Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of EV68-3C Protease
2.2. Protein Crystallization
2.3. Data Collection and Structure Determination
2.4. Inhibitor Modeling and Molecular Dynamics Simulations
2.5. Structural Analysis: Hydrogen Bonds and Van Der Waals Interactions
2.6. Enzyme Activity Assay to Determine KM
2.7. Enzyme Inhibition Assay to Determine IC50
3. Results
3.1. Characterization of 3C and 3C-Like Inhibitors for Activity Against EV68-3C Protease
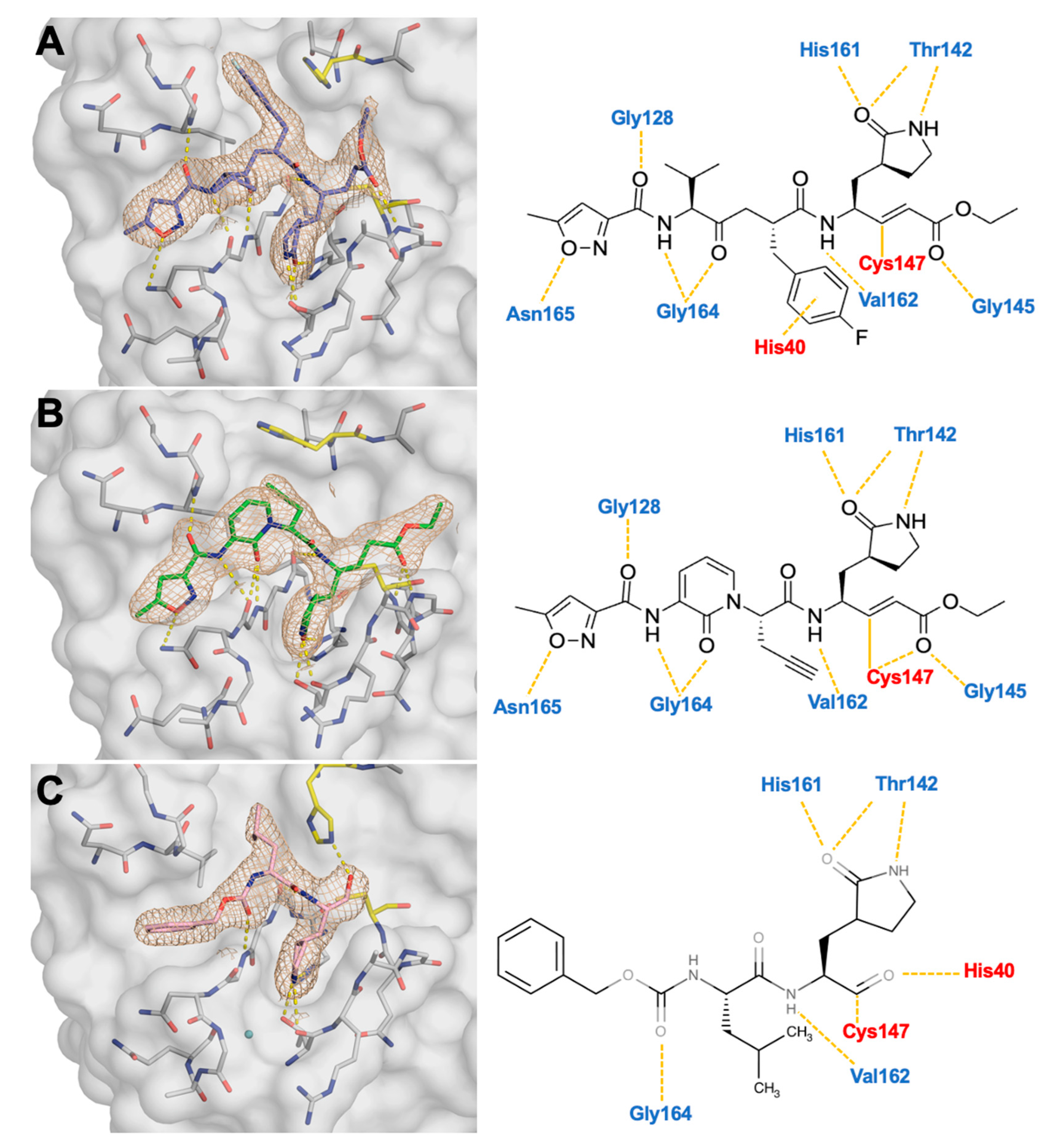
3.2. Co-Crystal Structures of EV68-3C Protease with the Inhibitor Bound Within the Active Site
3.3. Crystal Structures Reveal the Conformational Flexibility of His40 to Accommodate Inhibitors Within the Active Site
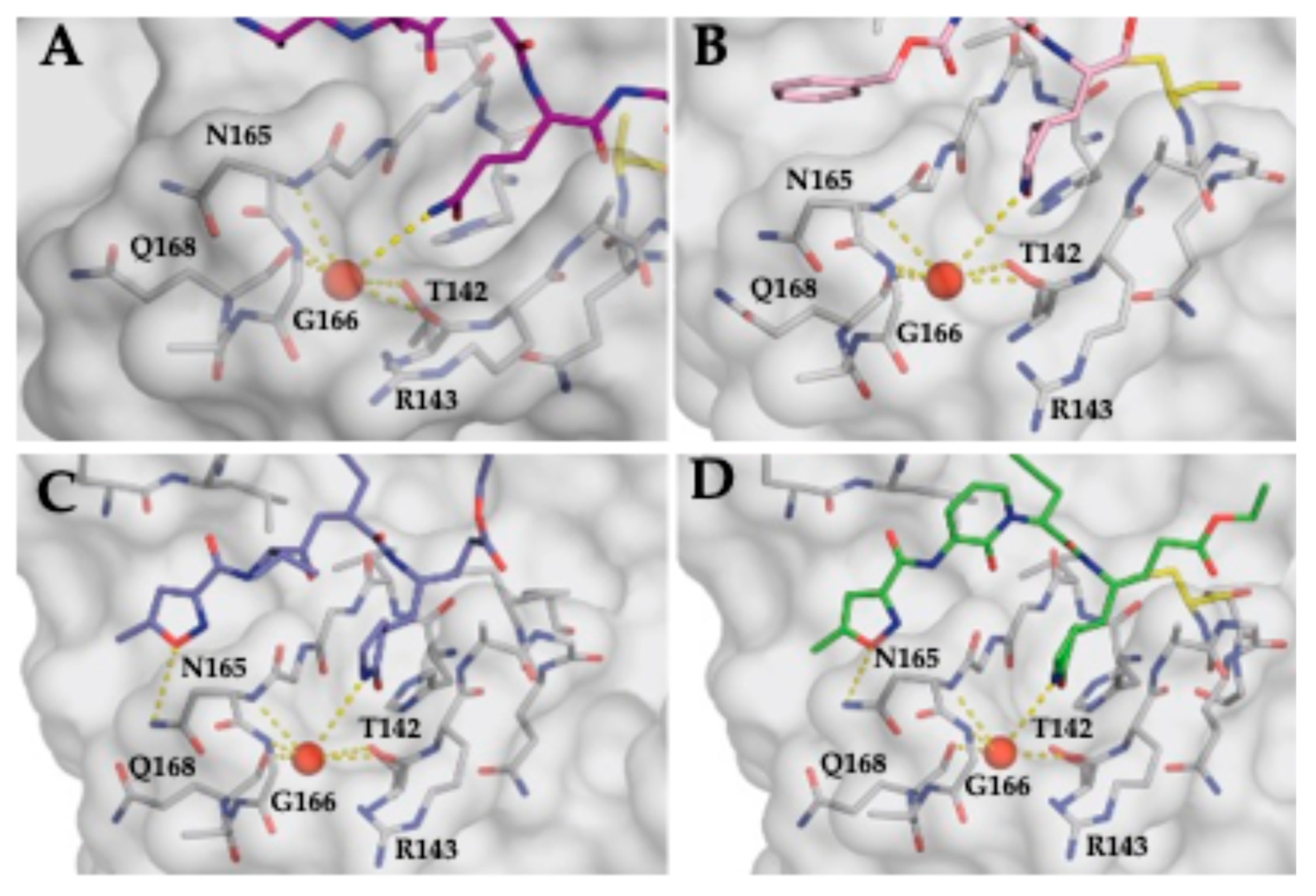
3.4. Stabilization of the S1 Pocket and a Key Structural Water Molecule in Ligand-Bound EV68-3C Structures
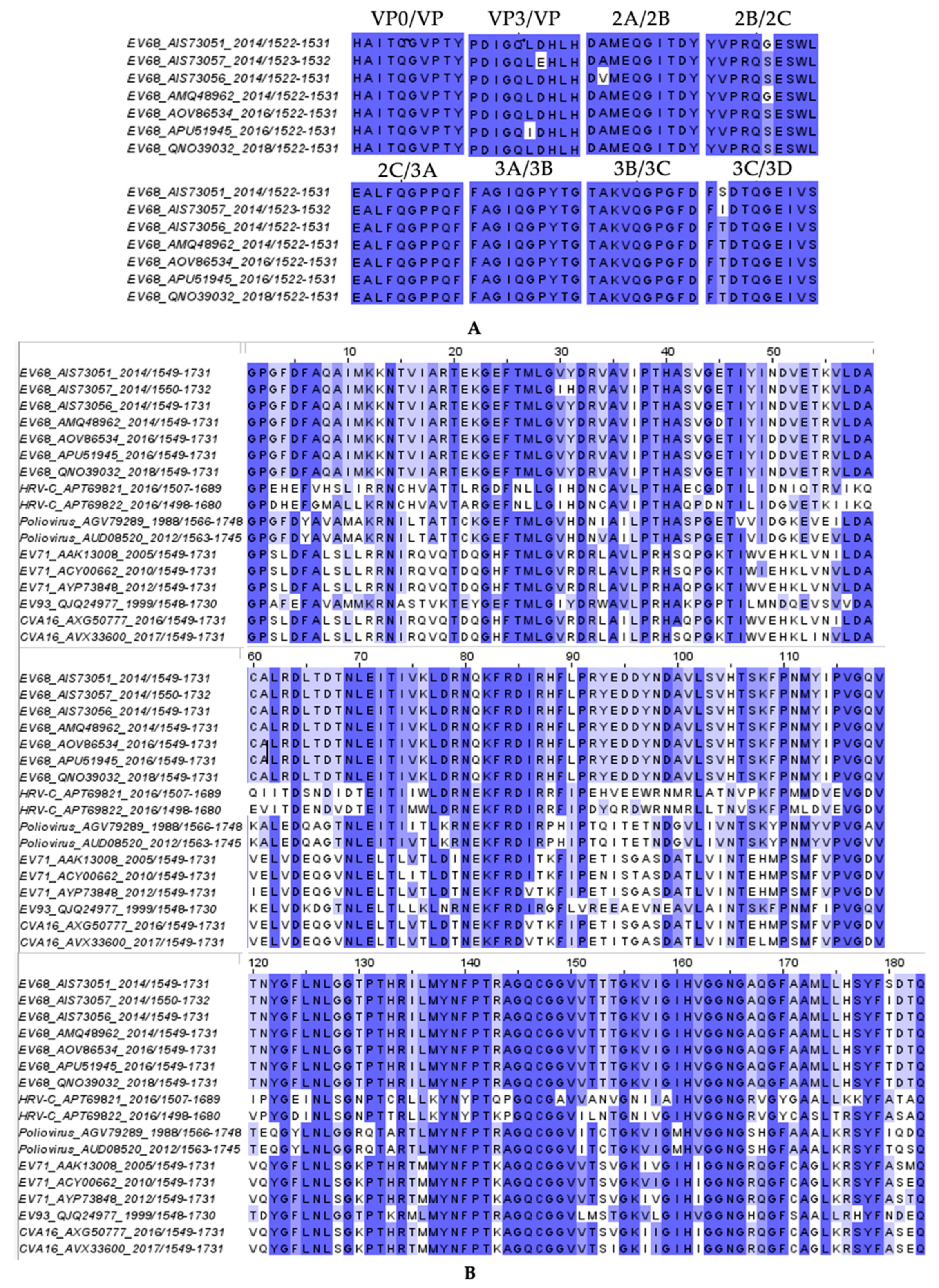
3.5. Substrate Envelope Analysis of Inhibitor Co-Crystal Structures Predicts Potential Sites of Resistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haaheim, L.R.; Pattison, J.R.; Whitley, R.J. Enteroviruses: Polioviruses, Coxsackieviruses, Echoviruses, and Newer Enteroviruses, 6th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Pons-Salort, M.; Parker, E.P.; Grassly, N.C. The epidemiology of non-polio enteroviruses: Recent advances and outstanding questions. Curr. Opin. Infect. Dis. 2015, 28, 479–487. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Strating, J.; van Kuppeveld, F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018, 16, 368–381. [Google Scholar] [CrossRef]
