Genome and Pathogenicity Analysis of an NADC30-like PRRSV Strain in China’s Xinjiang Province
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statements
2.2. Clinical Sample Collection and Detection
2.3. Virus Isolation
2.4. Genome Sequencing
2.5. Phylogenetic Analysis
2.6. Recombination Analysis
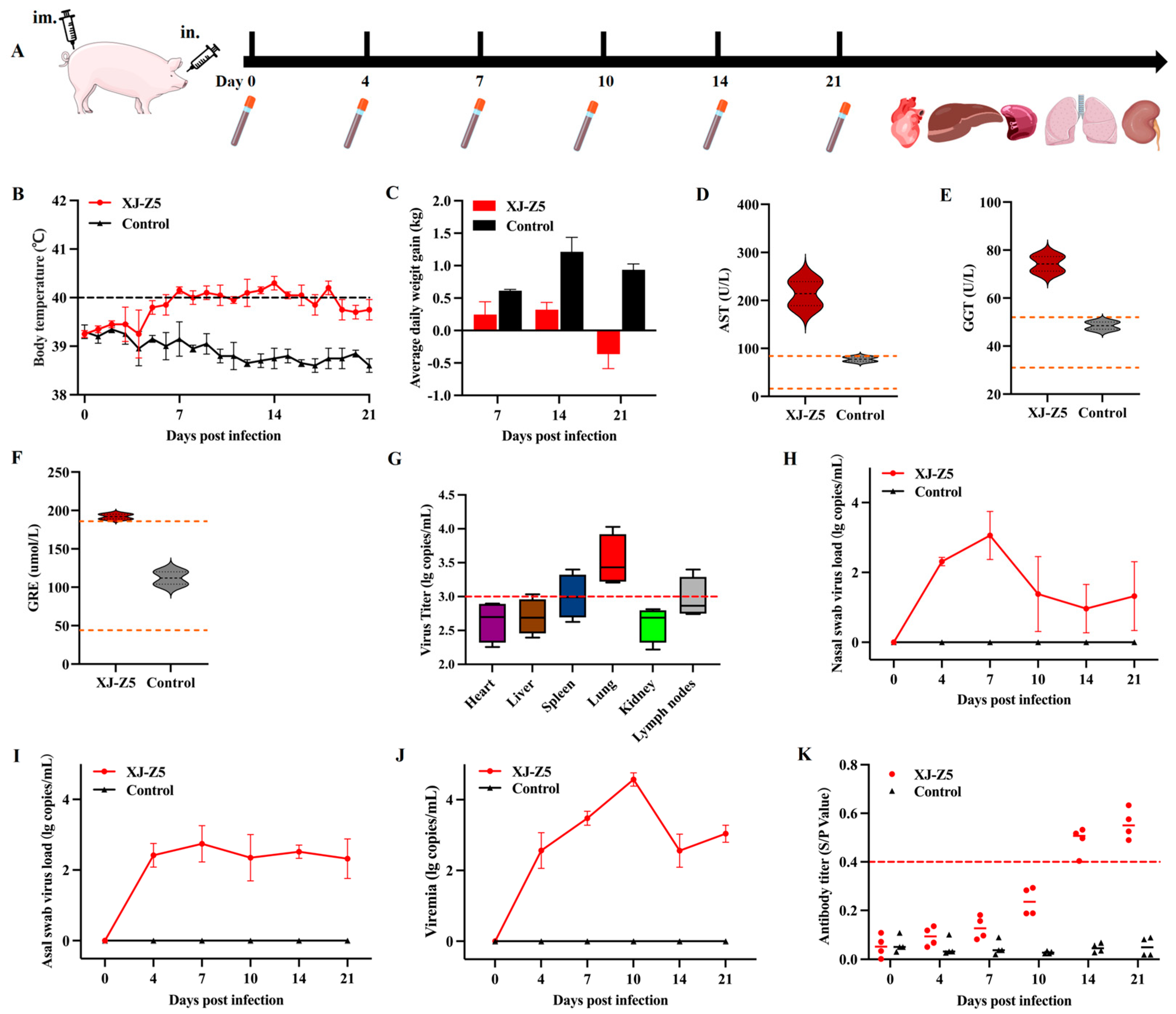
2.7. Animals and Experimental Design
2.8. Viral Load Detection
2.9. Blood Biochemical
2.10. Antibody Detection
2.11. Histopathology
2.12. Data Analysis
3. Results
3.1. Virus Isolation and Whole Genome Sequencing
3.2. Results of Phylogenetic Analysis
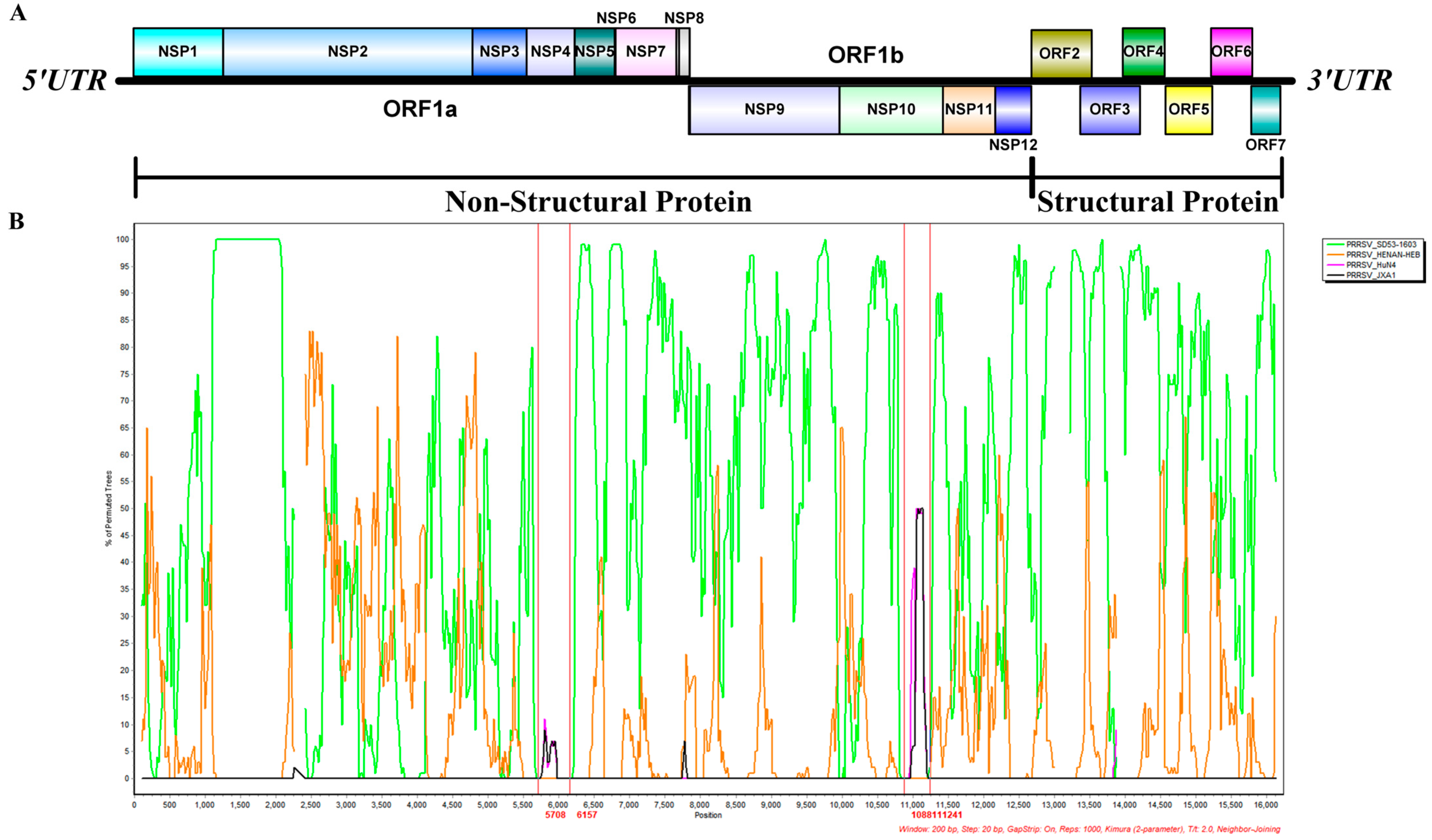
3.3. Results of Recombination Analysis
3.4. Results of Pathogenicity Analysis
3.5. Viral Load and Antibody Levels
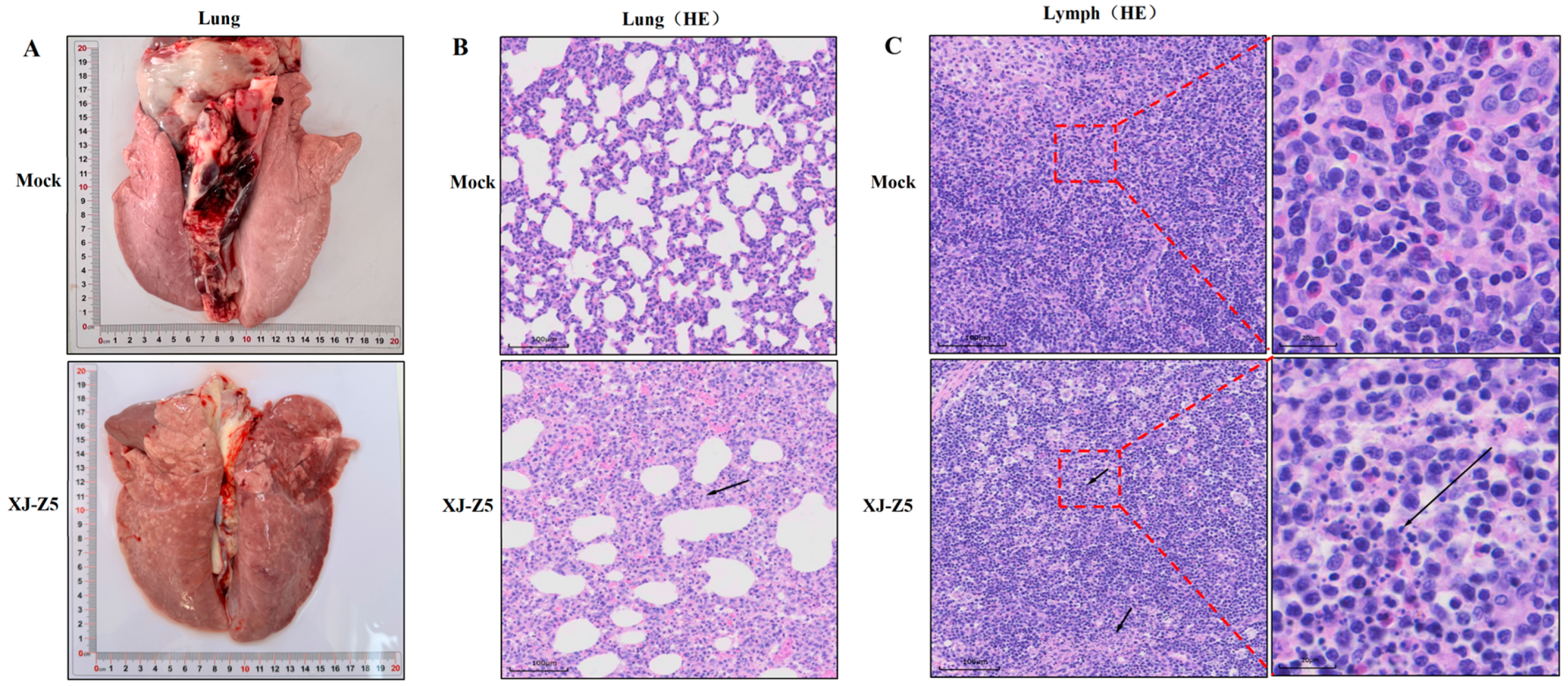
3.6. Results of Histopathology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruedas-Torres, I.; Rodríguez-Gómez, I.M.; Sánchez-Carvajal, J.M.; Larenas-Muñoz, F.; Pallarés, F.J.; Carrasco, L.; Gómez-Laguna, J. The jigsaw of PRRSV virulence. Vet. Microbiol. 2021, 260, 109168. [Google Scholar] [CrossRef]
- Stadejek, T.; Stankevicius, A.; Murtaugh, M.P.; Oleksiewicz, M.B. Molecular evolution of PRRSV in Europe: Current state of play. Vet. Microbiol. 2013, 165, 21–28. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, X.X.; Li, R.; Qiao, S.; Zhang, G. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: A molecular epidemiological perspective. Virol. J. 2018, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Leng, C.; Ding, Y.; Zhai, H.; Li, Z.; Xiang, L.; Zhang, W.; Liu, C.; Li, M.; Chen, J.; et al. Characterization of newly emerged NADC30-like strains of porcine reproductive and respiratory syndrome virus in China. Arch. Virol. 2019, 164, 401–411. [Google Scholar] [CrossRef]
