńska, M.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-encoded enzymes destroying bacterial cell membranes and walls, and their potential use as antimicrobial agents. Microbiol. Res. 2021, 248, 126746. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, J.G.; Guerreiro-Pereira, M.C.; Costa, S.F.; Sao-Jose, C.; Santos, M.A. Nisin-triggered activity of Lys44, the secreted endolysin from Oenococcus oeni phage fOg44. J. Bacteriol. 2008, 190, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Struck, D.K.; Deaton, J.; Wang, I.N.; Young, R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. USA 2004, 101, 6415–6420. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-encoded endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Gontijo, M.T.P.; Jorge, G.P.; Brocchi, M. Current status of endolysin-based treatments against gram-negative bacteria. Antibiotics 2021, 10, 1143. [Google Scholar] [CrossRef]
- Fong, K.; Wong, C.W.Y.; Wang, S.; Delaquis, P. How Broad Is Enough: The Host Range of Bacteriophages and Its Impact on the Agri-Food Sector. PHAGE 2021, 2, 83–91. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, X.; Ahmed, T.; Liu, M.; Wu, Z.; Luo, J.; Tian, Y.; Jiang, H.; Wang, Y.; Sun, G.; et al. Identification of genes involved in antifungal activity of Burkholderia seminalis against Rhizoctonia solani using Tn5 transposon mutation method. Pathogens 2020, 9, 797. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K. Working with bacteriophages: Common techniques and methodological approaches. Bacteriophages Biol. Appl. 2005, 1, 437–494. [Google Scholar]
- Birg, E.A.; Birg, E.A. Bacterial and Bacteriophage Genetics; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2000; p. i. [Google Scholar]
- Gencay, Y.E.; Birk, T.; Sorensen, M.C.H.; Brondsted, L. Methods for isolation, purification, and propagation of bacteriophages of Campylobacter jejuni. In Campylobacter Jejuni: Methods and Protocols; Butcher, J., Stintzi, A., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1512, pp. 19–28. [Google Scholar]
- Zhang, M.C.; Qian, J.H.; Xu, X.Y.; Ahmed, T.; Yang, Y.; Yan, C.Q.; Elsharkawy, M.M.; Hassan, M.M.; Alorabi, J.A.; Chen, J.P.; et al. Resistance of Xanthomonas oryzae pv. oryzae to lytic phage X2 by spontaneous mutation of lipopolysaccharide synthesis-related glycosyltransferase. Viruses 2022, 14, 1088. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.Q.; Yuan, Z.M.; Huang, Y. Disinfectants against SARS-CoV-2: A Review. Viruses 2022, 14, 1721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ning, H.; Lin, H.; She, J.; Wang, L.; Jing, Y.; Wang, J. Expansion of the plaquing host range and improvement of the absorption rate of a T5-like Salmonella phage by altering the long tail fibers. Appl. Environ. Microbiol. 2022, 88, e0089522. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W.; Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 2001; pp. 11803–12500. [Google Scholar]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 10. [Google Scholar] [CrossRef]
- Liu, Z.; Hou, S.; Rodrigues, O.; Wang, P.; Luo, D.; Munemasa, S.; Lei, J.; Liu, J.; Ortiz-Morea, F.A.; Wang, X.; et al. Phytocytokine signalling reopens stomata in plant immunity and water loss. Nature 2022, 605, 332–339. [Google Scholar] [CrossRef]
- Zhang, M.C.; Wang, Y.L.; Chen, J.; Hong, X.X.; Xu, X.Y.; Wu, Z.F.; Ahmed, T.; Loh, B.; Leptihn, S.; Hassan, S.; et al. Identification and characterization of a new type of holin-endolysin lysis cassette in Acidovorax oryzae phage AP1. Viruses 2022, 14, 167. [Google Scholar] [CrossRef]
