Molecular Detection, Seroprevalence and Biochemical Analysis of Lumpy Skin Disease Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Genomic DNA Extraction
2.3. Molecular Analysis
2.4. Nucleotide Sequencing
2.5. Serological Analysis
2.6. Biochemical Analysis
2.7. Statistical Analysis
3. Results
3.1. Molecular Analysis
3.2. Serological Analysis
3.3. Biochemical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Babiuk, S.; Bowden, T.R.; Boyle, D.B.; Wallace, D.B.; Kitching, R.P. Capripoxviruses: An emerging worldwide threat to sheep, goats and cattle. Transbound. Emerg. Dis. 2008, 55, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Mac Owan, K.D.S. Observations on the epizootiology of lumpy skin disease during the first year of its occurrence in Kenya. Bull. Epizoot. Dis. Afr. 1959, 7, 7–20. [Google Scholar]
- Tuppurainen, E.S.M.; Oura, C.A.L. Lumpy skin disease: An Emerging Threat to Europe, the Middle East and Asia. Transbound. Emerg. Dis. 2012, 59, 40–48. [Google Scholar] [CrossRef]
- Sudhakar, S.B.; Mishra, N.; Kalaiyarasu, S.; Jhade, S.K.; Hemadri, D.; Sood, R.; Singh, V.P. Lumpy skin disease (LSD) outbreaks in cattle in Odisha state, India in August 2019: Epidemiological features and molecular studies. Transbound. Emerg. Dis. 2020, 67, 2408–2422. [Google Scholar] [CrossRef]
- King, A.M.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus taxonomy. Classification and nomenclature of viruses. In Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2012; pp. 289–307. Available online: https://www.sciencedirect.com/book/9780123846846/virus-taxonomy (accessed on 27 January 2025).
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. Genome of lumpy skin disease virus. J. Virol. 2001, 75, 7122–7130. [Google Scholar] [CrossRef]
- Bianchini, J.; Simons, X.; Humblet, M.F.; Saegerman, C. Lumpy skin disease: A systematic review of mode of transmission, risk of emergence and risk entry pathway. Viruses 2023, 15, 1622. [Google Scholar] [CrossRef]