- Wei, W.; Guo, H.; Chang, J.; Yu, Y.; Liu, G.; Zhang, N.; Willard, S.H.; Zheng, S.; Yu, X.F. ICAM-5/Telencephalin Is a Functional Entry Receptor for Enterovirus D68. Cell Host Microbe 2016, 20, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Bove, R.; Rowles, W.; Carleton, M.; Olivera, E.; Sheehan, M.; Werdal, H.P.; Scott, R.; Axton, L.; Benson, L. Unmet Needs in the Evaluation, Treatment, and Recovery for 167 Children Affected by Acute Flaccid Myelitis Reported by Parents Through Social Media. Pediatr. Neurol. 2020, 102, 20–27. [Google Scholar] [CrossRef]
- Carballo, C.M.; Erro, M.G.; Sordelli, N.; Vazquez, G.; Mistchenko, A.S.; Cejas, C.; Rodriguez, M.; Cisterna, D.M.; Freire, M.C.; Contrini, M.M.; et al. Acute Flaccid Myelitis Associated with Enterovirus D68 in Children, Argentina, 2016. Emerg. Infect. Dis. 2019, 25, 573–576. [Google Scholar] [CrossRef]
- Dyda, A.; Stelzer-Braid, S.; Adam, D.; Chughtai, A.A.; MacIntyre, C.R. The association between acute flaccid myelitis (AFM) and Enterovirus D68 (EV-D68)—What is the evidence for causation? Euro Surveill. 2018, 23, 17-00310. [Google Scholar] [CrossRef]
- Murphy, O.C.; Messacar, K.; Benson, L.; Bove, R.; Carpenter, J.L.; Crawford, T.; Dean, J.; DeBiasi, R.; Desai, J.; Elrick, M.J.; et al. Acute flaccid myelitis: Cause, diagnosis, and management. Lancet 2021, 397, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Messacar, K.; Yang, M.L.; Maloney, J.A.; Lindwall, J.; Carry, T.; Kenyon, P.; Sillau, S.H.; Oleszek, J.; Tyler, K.L.; et al. Outcomes of Colorado children with acute flaccid myelitis at 1 year. Neurology 2017, 89, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Melicosta, M.E.; Dean, J.; Hagen, K.; Oppenheimer, K.; Porter, C.; Rybczynski, S.; Salorio, C.; Sadowsky, C. Acute flaccid myelitis: Rehabilitation challenges and outcomes in a pediatric cohort. J. Pediatr. Rehabil. Med. 2019, 12, 245–253. [Google Scholar] [CrossRef]
- Messacar, K.; Abzug, M.J.; Dominguez, S.R. 2014 outbreak of enterovirus D68 in North America. J. Med. Virol. 2016, 88, 739–745. [Google Scholar] [CrossRef]
