Role of Defective Interfering Particles in Complement-Mediated Lysis of Parainfluenza Virus-Infected Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells Lines, Viruses, and Infections
2.2. Generation of Defective Interfering Particle-Enriched Viral Stocks
2.3. Cytotoxicity and Cell Killing Assays
2.4. Flow Cytometry
2.5. Western Blotting
2.6. Quantitative-PCR
2.7. ELISA
2.8. Statistics
3. Results
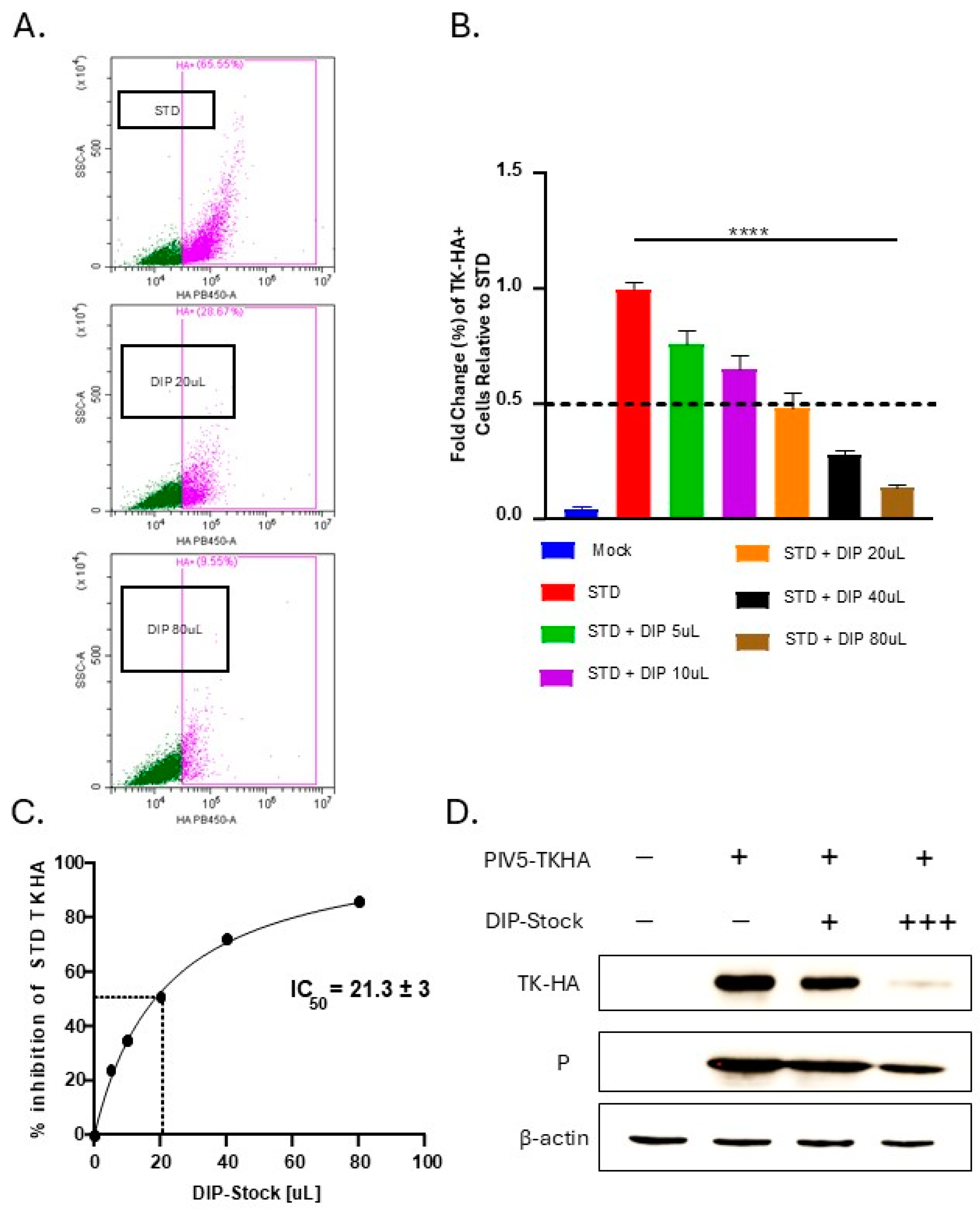