- Wu, J.; Abbas, H.M.K.; Li, J.L.; Yuan, Y.; Liu, Y.J.; Wang, G.Y.; Dong, W.B. Cell Membrane-Interrupting Antimicrobial Peptides from Isatis indigotica Fortune Isolated by a Bacillus subtilis Expression System. Biomolecules 2020, 10, 30. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Q.; Ma, Y.F.; Wang, Y.T.; Yang, H.X.; Shen, W.; Chen, X.Z. Transcription regulation mechanisms of bacteriophages Recent advances and future prospects. Bioengineered 2014, 5, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Bamford, D.H. Holin of bacteriophage lambda: Structural insights into a membrane lesion. Mol. Microbiol. 2008, 69, 781–783. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Xu, Z.H.; Shao, S.; Ding, Z.H.; Zhang, Y.X.; Wang, Q.Y.; Liu, X.H.; Liu, Q. Therapeutic efficacies of two newly isolated Edwardsiella phages against Edwardsiella piscicida infection. Microbiol. Res. 2022, 263, 127043. [Google Scholar] [CrossRef]
- Abedon, S.T.; Garcia, P.; Mullany, P.; Aminov, R. Editorial: Phage Therapy: Past, Presentand Future. Front. Microbiol. 2017, 8, 7. [Google Scholar] [CrossRef]
- Wakimoto, S. Classification of strains of Xanthomonas oryzae on the basis of their susceptibility against bacteriophages. Jpn. J. Phytopathol. 1960, 25, 193–198. [Google Scholar] [CrossRef]
- Inoue, Y.; Matsuura, T.; Ohara, T.; Azegami, K. Bacteriophage OP1, lytic for Xanthomonas oryzae pv. oryzae, changes its host range by duplication and deletion of the small domain in the deduced tail fiber gene. J. Gen. Plant Pathol. 2006, 72, 111–118. [Google Scholar] [CrossRef]
- Inoue, Y.; Matsuura, T.; Ohara, T.; Azegami, K. Sequence analysis of the genome of OP2, a lytic bacteriophage of Xanthomonas oryzae pv. oryzae. J. Gen. Plant Pathol. 2006, 72, 104–110. [Google Scholar] [CrossRef]
- Ramesh, N.; Manohar, P.; Eniyan, K.; Archana, L.; Athira, S.; Loh, B.; Ma, L.; Leptihn, S. A lysozyme murein hydrolase with broad-spectrum antibacterial activity from Enterobacter phage myPSH1140. Antimicrob. Agents Chemother. 2022, 66, e0050622. [Google Scholar] [CrossRef]
- Kashani, H.H.; Schmelcher, M.; Sabzalipoor, H.; Hosseini, E.S.; Moniri, R. Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: Current status of research and novel delivery strategies. Clin. Microbiol. Rev. 2018, 31, e00071-17. [Google Scholar] [CrossRef]
- Larpin, Y.; Oechslin, F.; Moreillon, P.; Resch, G.; Entenza, J.M.; Mancini, S. In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS ONE 2018, 13, e0192507. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.B.; Yan, Y.X.; Ji, W.H.; Du, B.; Meng, X.P.; Wang, H.G.; Sun, J.H. Characterization and determination of holin protein of Streptococcus suis bacteriophage SMP in heterologous host. Virol. J. 2012, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Arulandu, A.; Struck, D.K.; Swanson, S.; Sacchettini, J.C.; Young, R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 2005, 307, 113–117. [Google Scholar] [CrossRef]
- Lee, C.N.; Lin, J.W.; Chow, T.Y.; Tseng, Y.H.; Weng, S.F. A novel lysozyme from Xanthomonas oryzae phage phi Xo411 active against Xanthomonas and Stenotrophomonas. Protein Expr. Purif. 2006, 50, 229–237. [Google Scholar] [CrossRef]


| Plasmids | Description | Sources |
|---|---|---|
| pET28a | KmR; expression vector | Novagen (Madison, WI, USA) |
| pET28a-LysZP | KmR; recombinant expression vector with LysZP | This study |
| pET28a-HolZP | KmR; recombinant expression vector with HolZP | This study |
| pET28a-LysZP+HolZP | KmR; recombinant expression vector with LysZP and HolZP | This study |
| pET28a-LysZPΔTMD | KmR; expression vector recombinant expression vector with LysZP without TMD | This study |
| Primers Name | Sequences (5′-3′) | Description |
|---|---|---|
| Lys-F | GCAAATGGGTCGCGGATCCATGAGCGAATCCGCGAAG | Gene of Lys from ZP3 |
| Lys-R | GGAGCTCGAATTCGGATCCACGCGTCGCCATAATAGCAG | |
| Hol-F | GCAAATGGGTCGCGGATCCATGTCAATGCTGCTATTATGGCG | Gene of Hol from ZP3 |
| Hol-R | GGAGCTCGAATTCGGATCCCAGGGACGGAGGTACTGCTC | |