- Abutarbush, S.M.; Ababneh, M.M.; Zoubil, I.G.; Sheyab, O.M.; Zoubi, M.G.; Alekish, M.O.; Gharbat, R.J. Lumpy skin disease in Jordan: Disease emergence, clinical signs, complications and preliminary-associated economic losses. Transbound. Emerg. Dis. 2013, 62, 549–554. [Google Scholar] [CrossRef]
- Tuppurainen, E.; Alexandrov, T.; Beltrán-Alcrudo, D. Lumpy Skin Disease Field Manual—A Manual for Veterinarians; FAO Animal Production and Health Manual No. 20; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2017; pp. 1–5. [Google Scholar]
- Office International des Epizooties (OIE). Lumpy Skin Disease; Chapter 2.4.14 OIE Terrestrial Manual; Office International des Epizooties: Paris, France, 2010. [Google Scholar]
- Ireland, D.C.; Binepal, Y.S. Improved detection of capripoxvirus in biopsy samples by PCR. J. Virol. Methods 1998, 74, 1–7. [Google Scholar] [CrossRef]
- Ntombela, N.; Matsiela, M.; Zuma, S.; Hiralal, S.; Naicker, L.; Mokoena, N.; Khoza, T. Production of recombinant lumpy skin disease virus A27L and L1R proteins for qapplication in diagnostics and vaccine development. Vaccine 2023, 15, 100384. [Google Scholar]
- Sanganagouda, K.; Nagraja, K.; Sajjanar, B.; Kounin, S.; Gomes, A.R.; Pavithra, B.H.; Hegade, R. Molecular characterization, phylogenetic analysis and viral load quantification of lumpy skin disease virus in Cattle. Preprints 2023. [Google Scholar] [CrossRef]
- Varadarajan, M.T.; Karuppannan, A.K. Molecular diagnosis, phylogenetic analysis and identification of an unique SNP in the viral attachment protein (p32) gene of lumpy skin disease virus in tissue biopsies of Cattle in India. Preprints 2022. [Google Scholar] [CrossRef]
- Kumar, N.; Chander, Y.; Kumar, R.; Khandelwal, N.; Riyesh, T.; Chaudhary, K.; Tripathi, B.N. Isolation and characterization of lumpy skin disease virus from cattle in India. PLoS ONE 2021, 16, e0241022. [Google Scholar] [CrossRef]