- Zhou, L.; Han, J.; Yang, H. The evolution and diversity of porcine reproductive and respiratory syndrome virus in China. Vet. Microbiol. 2024, 298, 110252. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xing, J.; Hong, S.L.; Bollen, N.; Xu, S.; Li, Y.; Zhong, J.; Gao, X.; Zhu, D.; Liu, J.; et al. Untangling lineage introductions, persistence and transmission drivers of HP-PRRSV sublineage 8.7. Nat. Commun. 2024, 15, 8842. [Google Scholar] [CrossRef]
- Li, J.; Gong, W.; Mao, L.; Pan, X.; Wu, Q.; Guo, Y.; Jiang, J.; Tang, H.; Zhao, Y.; Cheng, L.; et al. Molecular epizootiology of porcine reproductive and respiratory syndrome virus in the Xinjiang Uygur Autonomous Region of China. Front. Microbiol. 2024, 15, 1419499. [Google Scholar] [CrossRef]
- Dokland, T. The structural biology of PRRSV. Virus Res. 2010, 154, 86–97. [Google Scholar] [CrossRef]
- Kappes, M.A.; Faaberg, K.S. PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity. Virology 2015, 479–480, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Tang, D.; Zeng, Z.; Wang, B.; Zhou, M.; Mao, Y.; Zhou, P.; He, S. Research progress on the molecular mechanism of immune escape of porcine reproductive and respiratory syndrome virus. Virology 2025, 602, 110298. [Google Scholar] [CrossRef]
- Yu, F.; Yan, Y.; Shi, M.; Liu, H.Z.; Zhang, H.L.; Yang, Y.B.; Huang, X.Y.; Gauger, P.C.; Zhang, J.; Zhang, Y.H.; et al. Phylogenetics, Genomic Recombination, and NSP2 Polymorphic Patterns of Porcine Reproductive and Respiratory Syndrome Virus in China and the United States in 2014-2018. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; Lager, K.M.; Golde, W.; Faaberg, K.S.; Sinkora, M.; Loving, C.; Zhang, Y.I. Porcine reproductive and respiratory syndrome (PRRS): An immune dysregulatory pandemic. Immunol. Res. 2014, 59, 81–108. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.G.; Dee, S.A. Porcine reproductive and respiratory syndrome virus. Theriogenology 2006, 66, 655–662. [Google Scholar] [CrossRef]