| Lys-hol-F | CTGCTATTATGGCGACGCGTATGTCAATGCTGCTATTATGGCG | Gene of Lys and Hol from ZP3 |
| Lys-hol-R | CATAATAGCAGCATTGACATACGCGTCGCCATAATAGCAG | |
| Lys-TMD-F | GCAAATGGGTCGCGGATCCATGAGCGAATCCGCGAAGCAGCAGGACCATGAGGGTCTCAGGTACTC | Gene of Lys without TMD from ZP3 |
| Strains | Sensitivity to Phage ZP3 | Origin | Species | Strains | Sensitivity to Phage ZP3 | Origin | Species |
|---|---|---|---|---|---|---|---|
| C2 | − | Guangdong, China | Xoo | Z2 | - | Zhejiang, China | Xoo |
| C4 | − | Guangdong, China | Xoo | Z3 | − | Zhejiang, China | Xoo |
| C8 | − | Guangdong, China | Xoo | Z4 | − | Zhejiang, China | Xoo |
| G1 | + | Guangdong, China | Xoo | L1 | − | Liaoning, China | Xoo |
| G2 | + | Guangdong, China | Xoo | L2 | − | Liaoning, China | Xoo |
| G3 | + | Guangdong, China | Xoo | L3 | − | Liaoning, China | Xoo |
| G4 | + | Guangdong, China | Xoo | L4 | − | Liaoning, China | Xoo |
| G5 | + | Guangdong, China | Xoo | L5 | − | Liaoning, China | Xoo |
| G6 | + | Guangdong, China | Xoo | L6 | − | Liaoning, China | Xoo |
| G8 | + | Guangdong, China | Xoo | LN4 | − | Liaoning, China | Xoo |
| G9 | + | Guangdong, China | Xoo | LN18 | − | Liaoning, China | Xoo |
| G10 | + | Guangdong, China | Xoo | ScYc-b | + | Sichuan, China | Xoo |
| G11 | − | Guangdong, China | Xoo | YN1 | − | Yunnan, China | Xoo |
| G12 | + | Guangdong, China | Xoo | YN7 | − | Yunnan, China | Xoo |
| G13 | + | Guangdong, China | Xoo | YN11 | − | Yunnan, China | Xoo |
| G14 | + | Guangdong, China | Xoo | YN18 | − | Yunnan, China | Xoo |
| G15 | − | Guangdong, China | Xoo | YN24 | + | Yunnan, China | Xoo |
| G16 | + | Guangdong, China | Xoo | HEN | + | Henan, China | Xoo |
| GD414 | − | Guangdong, China | Xoo | HEN11 | + | Henan, China | Xoo |
| Y1 | − | Zhejiang, China | Xoo | FuJ | + | Fujian, China | Xoo |
| Y2 | − | Zhejiang, China | Xoo | OS198 | + | Guangxi, China | Xoo |
| Y3 | + | Zhejiang, China | Xoo | GX1 | + | Guangxi, China | Xoo |
| Y4 | − | Zhejiang, China | Xoo | GX2 | + | Guangxi, China | Xoo |
| Y5 | − | Zhejiang, China | Xoo | GX3 | + | Guangxi, China | Xoo |
| Y6 | − | Zhejiang, China | Xoo | GX4 | + | Guangxi, China | Xoo |
| Y7 | + | Zhejiang, China | Xoo | Pxo99A | − | Philippines | Xoo |
| Y8 | + | Zhejiang, China | Xoo | RS105 | − | China | Xoc |
| T1 | + | Zhejiang, China | Xoo | RS-1 | − | Zhejiang, China | Ao |
| T173 | + | Zhejiang, China | Xoo | RS-2 | − | Zhejiang, China | Ao |
| Z1 | − | Zhejiang, China | Xoo | R456 | − | Zhejiang, China | Bs |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Xu, X.; Lv, L.; Luo, J.; Ahmed, T.; Alsakkaf, W.A.A.; Ali, H.M.; Bi, J.; Yan, C.; Gu, C.; et al. Genomic Characterization of Phage ZP3 and Its Endolysin LysZP with Antimicrobial Potential against Xanthomonas oryzae pv. oryzae. Viruses 2024, 16, 1450. https://doi.org/10.3390/v16091450
Zhang M, Xu X, Lv L, Luo J, Ahmed T, Alsakkaf WAA, Ali HM, Bi J, Yan C, Gu C, et al. Genomic Characterization of Phage ZP3 and Its Endolysin LysZP with Antimicrobial Potential against Xanthomonas oryzae pv. oryzae. Viruses. 2024; 16(9):1450. https://doi.org/10.3390/v16091450
Chicago/Turabian StyleZhang, Muchen, Xinyan Xu, Luqiong Lv, Jinyan Luo, Temoor Ahmed, Waleed A. A. Alsakkaf, Hayssam M. Ali, Ji’an Bi, Chengqi Yan, Chunyan Gu, and et al. 2024. "Genomic Characterization of Phage ZP3 and Its Endolysin LysZP with Antimicrobial Potential against Xanthomonas oryzae pv. oryzae" Viruses 16, no. 9: 1450. https://doi.org/10.3390/v16091450
APA StyleZhang, M., Xu, X., Lv, L., Luo, J., Ahmed, T., Alsakkaf, W. A. A., Ali, H. M., Bi, J., Yan, C., Gu, C., Shou, L., & Li, B. (2024). Genomic Characterization of Phage ZP3 and Its Endolysin LysZP with Antimicrobial Potential against Xanthomonas oryzae pv. oryzae. Viruses, 16(9), 1450. https://doi.org/10.3390/v16091450







