High-Throughput Oxford Nanopore Sequencing Unveils Complex Viral Population in Kansas Wheat: Implications for Sustainable Virus Management
Abstract
1. Introduction
2. Materials and Methods
2.1. Winter Wheat Field Survey
2.2. RNA Extraction and Nanopore Sequencing
2.3. Sequence Alignment and Diagnosis
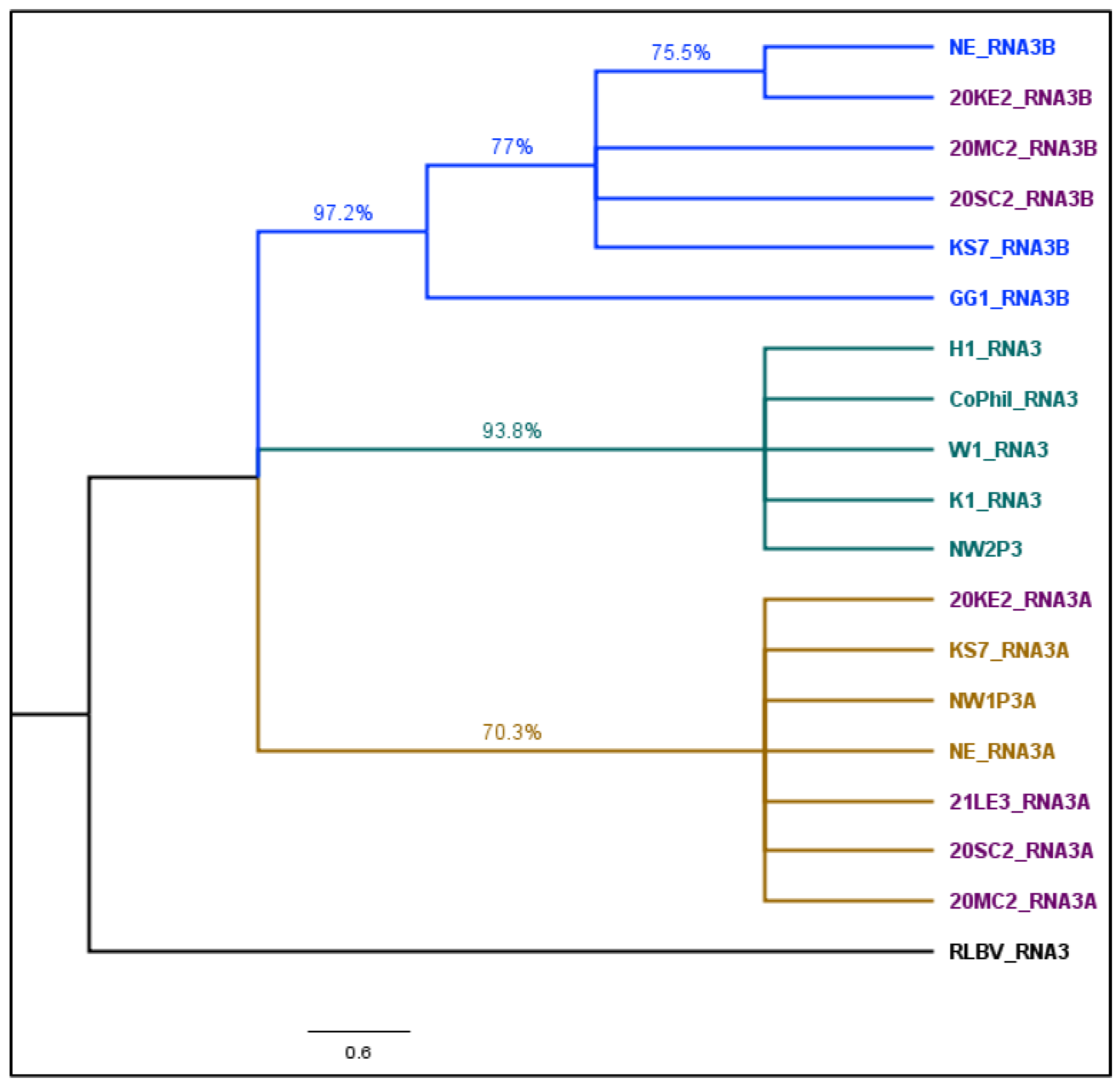
2.4. Phylogenetic Analysis
2.5. Recombinant Analysis
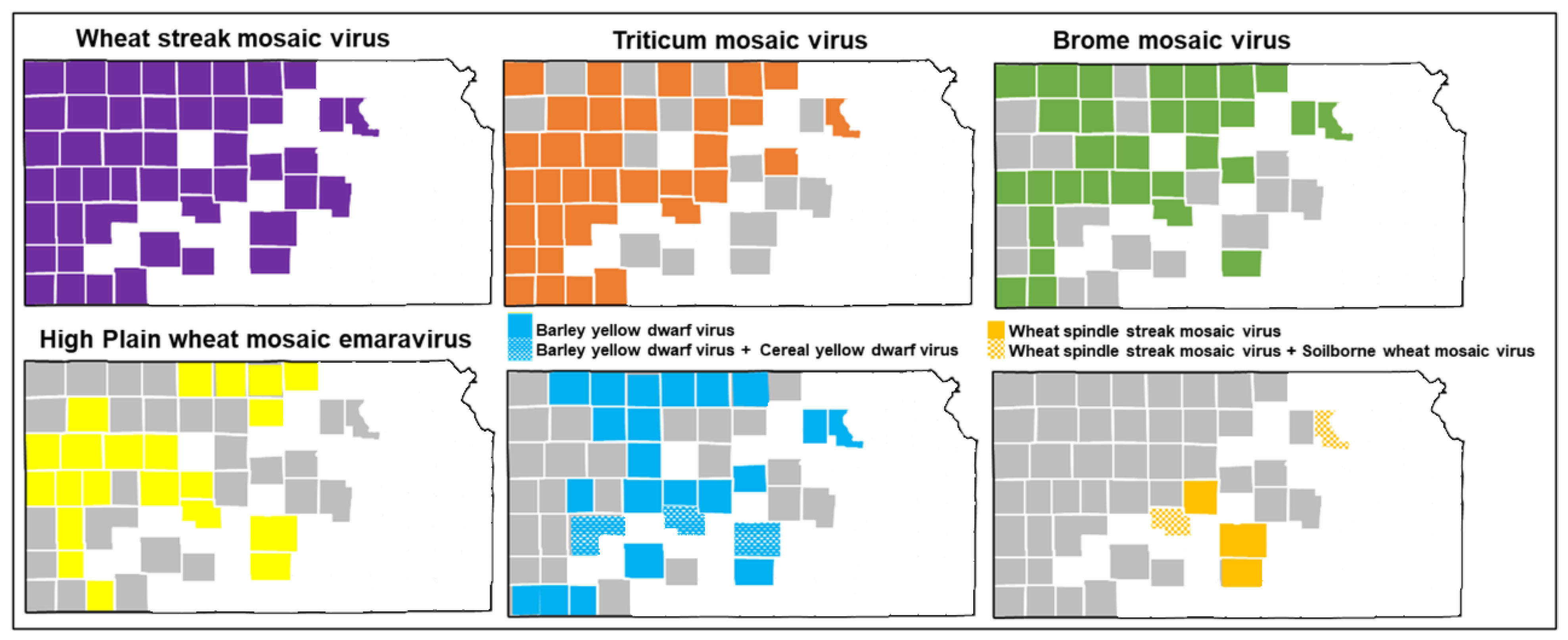
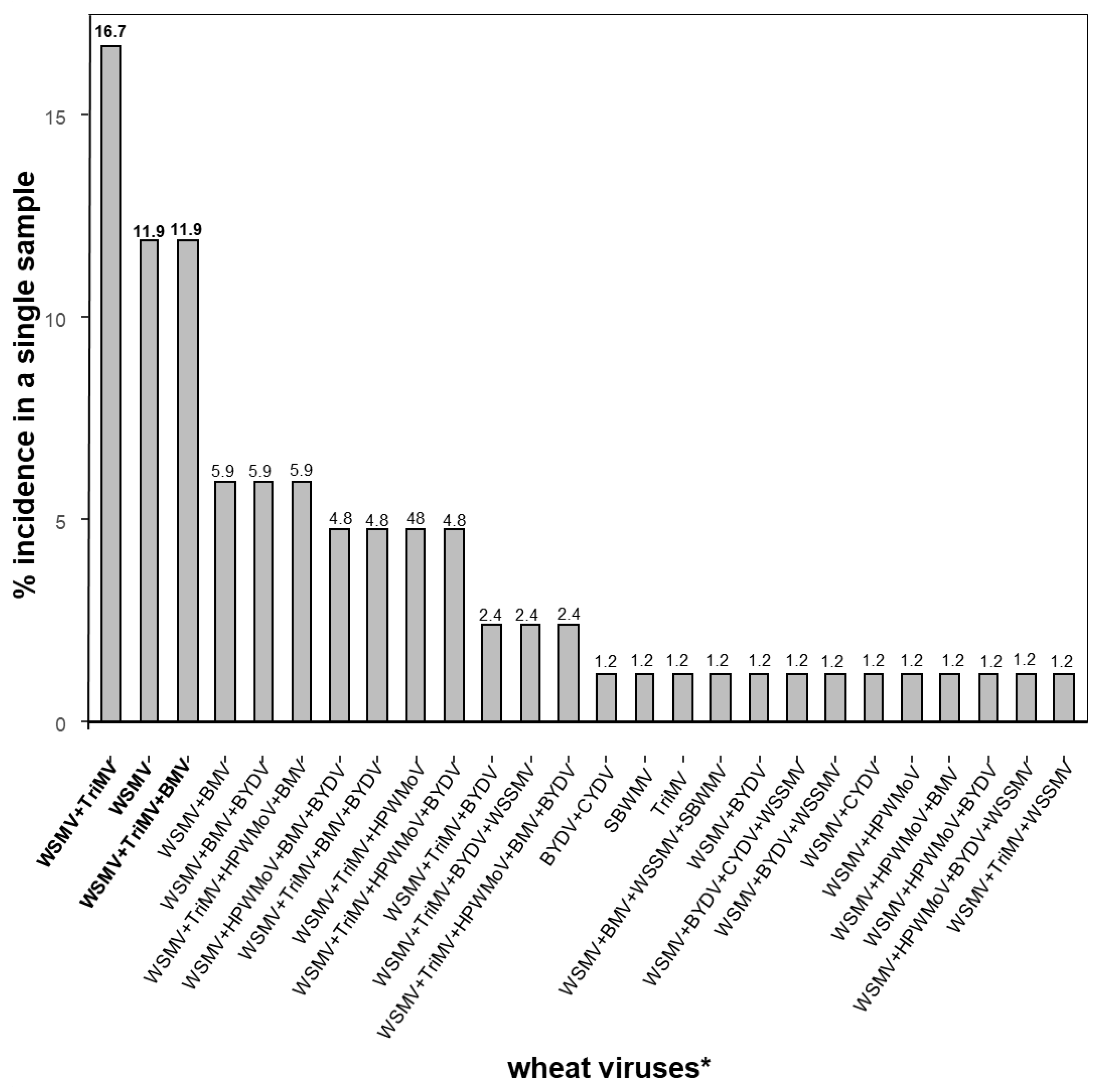
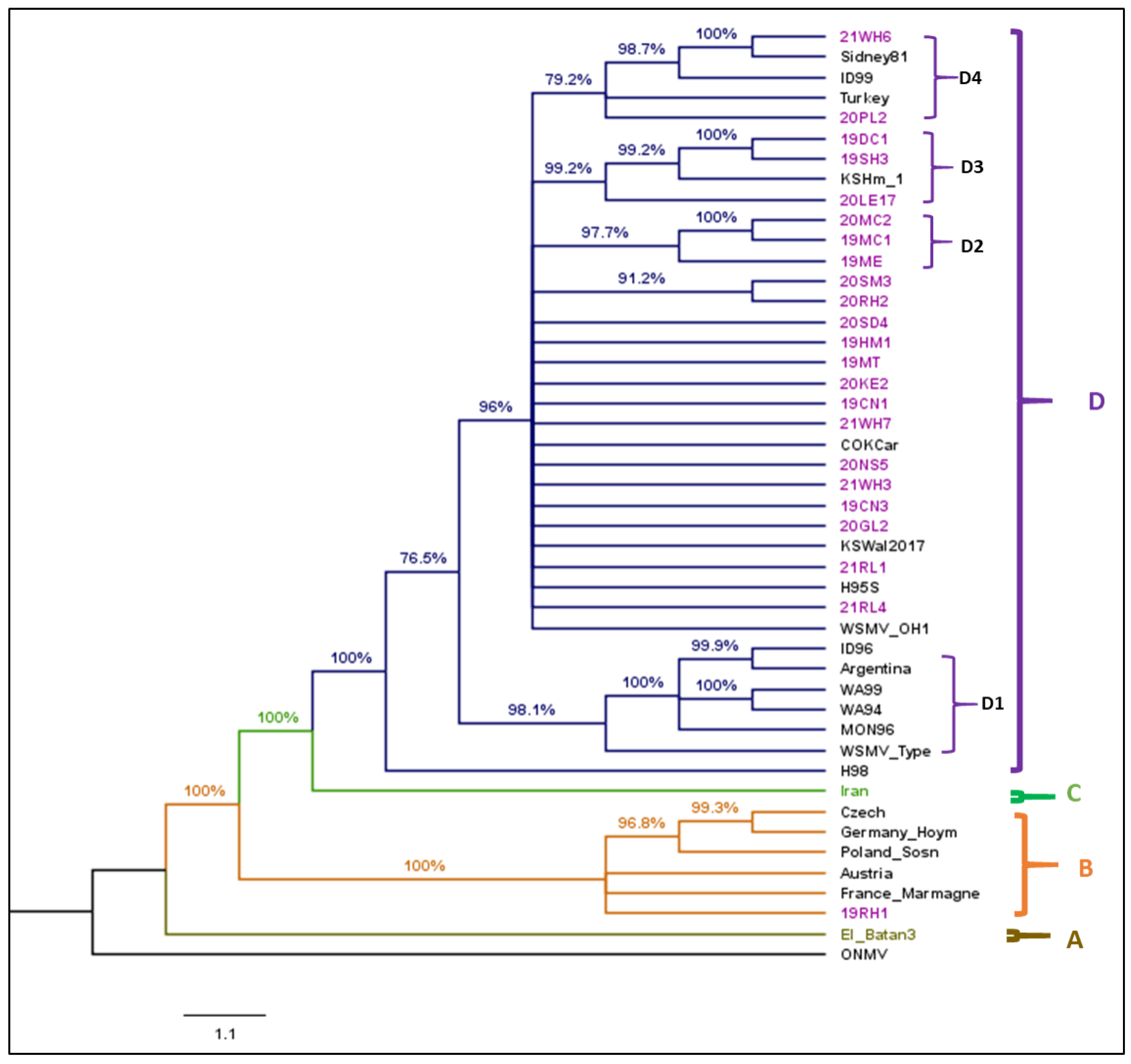
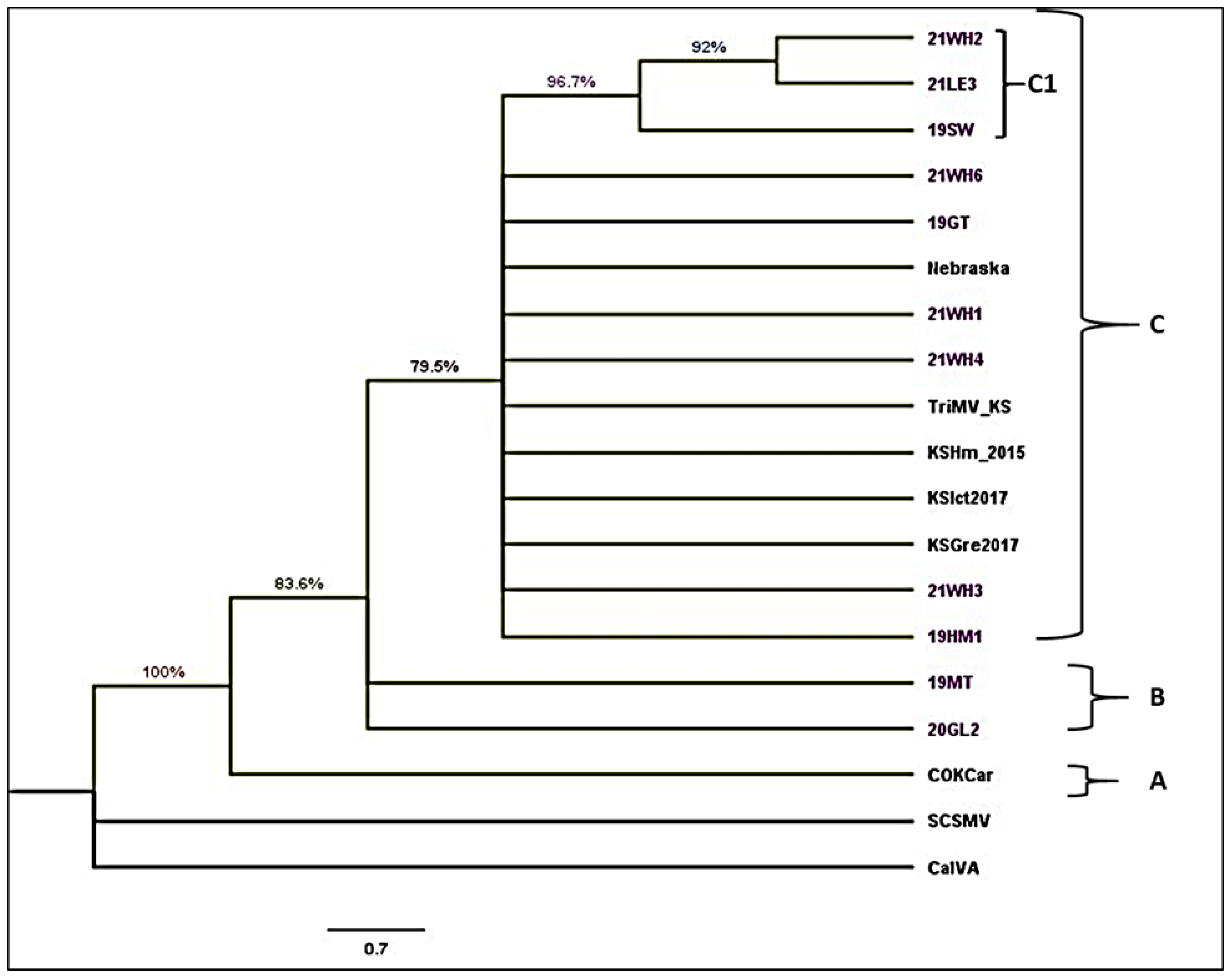
3. Results
3.1. Sequence Alignments
3.2. Recombinant Analysis of WSMV
3.3. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hollandbeck, F.G.; De Wolf, E.; Todd, T. Kansas Cooperative Plant Disease Survey Report, Preliminary 2020 Kansas Wheat Disease Loss Estimates. Kans. Dep. Agric. 2020. [Google Scholar]
- Hollandbeck, F.G.; De Wolf, E.; Todd, T. Kansas Cooperative Plant Disease Survey Report, Preliminary 2021 Kansas Wheat Disease Loss Estimates. Kans. Dep. Agric. 2021. [Google Scholar]
- Byamukama, E.; Wegulo, S.; Tatineni, S.; Hein, G.; Graybosch, R.; Baenziger, P.S.; French, R. Quantification of Yield Loss Caused by Triticum Mosaic Virus and Wheat Streak Mosaic Virus in Winter Wheat under Field Conditions. Plant Dis. 2014, 98, 127–133. [Google Scholar] [CrossRef]
- Campbell, L.; Heyne, E.; Gronau, D.; Niblett, C. Effect of Soilborne Wheat Mosaic Virus on Wheat Yield. Plant Dis. Report. 1975, 59, 472–476. [Google Scholar]
- Cunfer, B.M.; Demski, J.W.; Bays, D.C. Reduction in Plant Development, Yield, and Grain Quality Associated with Wheat Spindle Streak Mosaic Virus. Phytopathology 1988, 78, 198–204. [Google Scholar] [CrossRef]
- Choudhury, S.; Larkin, P.; Meinke, H.; Hasanuzzaman, M.; Johnson, P.; Zhou, M. Barley Yellow Dwarf Virus Infection Affects Physiology, Morphology, Grain Yield and Flour Pasting Properties of Wheat. Crop Pasture Sci. 2019, 70, 16–25. [Google Scholar] [CrossRef]
- Ordon, F.; Habekuss, A.; Kastirr, U.; Rabenstein, F.; Kühne, T. Virus Resistance in Cereals: Sources of Resistance, Genetics and Breeding. J. Phytopathol. 2009, 157, 535–545. [Google Scholar] [CrossRef]