- Molitor, T.W.; Bautista, E.M.; Choi, C.S. Immunity to PRRSV: Double-edged sword. Vet. Microbiol. 1997, 55, 265–276. [Google Scholar] [CrossRef]
- Tian, K. NADC30-Like Porcine Reproductive and Respiratory Syndrome in China. Open Virol. J. 2017, 11, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wang, X.; Liu, K.; Chen, G.; Zhang, K.; Liu, J.; Pang, Y.; Ren, T.; Qin, Y.; Ouyang, K.; et al. Recombination and Pathogenicity Analysis of NADC30-like and QYYZ-like PRRSV Strains in South China. Microb. Pathog. 2025, 200, 107351. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Samson, S.; Lord, É.; Makarenkov, V. SimPlot++: A Python application for representing sequence similarity and detecting recombination. Bioinformatics 2022, 38, 3118–3120. [Google Scholar] [CrossRef]
- Brar, M.S.; Shi, M.; Murtaugh, M.P.; Leung, F.C. Evolutionary diversification of type 2 porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2015, 96, 1570–1580. [Google Scholar] [CrossRef]
- Brockmeier, S.L.; Loving, C.L.; Vorwald, A.C.; Kehrli, M.E., Jr.; Baker, R.B.; Nicholson, T.L.; Lager, K.M.; Miller, L.C.; Faaberg, K.S. Genomic sequence and virulence comparison of four Type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. 2012, 169, 212–221. [Google Scholar] [CrossRef]
- Han, G.; Xu, H.; Wang, K.; He, F. Emergence of Two different recombinant PRRSV strains with low neutralizing antibody susceptibility in China. Sci. Rep. 2019, 9, 2490. [Google Scholar] [CrossRef]
- Ouyang, Y.; Du, Y.; Zhang, H.; Guo, J.; Sun, Z.; Luo, X.; Mei, X.; Xiao, S.; Fang, L.; Zhou, Y. Genetic Characterization and Pathogenicity of a Recombinant Porcine Reproductive and Respiratory Syndrome Virus Strain in China. Viruses 2024, 16, 993. [Google Scholar] [CrossRef]