- Halmandge, S.; Kasaralikar, V.; Ravindra, B.G.; Mallinath, K.C.; Rajendrakumar, T. Molecular Diagnosis of lumpy skin disease Outbreak in Cattle of Bidar, Karnataka. Indian J. Vet. Sci. Biotechnol. 2024, 20, 102–105. [Google Scholar]
- Geletu, U.S.; Musa, A.A.; Usmael, M.A.; Keno, M.S. Molecular Detection and Isolation of lumpy skin disease virus during an Outbreak in West Hararghe Zone, Eastern Ethiopia. Vet. Med. Int. 2024, 2024, 9487970. [Google Scholar] [CrossRef] [PubMed]
- Makoga, F.T.; Chang’a, J.S.; Meki, I.K.; Mayenga, C.; Settypalli, T.B.; Bitanyi, S.; Lamien, C.E. Detection and molecular characterization of lumpy skin disease and bovine papular stomatitis viruses in lumpy skin disease-suspected outbreaks in Tanzania. Virol. J. 2024, 21, 276. [Google Scholar] [CrossRef] [PubMed]
- SaiKumar, G.; Ramani Pushpa, R.N.; Supriya, A.R. Isolation and molecular characterization of lumpy skin disease virus from recent outbreaks in Andhra Pradesh, India. Pharma Innov. 2023, 12, 1250–1257. [Google Scholar]
- Prabhu, M.; Malmarugan, S.; Rajagunalan, S.; Balakrishnan, G.; Lakshmi Prasanth, T.; Ganapathi, P. Molecular detection of lumpy skin disease virus from Tamil Nadu, India. Pharma. Innov. J. 2022, 11, 75–78. [Google Scholar]
- Seerintra, T.; Saraphol, B.; Wankaew, S.; Piratae, S. Molecular identification and characterization of lumpy skin disease virus emergence from cattle in the northeastern part of Thailand. J. Vet. Sci. 2022, 23, e73. [Google Scholar] [CrossRef]
- Abera, Z.; Degefu, H.; Gari, G.; Kidane, M. Sero-prevalence of lumpy skin disease in selected districts of West Wollega zone, Ethiopia. BMC Vet. Res. 2015, 11, 135. [Google Scholar] [CrossRef]
- Ochwo, S.; VanderWaal, K.; Munsey, A.; Nkamwesiga, J.; Ndekezi, C.; Auma, E.; Mwiine, F.N. Seroprevalence and risk factors for lumpy skin disease virus seropositivity in cattle in Uganda. BMC Vet. Res. 2019, 15, 236. [Google Scholar] [CrossRef] [PubMed]