- Brakke, M. Virus Diseases of Wheat. Wheat Wheat Improv. 1987, 13, 585–624. [Google Scholar]
- Hodge, B.; Paul, P.; Stewart, L.R. Occurrence and High-Throughput Sequencing of Viruses in Ohio Wheat. Plant Dis. 2020, 104, 1789–1800. [Google Scholar] [CrossRef]
- Rotenberg, D.; Bockus, W.W.; Whitfield, A.E.; Hervey, K.; Baker, K.D.; Ou, Z.; Laney, A.G.; De Wolf, E.D.; Appel, J.A. Occurrence of Viruses and Associated Grain Yields of Paired Symptomatic and Nonsymptomatic Tillers in Kansas Winter Wheat Fields. Phytopathology 2016, 106, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Ranabhat, N.B.; Fellers, J.P.; Bruce, M.A.; Rupp, J.L.S. Brome Mosaic Virus Detected in Kansas Wheat Co-Infected with Other Common Wheat Viruses. Front. Plant Sci. 2023, 14, 1096249. [Google Scholar] [CrossRef] [PubMed]
- Fahim, M.; Mechanicos, A.; Ayala-Navarrete, L.; Haber, S.; Larkin, P. Resistance to Wheat Streak Mosaic Virus—A Survey of Resources and Development of Molecular Markers. Plant Pathol. 2012, 61, 425–440. [Google Scholar] [CrossRef]
- Friebe, B.; Qi, L.; Wilson, D.; Chang, Z.; Seifers, D.; Martin, T.; Fritz, A.; Gill, B. Wheat–Thinopyrum Intermedium Recombinants Resistant to Wheat Streak Mosaic Virus and Triticum Mosaic Virus. Crop Sci. 2009, 49, 1221–1226. [Google Scholar] [CrossRef]
- Haley, S.; Martin, T.; Quick, J.; Seifers, D.; Stromberger, J.; Clayshulte, S.; Clifford, B.; Peairs, F.; Rudolph, J.; Johnson, J.; et al. Registration of CO960293-2 Wheat Germplasm Resistant to Wheat Streak Mosaic Virus and Russian Wheat Aphid. (Registrations of Germplasm). Crop Sci. 2002, 42, 1381–1383. [Google Scholar] [CrossRef]
- Lu, H.; Price, J.; Devkota, R.; Rush, C.; Rudd, J. A Dominant Gene for Resistance to Wheat Streak Mosaic Virus in Winter Wheat Line CO960293-2. Crop Sci. 2011, 51, 5–12. [Google Scholar] [CrossRef]
- Graybosch, R.A.; Peterson, C.; Baenziger, P.S.; Baltensperger, D.D.; Nelson, L.A.; Jin, Y.; Kolmer, J.; Seabourn, B.; French, R.; Hein, G.; et al. Registration of ‘Mace’Hard Red Winter Wheat. J. Plant Regist. 2009, 3, 51–56. [Google Scholar] [CrossRef]
- Zhang, G.; Martin, T.J.; Fritz, A.K.; Miller, R.; Chen, M.-S.; Bowden, R.L.; Bai, G. Registration of ‘Joe’Hard White Winter Wheat. J. Plant Regist. 2016, 10, 283–286. [Google Scholar] [CrossRef]