- Maloney, J.A.; Mirsky, D.M.; Messacar, K.; Dominguez, S.R.; Schreiner, T.; Stence, N.V. MRI findings in children with acute flaccid paralysis and cranial nerve dysfunction occurring during the 2014 enterovirus D68 outbreak. AJNR Am. J. Neuroradiol. 2015, 36, 245–250. [Google Scholar] [CrossRef]
- Messacar, K.; Schreiner, T.L.; Van Haren, K.; Yang, M.; Glaser, C.A.; Tyler, K.L.; Dominguez, S.R. Acute flaccid myelitis: A clinical review of US cases 2012–2015. Ann. Neurol. 2016, 80, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Aliabadi, N.; Messacar, K.; Pastula, D.M.; Robinson, C.C.; Leshem, E.; Sejvar, J.J.; Nix, W.A.; Oberste, M.S.; Feikin, D.R.; Dominguez, S.R. Enterovirus D68 Infection in Children with Acute Flaccid Myelitis, Colorado, USA, 2014. Emerg. Infect. Dis. 2016, 22, 1387–1394. [Google Scholar] [CrossRef]
- Tan, J.; George, S.; Kusov, Y.; Perbandt, M.; Anemuller, S.; Mesters, J.R.; Norder, H.; Coutard, B.; Lacroix, C.; Leyssen, P.; et al. 3C protease of enterovirus 68: Structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 2013, 87, 4339–4351. [Google Scholar] [CrossRef]
- De Palma, A.M.; Vliegen, I.; De Clercq, E.; Neyts, J. Selective inhibitors of picornavirus replication. Med. Res. Rev. 2008, 28, 823–884. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Qi, J.; Chen, Z.; Xu, X.; Gao, F.; Lin, D.; Qian, W.; Liu, H.; Jiang, H.; Yan, J.; et al. Enterovirus 71 and coxsackievirus A16 3C proteases: Binding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J. Virol. 2011, 85, 10319–10331. [Google Scholar] [CrossRef] [PubMed]
- Binford, S.L.; Maldonado, F.; Brothers, M.A.; Weady, P.T.; Zalman, L.S.; Meador, J.W., 3rd; Matthews, D.A.; Patick, A.K. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 2005, 49, 619–626. [Google Scholar] [CrossRef]
- Matthews, D.A.; Dragovich, P.S.; Webber, S.E.; Fuhrman, S.A.; Patick, A.K.; Zalman, L.S.; Hendrickson, T.F.; Love, R.A.; Prins, T.J.; Marakovits, J.T.; et al. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. USA 1999, 96, 11000–11007. [Google Scholar] [CrossRef] [PubMed]