- Hailu, B.; Tolosa, T.; Gari, G.; Teklue, T.; Beyene, B. Estimated prevalence and risk factors associated with clinical lumpy skin disease in north-eastern Ethiopia. Prev. Vet. Med. 2014, 115, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Gari, G.; Grosbois, V.; Waret-Szkuta, A.; Babiuk, S.; Jacquiet, P.; Roger, F. Lumpy skin disease in Ethiopia: Seroprevalence study across different agro-climate zones. Acta Trop. 2012, 123, 101–106. [Google Scholar] [CrossRef]
- Molla, W.; Frankena, K.; Gari, G.; Kidane, M.; Shegu, D.; de Jong, M.C. Seroprevalence and risk factors of lumpy skin disease in Ethiopia. Prev. Vet. Med. 2018, 160, 99–104. [Google Scholar] [CrossRef]
- Pandey, G.; Pathak, C.R.; Sadaula, A.; Bastakoti, R.; Hamal, P.; Khanal, P.; Sapkota, P.; Pandeya, Y.R.; Paudel, S. Molecular and serological detection of lumpy skin disease in cattle of Western Chitwan, Nepal. In Proceedings of the 12th National Workshop on Livestock and Fisheries Research in Nepal, Western Chitwan, Nepal, 3–4 March 2021; Volume 3, p. 57. [Google Scholar]
- Troyo, A.; Calderón-Arguedas, O.; Fuller, D.O.; Solano, M.E.; Avendaño, A.; Arheart, K.L.; Beier, J.C. Seasonal profiles of Aedes aegypti (Diptera: Culicidae) larval habitats in an urban area of Costa Rica with a history of mosquito control. J. Soc. Vector Ecol. 2008, 33, 76. [Google Scholar] [CrossRef]
- Agag, B.L.; Hafiz, M.A.; Ragab, A.; Tawfik, A.; Mousa, H.L.; Shaker, M.; El-Danal, N. Changes in serum biochemical component of cattle suffering from LSD in Egypt. Egypt. J. Comp. Pathol. Clin. Pathol. 1989, 2, 9–25. [Google Scholar]
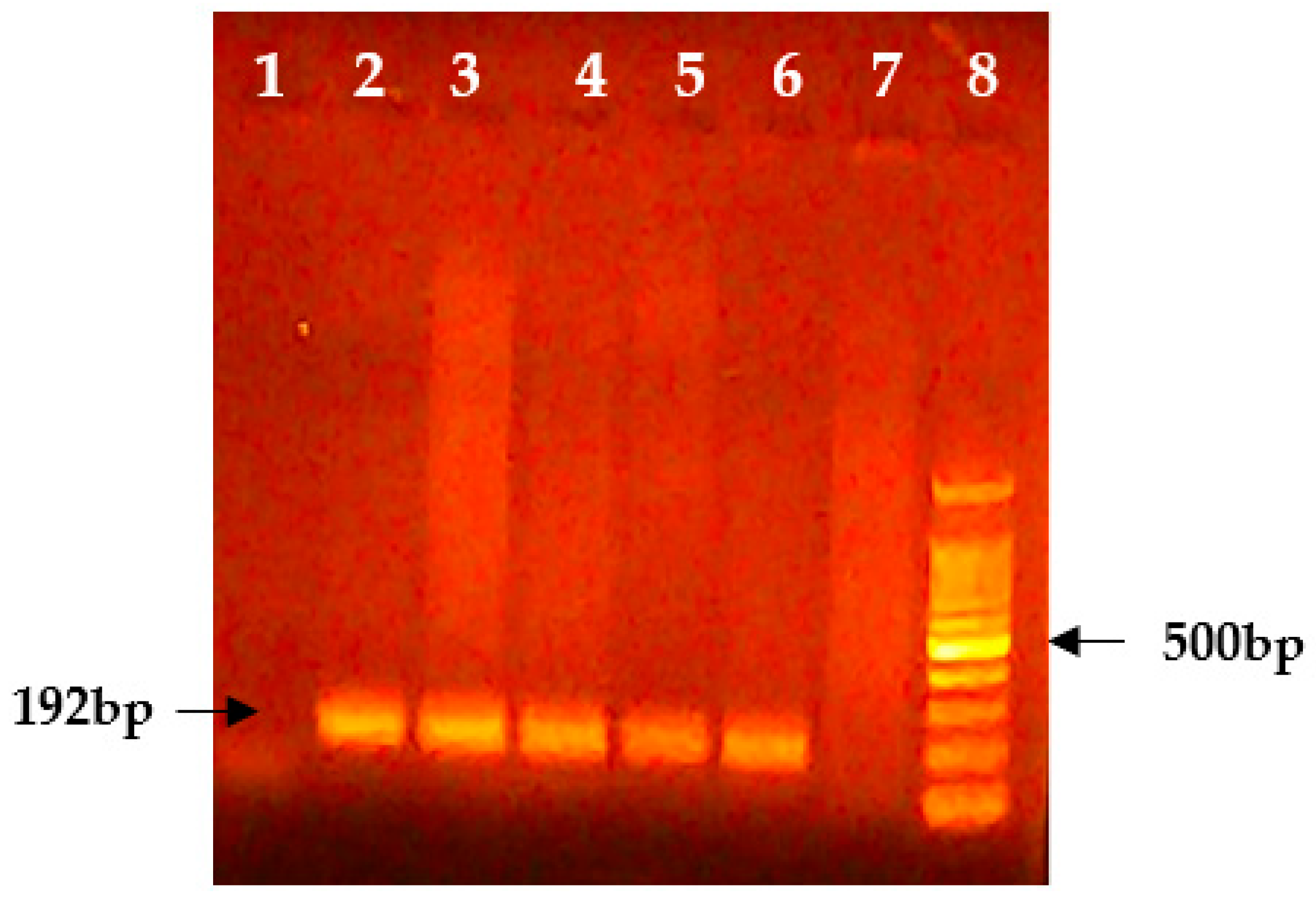
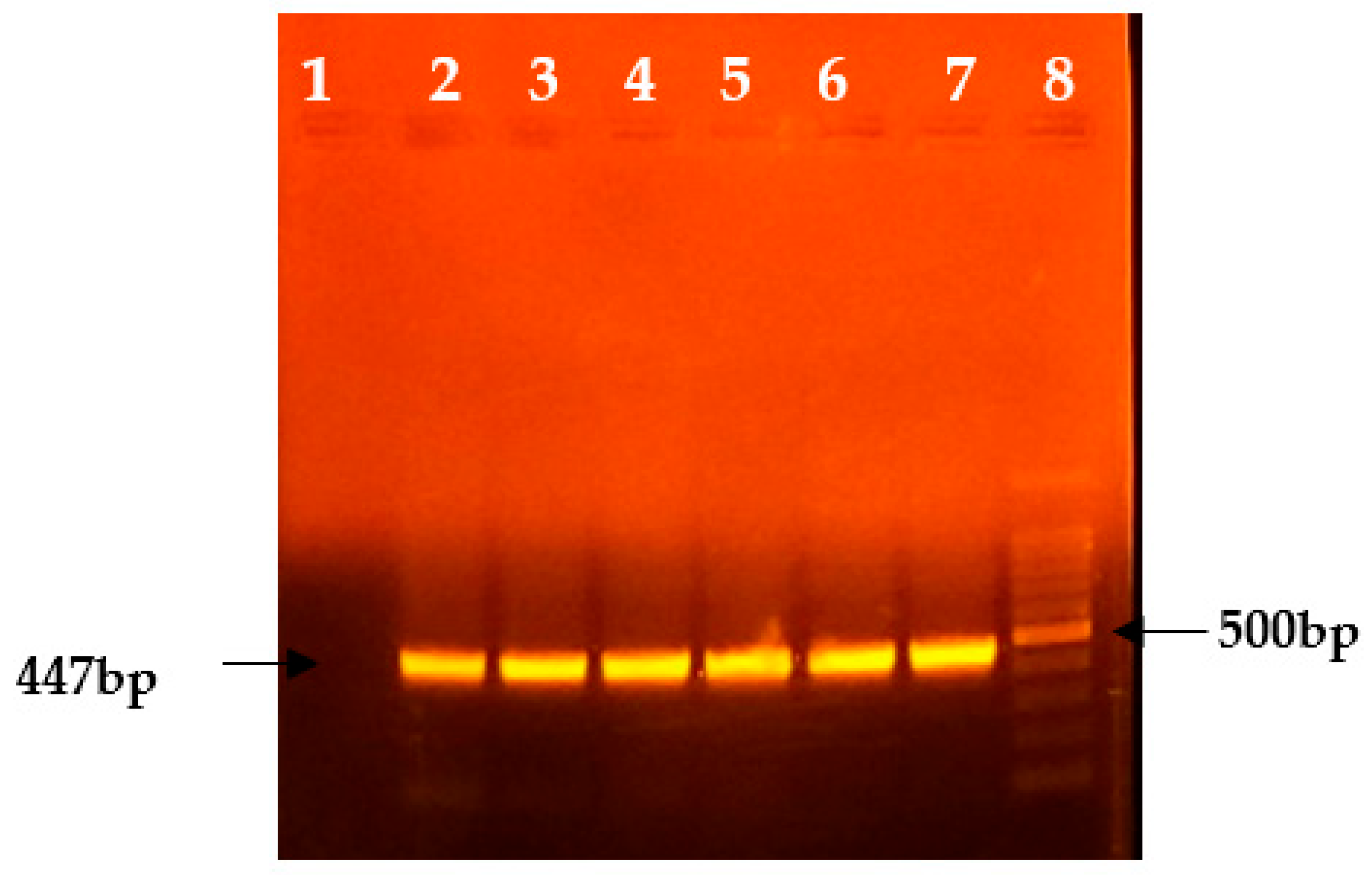
| S. No | Sample Type | Total | Positive | Negative |
|---|---|---|---|---|
| 1. | Blood | 116 | 00 (0.0%, 0.0–3.13%) | 116 (100%, 96.9–100%) |