- Chang, H.; Gao, X.; Wu, Y.; Wang, F.; Lai, M.; Zheng, J.; Qiu, Y.; He, Y.; Liang, X.; Yuan, K.; et al. Genomic and pathogenicity analysis of two novel highly pathogenic recombinant NADC30-like PRRSV strains in China, in 2023. Microbiol. Spectr. 2024, 12, e0036824. [Google Scholar] [CrossRef]
- Shi, C.; Liu, Y.; Ding, Y.; Zhang, Y.; Zhang, J. PRRSV receptors and their roles in virus infection. Arch. Microbiol. 2015, 197, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ye, M.; Sun, S.; Cao, Q.; Luo, J.; Wang, Y.; Zheng, W.; Meurens, F.; Chen, N.; Zhu, J. CD163-Expressing Porcine Macrophages Support NADC30-like and NADC34-like PRRSV Infections. Viruses 2022, 14, 2056. [Google Scholar] [CrossRef] [PubMed]
- Rowland, R.R.R.; Brandariz-Nuñez, A. Role of CD163 in PRRSV infection. Virology 2024, 600, 110262. [Google Scholar] [CrossRef]
- Zeller, M.A.; Chang, J.; Trevisan, G.; Main, R.G.; Gauger, P.C.; Zhang, J. Rapid PRRSV-2 ORF5-based lineage classification using Nextclade. Front Vet. Sci. 2024, 11, 1419340. [Google Scholar] [CrossRef] [PubMed]
- Yim-Im, W.; Anderson, T.K.; Paploski, I.A.D.; VanderWaal, K.; Gauger, P.; Krueger, K.; Shi, M.; Main, R.; Zhang, J. Refining PRRSV-2 genetic classification based on global ORF5 sequences and investigation of their geographic distributions and temporal changes. Microbiol. Spectr. 2023, 11, e0291623. [Google Scholar] [CrossRef]
- Xiao, Y.; Rouzine, I.M.; Bianco, S.; Acevedo, A.; Goldstein, E.F.; Farkov, M.; Brodsky, L.; Andino, R. RNA Recombination Enhances Adaptability and Is Required for Virus Spread and Virulence. Cell Host Microbe 2017, 22, 420. [Google Scholar] [CrossRef]