- Patick, A.K.; Brothers, M.A.; Maldonado, F.; Binford, S.; Maldonado, O.; Fuhrman, S.; Petersen, A.; Smith, G.J., 3rd; Zalman, L.S.; Burns-Naas, L.A.; et al. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 2005, 49, 2267–2275. [Google Scholar] [CrossRef]
- Lacroix, C.; George, S.; Leyssen, P.; Hilgenfeld, R.; Neyts, J. The enterovirus 3C protease inhibitor SG85 efficiently blocks rhinovirus replication and is not cross-resistant with rupintrivir. Antimicrob. Agents Chemother. 2015, 59, 5814–5818. [Google Scholar] [CrossRef]
- Sun, L.; Meijer, A.; Froeyen, M.; Zhang, L.; Thibaut, H.J.; Baggen, J.; George, S.; Vernachio, J.; van Kuppeveld, F.J.; Leyssen, P.; et al. Antiviral Activity of Broad-Spectrum and Enterovirus-Specific Inhibitors against Clinical Isolates of Enterovirus D68. Antimicrob. Agents Chemother. 2015, 59, 7782–7785. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lovell, S.; Tiew, K.C.; Mandadapu, S.R.; Alliston, K.R.; Battaile, K.P.; Groutas, W.C.; Chang, K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012, 86, 11754–11762. [Google Scholar] [CrossRef]
- Binford, S.L.; Weady, P.T.; Maldonado, F.; Brothers, M.A.; Matthews, D.A.; Patick, A.K. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 2007, 51, 4366–4373. [Google Scholar] [CrossRef]
- Fu, L.; Ye, F.; Feng, Y.; Yu, F.; Wang, Q.; Wu, Y.; Zhao, C.; Sun, H.; Huang, B.; Niu, P.; et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020, 11, 4417. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Musharrafieh, R.; Zheng, M.; Wang, J. Enterovirus D68 Antivirals: Past, Present, and Future. ACS Infect. Dis. 2020, 6, 1572–1586. [Google Scholar] [CrossRef] [PubMed]
- Patick, A.K.; Binford, S.L.; Brothers, M.A.; Jackson, R.L.; Ford, C.E.; Diem, M.D.; Maldonado, F.; Dragovich, P.S.; Zhou, R.; Prins, T.J.; et al. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 1999, 43, 2444–2450. [Google Scholar] [CrossRef]