- Haley, S.D.; Johnson, J.J.; Peairs, F.B.; Stromberger, J.A.; Heaton, E.E.; Seifert, S.A.; Kottke, R.A.; Rudolph, J.B.; Martin, T.J.; Bai, G.; et al. Registration of ‘Snowmass’ Wheat. J. Plant Regist. 2011, 5, 87–90. [Google Scholar] [CrossRef]
- Seifers, D.; Martin, T.; Harvey, T.; Haber, S.; Haley, S. Temperature Sensitivity and Efficacy of Wheat Streak Mosaic Virus Resistance Derived from CO960293 Wheat. Plant Dis. 2006, 90, 623–628. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seifers, D.; Martin, T.; Harvey, T.; Haber, S. Temperature-Sensitive Wheat Streak Mosaic Virus Resistance Identified in KS03HW12 Wheat. Plant Dis. 2007, 91, 1029–1033. [Google Scholar] [CrossRef]
- Danilova, T.V.; Zhang, G.; Liu, W.; Friebe, B.; Gill, B.S. Homoeologous Recombination-Based Transfer and Molecular Cytogenetic Mapping of a Wheat Streak Mosaic Virus and Triticum Mosaic Virus Resistance Gene Wsm3 from Thinopyrum Intermedium to Wheat. Theor. Appl. Genet. 2017, 130, 549–556. [Google Scholar] [CrossRef]
- Liu, W.; Seifers, D.; Qi, L.; Friebe, B.; Gill, B. A Compensating Wheat–Thinopyrum Intermedium Robertsonian Translocation Conferring Resistance to Wheat Streak Mosaic Virus and Triticum Mosaic Virus. Crop Sci. 2011, 51, 2382–2390. [Google Scholar] [CrossRef]
- Phannareth, T.; Nunziata, S.O.; Stulberg, M.J.; Galvez, M.E.; Rivera, Y. Comparison of Nanopore Sequencing Protocols and Real-Time Analysis for Phytopathogen Diagnostics. Plant Health Prog. 2020, 22, 31–36. [Google Scholar] [CrossRef]
- Bronzato Badial, A.; Sherman, D.; Stone, A.; Gopakumar, A.; Wilson, V.; Schneider, W.; King, J. Nanopore Sequencing as a Surveillance Tool for Plant Pathogens in Plant and Insect Tissues. Plant Dis. 2018, 102, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- Fellers, J.P.; Webb, C.; Fellers, M.C.; Shoup Rupp, J.; De Wolf, E. Wheat Virus Identification within Infected Tissue Using Nanopore Sequencing Technology. Plant Dis. 2019, 103, 2199–2203. [Google Scholar] [CrossRef] [PubMed]