- Molenkamp, R.; Greve, S.; Spaan, W.J.; Snijder, E.J. Efficient homologous RNA recombination and requirement for an open reading frame during replication of equine arteritis virus defective interfering RNAs. J. Virol. 2000, 74, 9062–9070. [Google Scholar] [CrossRef]
- Tang, J.; Hung, Y.F.; Yoo, D. Genomic RNA recombination of porcine reproductive and respiratory syndrome virus and other arteriviruses. Virology 2025, 601, 110284. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Xia, D.S.; Luo, L.Z.; An, T.Q. Recombination of Porcine Reproductive and Respiratory Syndrome Virus: Features, Possible Mechanisms, and Future Directions. Viruses 2024, 16, 929. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Li, Y.; Zhang, G.; Du, C.; Zhou, Y.; Jiang, D.; Luo, Y.; Yao, X.; Yang, Z.; Ren, M.; et al. Isolation, identification, recombination analysis and pathogenicity experiment of a PRRSV recombinant strain in Sichuan Province, China. Front Microbiol. 2024, 15, 1362471. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, Y.; Wang, Z.; Li, L.; Liu, L.; Liu, Y. The Time Sequences of Respiratory and Rectal Viral Shedding in Patients With Coronavirus Disease 2019. Gastroenterology 2020, 159, 1158–1160.e1152. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef]
- Ma, X.; Wang, P.; Zhang, R.; Zhao, Y.; Wu, Y.; Luo, C.; Zeshan, B.; Yang, Z.; Qiu, L.; Zhou, Y.; et al. A NADC30-like PRRSV causes serious intestinal infections and tropism in piglets. Vet. Microbiol. 2022, 268, 109397. [Google Scholar] [CrossRef]


| Primers | Sequences (5′-3′) | Amplification Position | Size (bp) |
|---|---|---|---|
| NSP2 | F: ATGTTGTGCTTCCTGGGGTTG | 2205–3225 | 628 (NADC30-like strain), 931 (HP-PRRSV), 1021 (PRRSV) |
| R: CTTGACAGGGAGCTGCTTGA | |||
| qRT-PCR | F: AAACCAGTCCAGAGGCAAGG | ORF 7 | 81 |
| R: TCAGTCGCAAGAGGGAATG | |||
| P1 | F: ATGACGTATAGGTGTTGGCTCTATGC | 1–2167 | 2167 |