- Rhoden, E.; Liu, H.M.; Wang-Chern, S.W.; Oberste, M.S. Anti-poliovirus activity of protease inhibitor AG-7404, and assessment of in vitro activity in combination with antiviral capsid inhibitor compounds. Antivir. Res. 2013, 98, 186–191. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Cully, M. A tale of two antiviral targets—And the COVID-19 drugs that bind them. Nat. Rev. Drug Discov. 2022, 21, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Holland, J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Zhang, Y.; Scheuermann, R.H. Epidemiology and Sequence-Based Evolutionary Analysis of Circulating Non-Polio Enteroviruses. Microorganisms 2020, 8, 1856. [Google Scholar] [CrossRef]
- Azzolino, V.N.; Shaqra, A.M.; Ali, A.; Kurt Yilmaz, N.; Schiffer, C.A. Elucidating the Substrate Envelope of Enterovirus 68-3C Protease: Structural Basis of Specificity and Potential Resistance. Viruses 2024, 16, 1419. [Google Scholar] [CrossRef] [PubMed]
- King, N.M.; Prabu-Jeyabalan, M.; Nalivaika, E.A.; Schiffer, C.A. Combating susceptibility to drug resistance: Lessons from HIV-1 protease. Chem. Biol. 2004, 11, 1333–1338. [Google Scholar] [CrossRef][Green Version]
- Romano, K.P.; Ali, A.; Royer, W.E.; Schiffer, C.A. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. USA 2010, 107, 20986–20991. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Bandaranayake, R.M.; Cai, Y.; King, N.M.; Kolli, M.; Mittal, S.; Murzycki, J.F.; Nalam, M.N.L.; Nalivaika, E.A.; Ozen, A.; et al. Molecular Basis for Drug Resistance in HIV-1 Protease. Viruses 2010, 2, 2509–2535. [Google Scholar] [CrossRef] [PubMed]