- Burrows, M.; Franc, G.; Rush, C.; Blunt, T.; Ito, D.; Kinzer, K.; Olson, J.; O’Mara, J.; Price, J.; Tande, C.; et al. Occurrence of Viruses in Wheat in the Great Plains Region, 2008. Plant Health Prog. 2009, 10, 14. [Google Scholar] [CrossRef]
- Redila, C.D.; Phipps, S.; Nouri, S. Full Genome Evolutionary Studies of Wheat Streak Mosaic-Associated Viruses Using High-Throughput Sequencing. Front. Microbiol. 2021, 12, 699078. [Google Scholar] [CrossRef] [PubMed]
- Wommack, K.E.; Bhavsar, J.; Ravel, J. Metagenomics: Read Length Matters. Appl. Environ. Microbiol. 2008, 74, 1453–1463. [Google Scholar] [CrossRef]
- Roux, S.; Emerson, J.B.; Eloe-Fadrosh, E.A.; Sullivan, M.B. Benchmarking Viromics: An in Silico Evaluation of Metagenome-Enabled Estimates of Viral Community Composition and Diversity. PeerJ 2017, 5, e3817. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Braidwood, L.; Müller, S.Y.; Baulcombe, D. Extensive Recombination Challenges the Utility of Sugarcane Mosaic Virus Phylogeny and Strain Typing. Sci. Rep. 2019, 9, 20067. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A Computer Program for Analyzing Recombination in, and Removing Signals of Recombination from, Nucleotide Sequence Datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Rybicki, E. RDP: Detection of Recombination amongst Aligned Sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Posada, D.; Crandall, K.; Williamson, C. A Modified Bootscan Algorithm for Automated Identification of Recombinant Sequences and Recombination Breakpoints. AIDS Res. Hum. Retroviruses 2005, 21, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible Emergence of New Geminiviruses by Frequent Recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Analyzing the Mosaic Structure of Genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.M.; Ratmann, O.; Boni, M.F. Improved Algorithmic Complexity for the 3SEQ Recombination Detection Algorithm. Mol. Biol. Evol. 2018, 35, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Evaluation of Methods for Detecting Recombination from DNA Sequences: Computer Simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-Scanning: A Monte Carlo Procedure for Assessing Signals in Recombinant Sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Postler, T.S.; Rubino, L.; Adriaenssens, E.M.; Dutilh, B.E.; Harrach, B.; Junglen, S.; Kropinski, A.M.; Krupovic, M.; Wada, J.; Crane, A.; et al. Guidance for Creating Individual and Batch Latinized Binomial Virus Species Names. J. Gen. Virol. 