| R: AGCTTTCTCAAGCCTAGCCAAGC | |||
| P2 | F: GGGTTTGACCCTGCCTGCCTTGA | 1823–3762 | 1940 |
| R: AAACTCACAAGCAGTGCCGACTG | |||
| P3 | F: CTGCTGGCTGGCTTTTGCTGTTG | 3694–5839 | 2146 |
| R: CCTCCTTCCAGTTCGGGTTTGGC | |||
| P4 | F: CGCCCTTCAGGCCAGTTTTGTAA | 5656–7816 | 2160 |
| R: CGCTAGGGGTCTTGTAAGGTATGTC | |||
| P5 | F: AGCGGCAAACCTGAACGGGTAAAAG | 7741–9939 | 2199 |
| R: AAGCCTCAGGACATCAAGATGATTG | |||
| P6 | F: AAAGCTTTGGGCACGTGTCGGTTCA | 9769–11,877 | 2109 |
| R: AACGGCAGGGCGCGGACGGAGTATC | |||
| P7 | F: CATCGCCGGATGGTTGGTGGTACTT | 11,804–13,548 | 1744 |
| R: GCCATTCAGCTCACATATCGTCAGG | |||
| P8 | F: GATATGTTGGGGAAATGCTTGACCG | 13,392–15,016 | 1623 |
| R: TTAATTACGGCCGCATGGTTCTC |
| Recombined Virus | Parental Virus | Breakpoints a | Score for the Seven Detection Methods in RDP5 b | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Major | Minor | Region | Begin | End | RDP | GENECONV | BoostScan | MaxChi | Chimaera | SiScan | 3Seq | |
| XJ-Z5 | HENAN-HEB | JXA1 | NSP4 | 5708 | 6157 | 3.568 × 10−33 | 1.128 × 10−35 | 1.917 × 10−23 | 2.593 × 10−10 | 2.003 × 10−9 | 6.947 × 10−11 | 2.183 × 10−3 |
| SD53-1603 | HuN4 | NSP10~11 | 10,881 | 11,241 | 4.840 × 10−23 | 1.992 × 10−18 | 9.657 × 10−24 | 1.901 × 10−11 | 1.039 × 10−8 | 2.665 × 10−2 | - | |
| XJ-Z5 | HENAN-HEB (%) | JXA1 (%) | SD53–1603 (%) | HuN4 (%) |
|---|---|---|---|---|
| Complete (15,015) a | 89.31 | 85.72 | 92.07 b | 85.5 |
| ORF1a (191–7309) | 87.21 | 84.10 | 89.60 | 84.18 |
| NSP1(191–1336) | 89.27 | 82.00 | 88.84 | 82.35 |
| NSP2 (1337–4528) | 85.45 | 82.34 | 87.87 | 82.25 |
| NSP3 (4529–5221) | 90.32 | 82.28 | 88.6 | 82.55 |
| NSP4 (5222–5834) | 81.32 | 92.54 | 85.15 | 92.18 |
| NSP5 (5835–6346) | 89.84 | 85.05 | 92.59 | 85.80 |
| NSP6 (6347–6394) | 85.88 | 100.00 | 95.83 | 99.46 |