- Nalam, M.N.; Ali, A.; Reddy, G.S.; Cao, H.; Anjum, S.G.; Altman, M.D.; Yilmaz, N.K.; Tidor, B.; Rana, T.M.; Schiffer, C.A. Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance. Chem. Biol. 2013, 20, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Matthew, A.N.; Leidner, F.; Lockbaum, G.J.; Henes, M.; Zephyr, J.; Hou, S.; Rao, D.N.; Timm, J.; Rusere, L.N.; Ragland, D.A.; et al. Drug Design Strategies to Avoid Resistance in Direct-Acting Antivirals and Beyond. Chem. Rev. 2021, 121, 3238–3270. [Google Scholar] [CrossRef]
- Ozen, A.; Prachanronarong, K.; Matthew, A.N.; Soumana, D.I.; Schiffer, C.A. Resistance outside the substrate envelope: Hepatitis C NS3/4A protease inhibitors. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Matthew, A.N.; Zephyr, J.; Nageswara Rao, D.; Henes, M.; Kamran, W.; Kosovrasti, K.; Hedger, A.K.; Lockbaum, G.J.; Timm, J.; Ali, A.; et al. Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors. mBio 2020, 11, e00172-20. [Google Scholar] [CrossRef] [PubMed]
- Shaqra, A.M.; Zvornicanin, S.N.; Huang, Q.Y.J.; Lockbaum, G.J.; Knapp, M.; Tandeske, L.; Bakan, D.T.; Flynn, J.; Bolon, D.N.A.; Moquin, S.; et al. Defining the substrate envelope of SARS-CoV-2 main protease to predict and avoid drug resistance. Nat. Commun. 2022, 13, 3556. [Google Scholar] [CrossRef] [PubMed]
- Romano, K.P.; Ali, A.; Aydin, C.; Soumana, D.; Ozen, A.; Deveau, L.M.; Silver, C.; Cao, H.; Newton, A.; Petropoulos, C.J.; et al. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog. 2012, 8, e1002832. [Google Scholar] [CrossRef]
- Prabu-Jeyabalan, M.; Nalivaika, E.; Schiffer, C.A. Substrate shape determines specificity of recognition for HIV-1 protease: Analysis of crystal structures of six substrate complexes. Structure 2002, 10, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Brunger, A.T. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 1992, 355, 472–475. [Google Scholar] [CrossRef]
- Schrödinger Release 2019-2. Desmond Molecular Dynamics System, D.E. Shaw Research; Schrödinger: New York, NY, USA, 2019.