2022, 103, 001800. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Siddell, S.G.; Mushegian, A.R.; Walker, P.J.; Lefkowitz, E.J.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dutilh, B.E.; García, M.L.; Junglen, S.; et al. Differentiating between Viruses and Virus Species by Writing Their Names Correctly. Arch. Virol. 2022, 167, 1231–1234. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Davison, A.J.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; et al. Changes to Virus Taxonomy and to the International Code of Virus Classification and Nomenclature Ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2021, 166, 2633–2648. [Google Scholar] [CrossRef] [PubMed]
- Syller, J. Facilitative and Antagonistic Interactions between Plant Viruses in Mixed Infections. Mol. Plant Pathol. 2012, 13, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Sanfaçon, H. Grand Challenge in Plant Virology: Understanding the Impact of Plant Viruses in Model Plants, in Agricultural Crops, and in Complex Ecosystems. Front. Microbiol. 2017, 8, 860. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Alexander, J.; Qu, F. Differential Synergistic Interactions among Four Different Wheat-Infecting Viruses. Front. Microbiol. 2021, 12, 800318. [Google Scholar] [CrossRef] [PubMed]
- Redinbaugh, M.G.; Stewart, L.R. Maize Lethal Necrosis: An Emerging, Synergistic Viral Disease. Annu. Rev. Virol. 2018, 5, 301–322. [Google Scholar] [CrossRef] [PubMed]
- Pita, J.; Fondong, V.; Sangare, A.; Otim-Nape, G.; Ogwal, S.; Fauquet, C. Recombination, Pseudorecombination and Synergism of Geminiviruses Are Determinant Keys to the Epidemic of Severe Cassava Mosaic Disease in Uganda. J. Gen. Virol. 2001, 82, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Seifers, D.L.; Martin, T.; Harvey, T.L.; Fellers, J.P.; Michaud, J. Identification of the Wheat Curl Mite as the Vector of Triticum Mosaic Virus. Plant Dis. 2009, 93, 25–29. [Google Scholar] [CrossRef]