| 2. | Swab | 47 | 03 (6.4%, 1.3–17.5%) | 44 (93.6%, 82.5–98.7%) |
| 3. | Tissue | 26 | 18 (69.2%, 48.2–85.7%) | 08 (30.8%, 14.3–51.8%) |
| S. No | Sample Type | Total | Positive | Negative |
|---|---|---|---|---|
| 1. | Blood | 00 | 00 | 00 |
| 2. | Swab | 03 | 02 (66.7%, 9.4–99.2%) | 01 (33.3%, 0.8–90.6%) |
| 3. | Tissue | 18 | 14 (77.8%, 52.4–93.6%) | 04 (22.2%, 6.4–47.6%) |
| Parameters | Sample | Seropositive | Seronegative | χ2 Value | p-Value | |
|---|---|---|---|---|---|---|
| Breed | HFC | 22 | 7 | 15 | 9.552 | p > 0.05 |
| JC | 58 | 15 | 43 | |||
| ND | 62 | 12 | 50 | |||
| OC | 8 | 0 | 8 | |||
| Sahiwal | 17 | 1 | 16 | |||
| Gir | 17 | 1 | 16 | |||
| Total | 184 | 36 | 148 | |||
| Age | <2 years | 52 | 10 | 42 | 3.01 | p > 0.05 |
| 2–4 years | 57 | 10 | 47 | |||
| >4 years | 75 | 16 | 59 | |||
| Total | 184 | 36 | 148 | |||
| Sex | Male | 49 | 10 | 39 | 8.38 | p < 0.05 |
| Female | 135 | 26 | 109 | |||
| Total | 184 | 36 | 148 | |||
| Immunization statusS | Immunized | 37 | 0 | 37 | 9.76 | p < 0.05 |
| Non-immunized | 147 | 36 | 111 | |||
| Total | 184 | 36 | 148 | |||
| S. No. | Sample | Creatinine (mg/dL) | SGPT (U/L) | SGOT (U/L) | BUN (mg/dL) | Total Protien (g/dL) | Albu. (g/dL) | Globu. (g/dL) | A: G (g/dL) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | S-02 | 0.96 | 90.17 | 99.01 | 29.63 | 6.87 | 5.36 | 1.51 | 3.550 |
| 2 | S-04 | 0.93 | 33.59 | 49.5 | 21.11 | 7.45 | 3.57 | 3.88 | 0.920 |
| 3 | S-09 | 1.26 | 76.02 | 83.1 | 15.69 | 7.81 | 4.83 | 2.98 | 1.621 |
| 4 | S-42 | 1.37 | 160.9 | 176.8 | 17.45 | 5.17 | 4.36 | 0.81 | 5.383 |
| 5 | NS-01 | 2.84 | N/A | N/A | 21.92 | 8.19 | 4.82 | 3.37 | 1.430 |
| 6 | NS-02 | 1.55 | 270.5 | 272.3 | 13.26 | 6.76 | 4.76 | 2 | 2.380 |
| Mean of Positive Samples | 1.485 | 126.236 | 136.142 | 19.843 | 7.042 | 4.617 | 2.425 | 2.547 | |
| Normal Values of Healthy Cattles | 0.7–1.1 | 11.0–40.0 | 78–132 | 10.0–26.0 | 5.9–7.7 | 2.7–4.3 | 2.5–4.1 | 0.6–1.6 | |