- Lockbaum, G.J.; Henes, M.; Lee, J.M.; Timm, J.; Nalivaika, E.A.; Thompson, P.R.; Kurt Yilmaz, N.; Schiffer, C.A. Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode. Biochemistry 2021, 60, 2925–2931. [Google Scholar] [CrossRef]
- Henes, M.; Lockbaum, G.J.; Kosovrasti, K.; Leidner, F.; Nachum, G.S.; Nalivaika, E.A.; Lee, S.K.; Spielvogel, E.; Zhou, S.; Swanstrom, R.; et al. Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance. ACS Chem. Biol. 2019, 14, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Leidner, F.; Kurt Yilmaz, N.; Paulsen, J.; Muller, Y.A.; Schiffer, C.A. Hydration Structure and Dynamics of Inhibitor-Bound HIV-1 Protease. J. Chem. Theory Comput. 2018, 14, 2784–2796. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.; Sondergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2019-2. Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2019.
- Leidner, F.; Kurt Yilmaz, N.; Schiffer, C.A. Deciphering Complex Mechanisms of Resistance and Loss of Potency through Coupled Molecular Dynamics and Machine Learning. J. Chem. Theory Comput. 2021, 17, 2054–2064. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Aydin, C.; Gildemeister, R.; Romano, K.P.; Cao, H.; Ozen, A.; Soumana, D.; Newton, A.; Petropoulos, C.J.; Huang, W.; et al. Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance. ACS Chem. Biol. 2013, 8, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Lockbaum, G.J.; Henes, M.; Talledge, N.; Rusere, L.N.; Kosovrasti, K.; Nalivaika, E.A.; Somasundaran, M.; Ali, A.; Mansky, L.M.; Kurt Yilmaz, N.; et al. Inhibiting HTLV-1 Protease: A Viable Antiviral Target. ACS Chem. Biol. 2021, 16, 529–538. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Dragovich, P.S.; Prins, T.J.; Zhou, R.; Fuhrman, S.A.; Patick, A.K.; Matthews, D.A.; Ford, C.E.; Meador, J.W., 3rd; Ferre, R.A.; Worland, S.T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 3. Structure-activity studies of ketomethylene-containing peptidomimetics. J. Med. Chem. 1999, 42, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Dragovich, P.S.; Prins, T.J.; Zhou, R.; Johnson, T.O.; Hua, Y.; Luu, H.T.; Sakata, S.K.; Brown, E.L.; Maldonado, F.C.; Tuntland, T.; et al. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 8. Pharmacological optimization of orally bioavailable 2-pyridone-containing peptidomimetics. J. Med. Chem. 2003, 46, 4572–4585. [Google Scholar] [CrossRef] [PubMed]
- Dragovich, P.S.; Prins, T.J.; Zhou, R.; Webber, S.E.; Marakovits, J.T.; Fuhrman, S.A.; Patick, A.K.; Matthews, D.A.; Lee, C.A.; Ford, C.E.; et al. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J. Med. Chem. 1999, 42, 1213–1224. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Webber, S.E.; Babine, R.E.; Fuhrman, S.A.; Patick, A.K.; Matthews, D.A.; Lee, C.A.; Reich, S.H.; Prins, T.J.; Marakovits, J.T.; et al. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure-activity studies. J. Med. Chem. 1998, 41, 2806–2818. [Google Scholar] [CrossRef] [PubMed]
- Dragovich, P.S.; Webber, S.E.; Babine, R.E.; Fuhrman, S.A.; Patick, A.K.; Matthews, D.A.; Reich, S.H.; Marakovits, J.T.; Prins, T.J.; Zhou, R.; et al. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 2. Peptide structure-activity studies. J. Med. Chem. 1998, 41, 2819–2834. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Zhou, R.; Skalitzky, D.J.; Fuhrman, S.A.; Patick, A.K.; Ford, C.E.; Meador, J.W., 3rd; Worland, S.T. Solid-phase synthesis of irreversible human rhinovirus 3C protease inhibitors. Part 1: Optimization of tripeptides incorporating N-terminal amides. Bioorg. Med. Chem. 1999, 7, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Cai, Q.; Chen, C.; Li, N.; Peng, X.; Cai, Y.; Yin, K.; Chen, X.; Wang, X.; Zhang, R.; et al. Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Costenaro, L.; Kaczmarska, Z.; Arnan, C.; Janowski, R.; Coutard, B.; Sola, M.; Gorbalenya, A.E.; Norder, H.; Canard, B.; Coll, M. Structural basis for antiviral inhibition of the main protease, 3C, from human enterovirus 93. J. Virol. 2011, 85, 10764–10773. [Google Scholar] [CrossRef]
- Mosimann, S.C.; Cherney, M.M.; Sia, S.; Plotch, S.; James, M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997, 273, 1032–1047. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Fan, K.; Chen, Z.; Sun, Y.; Hou, H.; Zhu, L. Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design. Virol. Sin. 2020, 35, 445–454. [Google Scholar] [CrossRef]

| Inhibitor PDB ID | AG-7088 8W3C | AG-7404 8W3M | GC-376 8W3T |
|---|---|---|---|
| Data Collection | |||
| Location | NLS-II, Synchrotron | NLS-II, Synchrotron | NLS-II, Synchrotron |
| Resolution Range (Å) | 32.12–1.97 (2.04–1.97) | 32.16–1.97 (2.06–1.97) | 32.07–1.98 (2.05–1.98) |
| Space Group | P 31 2 1 | P 31 2 1 | P 31 2 1 |
| a, b, c, (Å) | 56.431, 56.431, 170.482 | 56.491, 56.491, 170.72 | 56.174, 56.174, 170.629 |
| Alpha, Beta, Gamma (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Total Reflections | 46,210 (4560) | 46,334 (3768) | 45,125 (4362) |
| Unique Reflections | 22,964 (2671) | 23,046 (2663) | 22,473 (2668) |
| Multiplicity | 2 (2.0) | 2 (2.0) | 2 (2.0) |
| Completeness (%) | 99.29 (99.04) | 99.31 (98.80) | 99.46 (99.36) |
| (Average I)/sigma | 16 (4.3) | 15 (4) | 14 (4) |
| Wilson B-Factor | 36.97 | 37.33 | 39.51 |
| Rmerge | 0.018 (0.094) | 0.023 (0.141) | 0.029 (0.405) |
| CC1/2 | 1.000 (0.993) | 0.999 (0.991) | 0.999 (0.917) |
| Refinement * | |||
| Rfactor | 0.2131 (0.3553) | 0.2056 (0.3918) | 0.1993 (0.3378) |
| Rfree | 0.2363 (0.5142) | 0.2385 (0.4589) | 0.2348 (0.4214) |
| RMSD in: | |||
| Bond Lengths (Å) | 0.007 | 0.008 | 0.010 |
| Bond Angles (°) | 1.03 | 0.981 | 1.16 |
| Ramachandran: | |||
| Favored (%) | 97.81 | 95.63 | 97.85 |
| Allowed (%) | 2.19 | 4.37 | 2.15 |
| Outliers (%) | 0 | 0 | 0 |
| Rotamer outliers (%) | 0 | 0 | 0 |
| B-Factors: | |||
| Average | 46.94 | 46.53 | 47.76 |
| Macromolecules | 46.19 | 45.61 | 47.68 |
| Solvent | 53.64 | 53.46 | 55.86 |
| Inhibitor | IC50 (μM) |
|---|---|
| rupintrivir (AG7088) | 0.0078 ± 0.0006 |
| AG7404 | 0.0235 ± 0.0011 |
| GC-376 | 0.194 ± 0.019 |
| PF-00835231 | 1.8 ± 0.2 |
| nirmatrelvir (PF-07321332) | >10 μM |
| ensitrelvir | >10 μM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzolino, V.N.; Shaqra, A.M.; Ali, A.; Kurt Yilmaz, N.; Schiffer, C.A. Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease. Viruses 2025, 17, 75. https://doi.org/10.3390/v17010075
Azzolino VN, Shaqra AM, Ali A, Kurt Yilmaz N, Schiffer CA. Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease. Viruses. 2025; 17(1):75. https://doi.org/10.3390/v17010075
Chicago/Turabian StyleAzzolino, Vincent N., Ala M. Shaqra, Akbar Ali, Nese Kurt Yilmaz, and Celia A. Schiffer. 2025. "Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease" Viruses 17, no. 1: 75. https://doi.org/10.3390/v17010075
APA StyleAzzolino, V. N., Shaqra, A. M., Ali, A., Kurt Yilmaz, N., & Schiffer, C. A. (2025). Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease. Viruses, 17(1), 75. https://doi.org/10.3390/v17010075