- Tatineni, S.; Graybosch, R.A.; Hein, G.L.; Wegulo, S.N.; French, R. Wheat Cultivar-Specific Disease Synergism and Alteration of Virus Accumulation during Co-Infection with Wheat Streak Mosaic Virus and Triticum Mosaic Virus. Phytopathology 2010, 100, 230–238. [Google Scholar] [CrossRef]
- Seifers, D.L.; Harvey, T.L.; Louie, R.; Gordon, D.; Martin, T. Differential Transmission of Isolates of the High Plains Virus by Different Sources of Wheat Curl Mites. Plant Dis. 2002, 86, 138–142. [Google Scholar] [CrossRef]
- Wegulo, S.; Byamukama, E.; Tatineni, S.; Hein, G.; Graybosch, R.; Baenziger, P. Effects of Single and Double Infections of Winter Wheat by Triticum Mosaic Virus and Wheat Streak Mosaic Virus on Grain Yield and Yield Components. In Proceedings of the PHYTOPATHOLOGY; AMER PHYTOPATHOLOGICAL SOC 3340 PILOT KNOB ROAD, St. Paul, MN, USA, 14 June 2012; Volume 102, p. 134. [Google Scholar]
- Byamukama, E.; Tatineni, S.; Hein, G.L.; Graybosch, R.A.; Baenziger, P.S.; French, R.; Wegulo, S.N. Effects of Single and Double Infections of Winter Wheat by Triticum Mosaic Virus and Wheat Streak Mosaic Virus on Yield Determinants. Plant Dis. 2012, 96, 859–864. [Google Scholar] [CrossRef]
- Filloux, D.; Fernandez, E.; Loire, E.; Claude, L.; Galzi, S.; Candresse, T.; Winter, S.; Jeeva, M.; Makeshkumar, T.; Martin, D.P.; et al. Nanopore-Based Detection and Characterization of Yam Viruses. Sci. Rep. 2018, 8, 17879. [Google Scholar] [CrossRef] [PubMed]
- Sahlin, K.; Medvedev, P. Error Correction Enables Use of Oxford Nanopore Technology for Reference-Free Transcriptome Analysis. Nat. Commun. 2021, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Kumssa, T.; Rupp, J.; Fellers, M.; Fellers, J.; Zhang, G. An Isolate of Wheat Streak Mosaic Virus from Foxtail Overcomes Wsm2 Resistance in Wheat. Plant Pathol. 2019, 68, 783–789. [Google Scholar] [CrossRef]