| Result (Mean ± SD) | |||||
|---|---|---|---|---|---|
| Parameters | Positive Samples | 95% CI Values | Negative Samples | 95% CI Values | p-Value |
| Creatinine | 1.48 ± 0.705 | 74–223% | 1.13 ± 0.307 | 80–145% | 0.4476 |
| SGPT | 126.23 ± 92.8 | 288–22,358% | 49.5 ± 20.8 | 2767–7132% | 0.2197 |
| SGOT | 136.14 ± 89.3 | 4243–22,984% | 85.45 ± 37.5 | 4609–12,480% | 0.3967 |
| BUN | 26.6 ± 16.0 | 1376–2591% | 19.8 ± 5.80 | 1371–2588% | 0.367 |
| Total protein | 7.04 ± 1.07 | 592–816% | 5.39 ± 2.11 | 317–760% | 0.1489 |
| Albumin | 4.62 ± 0.604 | 3.98–5.25% | 3.98 ± 0.642 | 330–465% | 0.1864 |
| Globulin | 2.42 ± 1.18 | 118–366% | 1.41 ± 1.65 | 32–314% | 0.3155 |
| A: G | 6.32 ± 4.63 | 80–429% | 2.55 ± 1.66 | 80–429% | 0.1025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, V.; Pravalika, A.; Pandey, M.K.; Mareddy, V.; Jain, A.K.; Singh, A.; Nayak, A.; Tripathi, S.; Rajoriya, S. Molecular Detection, Seroprevalence and Biochemical Analysis of Lumpy Skin Disease Virus. Viruses 2025, 17, 293. https://doi.org/10.3390/v17030293
Gupta V, Pravalika A, Pandey MK, Mareddy V, Jain AK, Singh A, Nayak A, Tripathi S, Rajoriya S. Molecular Detection, Seroprevalence and Biochemical Analysis of Lumpy Skin Disease Virus. Viruses. 2025; 17(3):293. https://doi.org/10.3390/v17030293
Chicago/Turabian StyleGupta, Vandana, Annapureddy Pravalika, Megha Katare Pandey, Vineetha Mareddy, Anand Kumar Jain, Akansha Singh, Anju Nayak, Swati Tripathi, and Shweta Rajoriya. 2025. "Molecular Detection, Seroprevalence and Biochemical Analysis of Lumpy Skin Disease Virus" Viruses 17, no. 3: 293. https://doi.org/10.3390/v17030293
APA StyleGupta, V., Pravalika, A., Pandey, M. K., Mareddy, V., Jain, A. K., Singh, A., Nayak, A., Tripathi, S., & Rajoriya, S. (2025). Molecular Detection, Seroprevalence and Biochemical Analysis of Lumpy Skin Disease Virus. Viruses, 17(3), 293. https://doi.org/10.3390/v17030293