| NSP7 (6395–7171) | 90.59 | 80.47 | 94.59 | 81.19 |
| NSP8 (7172–7306) | 88.89 | 88.15 | 94.81 | 86.67 |
| ORF1b (7297–11,679) | 92.4 | 87.00 | 94.27 | 86.86 |
| NSP9 (7310–9221) | 92.83 | 86.54 | 95.71 | 86.22 |
| NSP10 (9226–10,548) | 91.38 | 85.22 | 93.88 | 85.22 |
| NSP11 (10,549–11,217) | 91.92 | 90.87 | 91.02 | 91.02 |
| NSP12 (11,218–11,679) | 94.63 | 89.15 | 94.36 | 88.89 |
| ORF2 (11,681–12,451) | 91.51 | 86.57 | 95.46 | 86.57 |
| ORF3 (12,304–13,068) | 85.6 | 85.49 | 95.18 | 85.49 |
| ORF4 (12,849–13,385) | 93.3 | 84.57 | 96.65 | 84.92 |
| ORF5 (13,396–13,995) | 91.54 | 86.48 | 95.67 | 86.64 |
| ORF6 (13,980–14,504) | 93.33 | 88.59 | 95.24 | 88.59 |
| ORF7 (14,494–14,865) | 90.59 | 86.29 | 94.09 | 86.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Zhang, W.; Qiao, Y.; Wang, W.; Zhang, W.; Wang, Y.; Yi, J.; Zhang, H.; Ma, Z.; Chen, C. Genome and Pathogenicity Analysis of an NADC30-like PRRSV Strain in China’s Xinjiang Province. Viruses 2025, 17, 379. https://doi.org/10.3390/v17030379
Li H, Zhang W, Qiao Y, Wang W, Zhang W, Wang Y, Yi J, Zhang H, Ma Z, Chen C. Genome and Pathogenicity Analysis of an NADC30-like PRRSV Strain in China’s Xinjiang Province. Viruses. 2025; 17(3):379. https://doi.org/10.3390/v17030379
Chicago/Turabian StyleLi, Honghuan, Wei Zhang, Yanjie Qiao, Wenxing Wang, Wenxiang Zhang, Yueli Wang, Jihai Yi, Huan Zhang, Zhongchen Ma, and Chuangfu Chen. 2025. "Genome and Pathogenicity Analysis of an NADC30-like PRRSV Strain in China’s Xinjiang Province" Viruses 17, no. 3: 379. https://doi.org/10.3390/v17030379
APA StyleLi, H., Zhang, W., Qiao, Y., Wang, W., Zhang, W., Wang, Y., Yi, J., Zhang, H., Ma, Z., & Chen, C. (2025). Genome and Pathogenicity Analysis of an NADC30-like PRRSV Strain in China’s Xinjiang Province. Viruses, 17(3), 379. https://doi.org/10.3390/v17030379