- Rabenstein, F.; Seifers, D.L.; Schubert, J.; French, R.; Stenger, D.C. Phylogenetic Relationships, Strain Diversity and Biogeography of Tritimoviruses. J. Gen. Virol. 2002, 83, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Stenger, D.C.; Seifers, D.L.; French, R. Patterns of Polymorphism in Wheat Streak Mosaic Virus: Sequence Space Explored by a Clade of Closely Related Viral Genotypes Rivals That between the Most Divergent Strains. Virology 2002, 302, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Murray, T.D. Genetic Variation of Wheat Streak Mosaic Virus in the United States Pacific Northwest. Phytopathology 2013, 103, 98–104. [Google Scholar] [CrossRef]
- Cervera, M.T.; Riechmann, J.L.; Martín, M.T.; García, J.A. 3′-Terminal Sequence of the Plum Pox Virus PS and Ŏ6 Isolates: Evidence for RNA Recombination within the Potyvirus Group. J. Gen. Virol. 1993, 74, 329–334. [Google Scholar] [CrossRef]
- Glais, L.; Tribodet, M.; Kerlan, C. Genomic Variability in Potato Potyvirus Y (PVY): Evidence That PVYNW and PVYNTN Variants Are Single to Multiple Recombinants between PVYO and PVYN Isolates. Arch. Virol. 2002, 147, 363–378. [Google Scholar] [CrossRef]
- Revers, F.; Le Gall, O.; Candresse, T.; Le Romancer, M.; Dunez, J. Frequent Occurrence of Recombinant Potyvirus Isolates. J. Gen. Virol. 1996, 77, 1953–1965. [Google Scholar] [CrossRef] [PubMed]
- Desbiez, C.; Lecoq, H. Evidence for Multiple Intraspecific Recombinants in Natural Populations of Watermelon Mosaic Virus (WMV, Potyvirus). Arch. Virol. 2008, 153, 1749–1754. [Google Scholar] [CrossRef]
- Stewart, L.R. Sequence Diversity of Wheat Mosaic Virus Isolates. Virus Res. 2016, 213, 299–303. [Google Scholar] [CrossRef]
- Tatineni, S.; McMechan, A.J.; Wosula, E.N.; Wegulo, S.N.; Graybosch, R.A.; French, R.; Hein, G.L. An Eriophyid Mite-Transmitted Plant Virus Contains Eight Genomic RNA Segments with Unusual Heterogeneity in the Nucleocapsid Protein. J. Virol. 2014, 88, 11834–11845. [Google Scholar] [CrossRef]
- Cadle-Davidson, L.; Gray, S. Soilborne Wheat Mosaic Virus. Plant Health Instr. 2006, 90, 1039–1044. [Google Scholar]
| Common Name | ||||||||
|---|---|---|---|---|---|---|---|---|
| Category | Wheat streak mosaic virus | Triticum mosaic virus | Brome mosaic virus | High Plain wheat mosaic emaravirus | Barley yellow dwarf virus | Cereal yellow dwarf virus | Wheat spindle streak mosaic virus | Soilborne wheat mosaic virus |
| Realm | Riboviria | Riboviria | Riboviria | Riboviria | Riboviria | Riboviria | Riboviria | Riboviria |
| Kingdom | Orthornavirae | Orthornavirae | Orthornavirae | Orthornavirae | Orthornavirae | Orthornavirae | Orthornavirae | Orthornavirae |
| Phylum | Pisuviricota | Pisuviricota | Kitrinoviricota | Negarnaviricota | Kitrinoviricota | Kitrinoviricota | Pisuviricota | Kitrinoviricota |
| Class | Stelpaviricetes | Stelpaviricetes | Alsuviricetes | Bunyaviricetes | Tolucaviricetes | Tolucaviricetes | Stelpaviricetes | Alsuviricetes |
| Order | Patatavirales | Patatavirales | Martellivirales | Elliovirales | Tolivirales | Tolivirales | Patatavirales | Martellivirales |
| Family | Potyviridae | Potyviridae | Bromoviridae | Fimoviridae | Tombusviridae | Tombusviridae | Potyviridae | Virgaviridae |
| Genus | Tritimovirus | Poacevirus | Bromovirus | Emaravirus | Luteovirus | Luteovirus | Bymovirus | Furovirus |
| Species | Tritimovirus tritici | Poacevirus tritici | Bromovirus BMV | Emaravirus tritici | Luteovirus pavhordei | Luteovirus sgvhordei | Bymovirus tritici | Furovirus tritici |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranabhat, N.B.; Fellers, J.P.; Bruce, M.A.; Rupp, J.L.S. High-Throughput Oxford Nanopore Sequencing Unveils Complex Viral Population in Kansas Wheat: Implications for Sustainable Virus Management. Viruses 2025, 17, 126. https://doi.org/10.3390/v17010126
Ranabhat NB, Fellers JP, Bruce MA, Rupp JLS. High-Throughput Oxford Nanopore Sequencing Unveils Complex Viral Population in Kansas Wheat: Implications for Sustainable Virus Management. Viruses. 2025; 17(1):126. https://doi.org/10.3390/v17010126
Chicago/Turabian StyleRanabhat, Nar B., John P. Fellers, Myron A. Bruce, and Jessica L. Shoup Rupp. 2025. "High-Throughput Oxford Nanopore Sequencing Unveils Complex Viral Population in Kansas Wheat: Implications for Sustainable Virus Management" Viruses 17, no. 1: 126. https://doi.org/10.3390/v17010126
APA StyleRanabhat, N. B., Fellers, J. P., Bruce, M. A., & Rupp, J. L. S. (2025). High-Throughput Oxford Nanopore Sequencing Unveils Complex Viral Population in Kansas Wheat: Implications for Sustainable Virus Management. Viruses, 17(1), 126. https://doi.org/10.3390/v17010126