3.1. Generation of DIP-Enriched PIV5 Stocks and RNA Sequence Analysis of DVG Structures
3.2. PIV5 DIPs Suppress STD Virus Gene Expression and Reduce Expression of PIV5 Surface Glycoproteins During Co-Infection
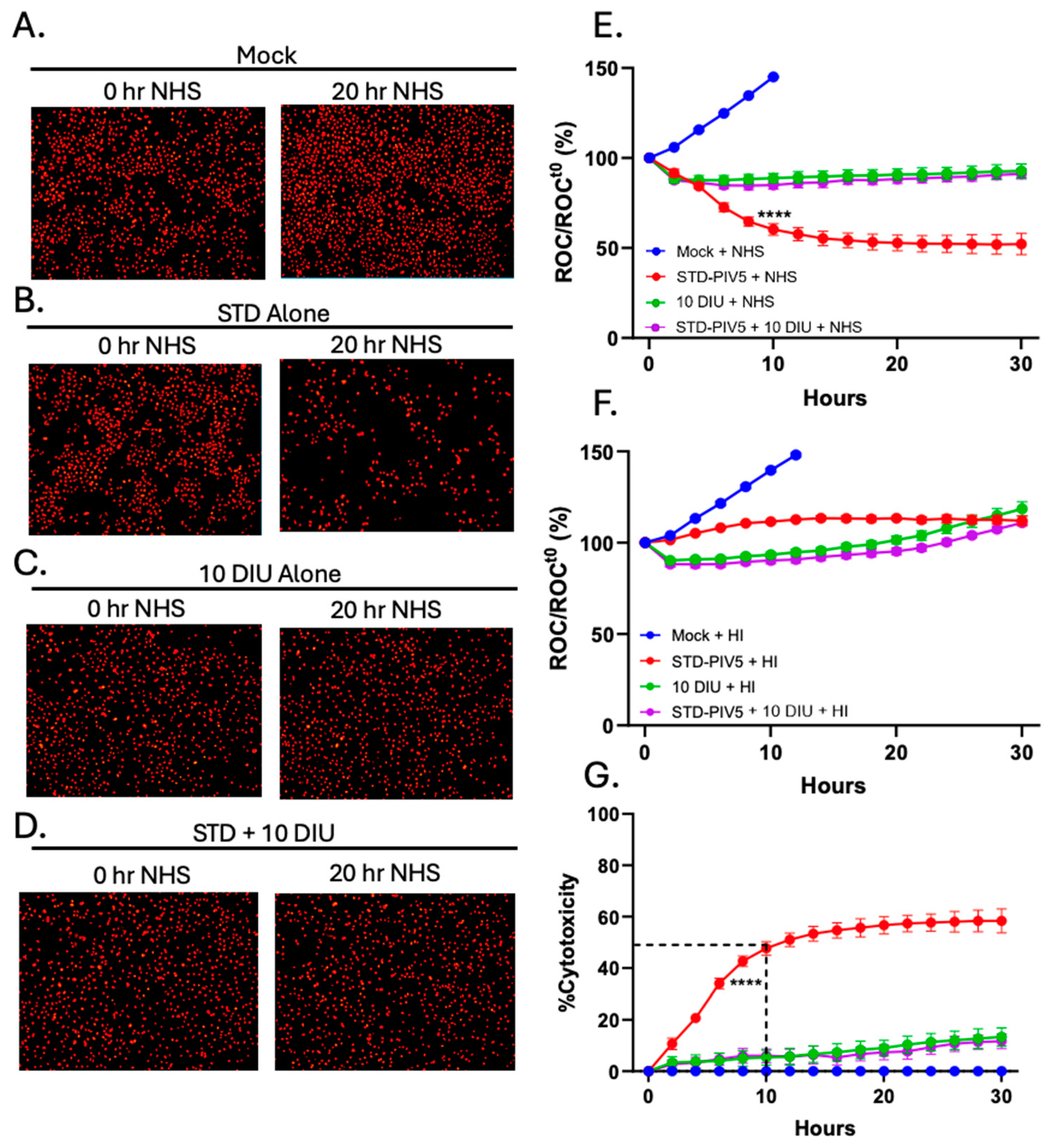
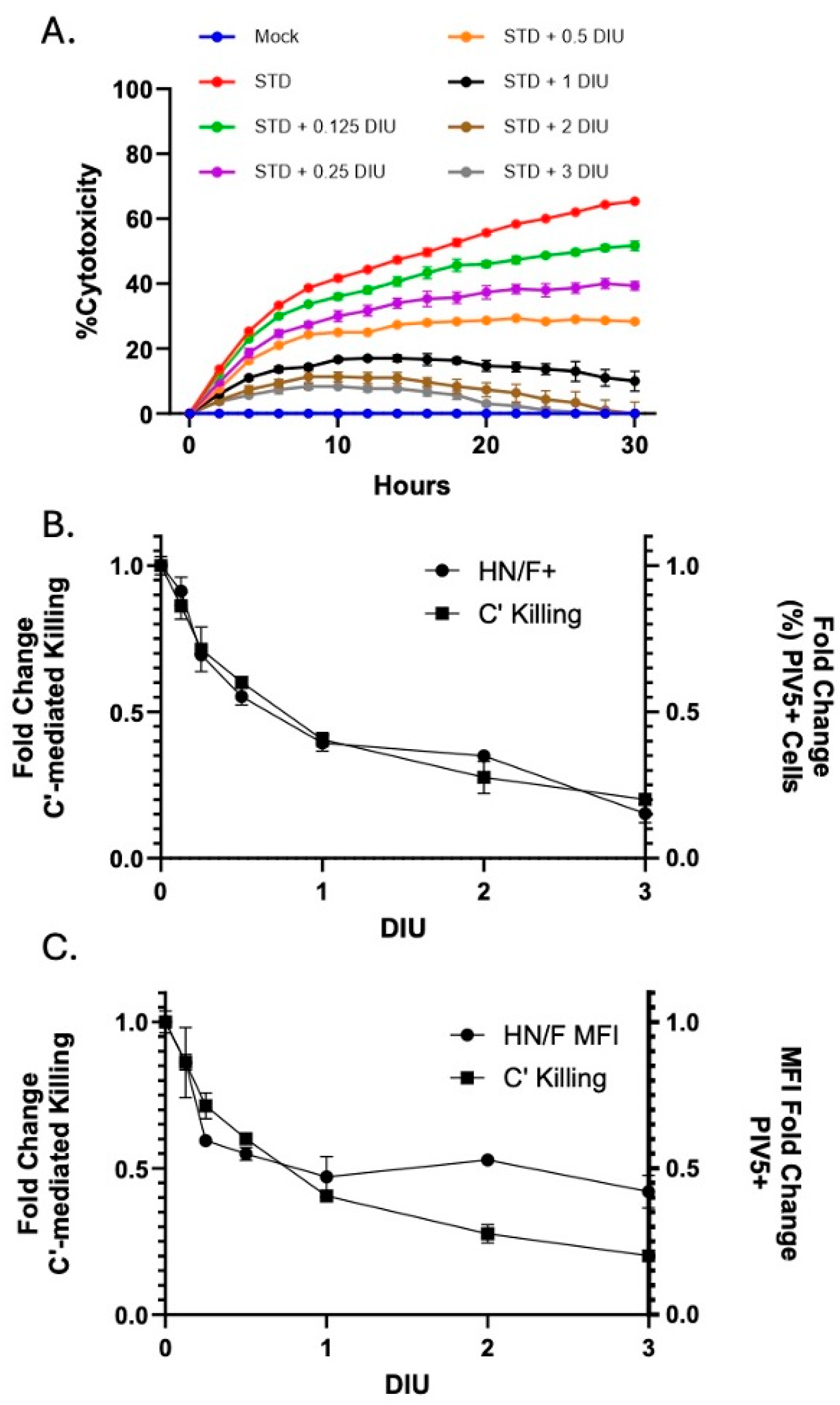
3.3. Infection with DIP-Enriched Stocks Reduces C′-Mediated Killing of Cells Infected with STD PIV5
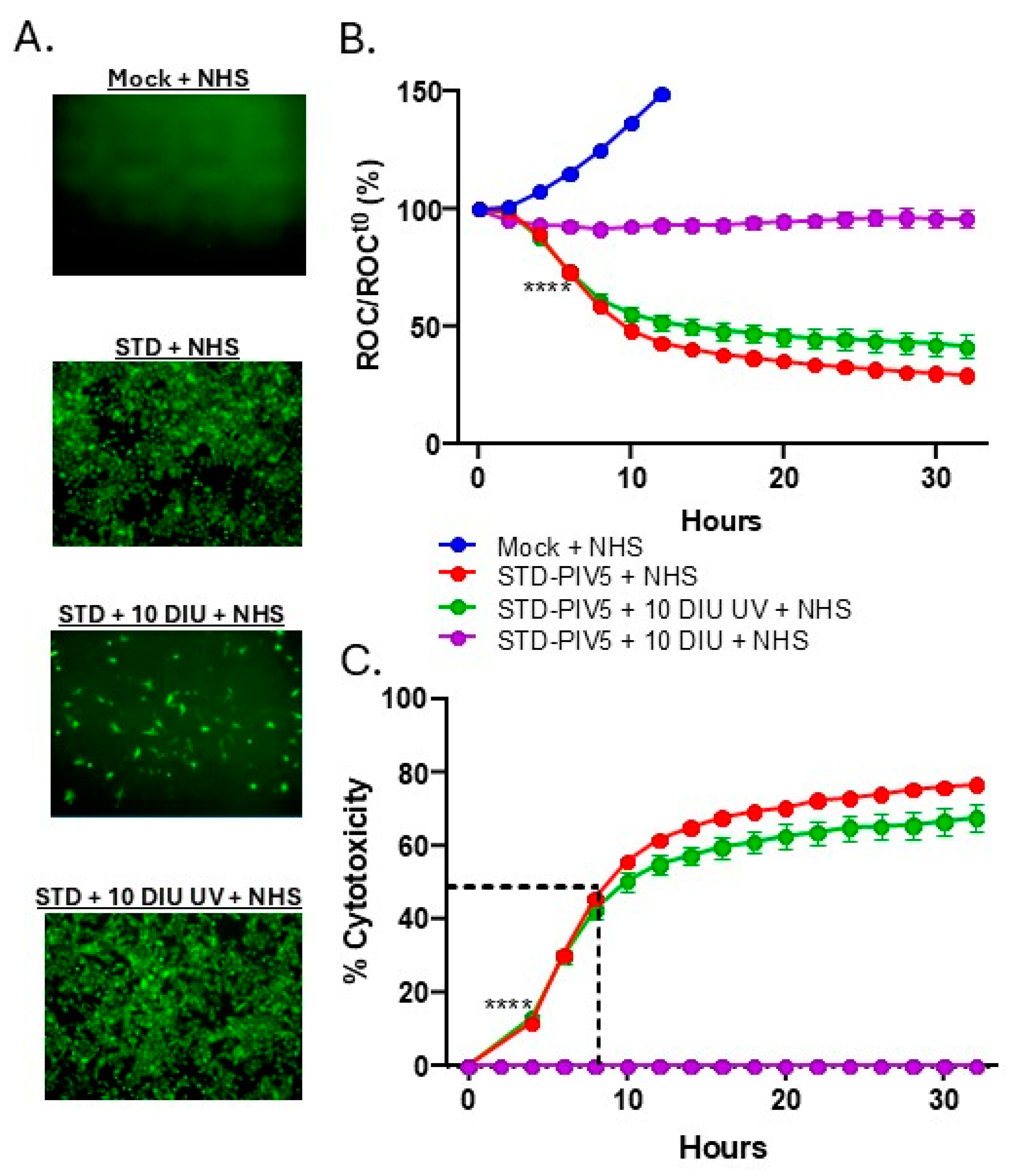
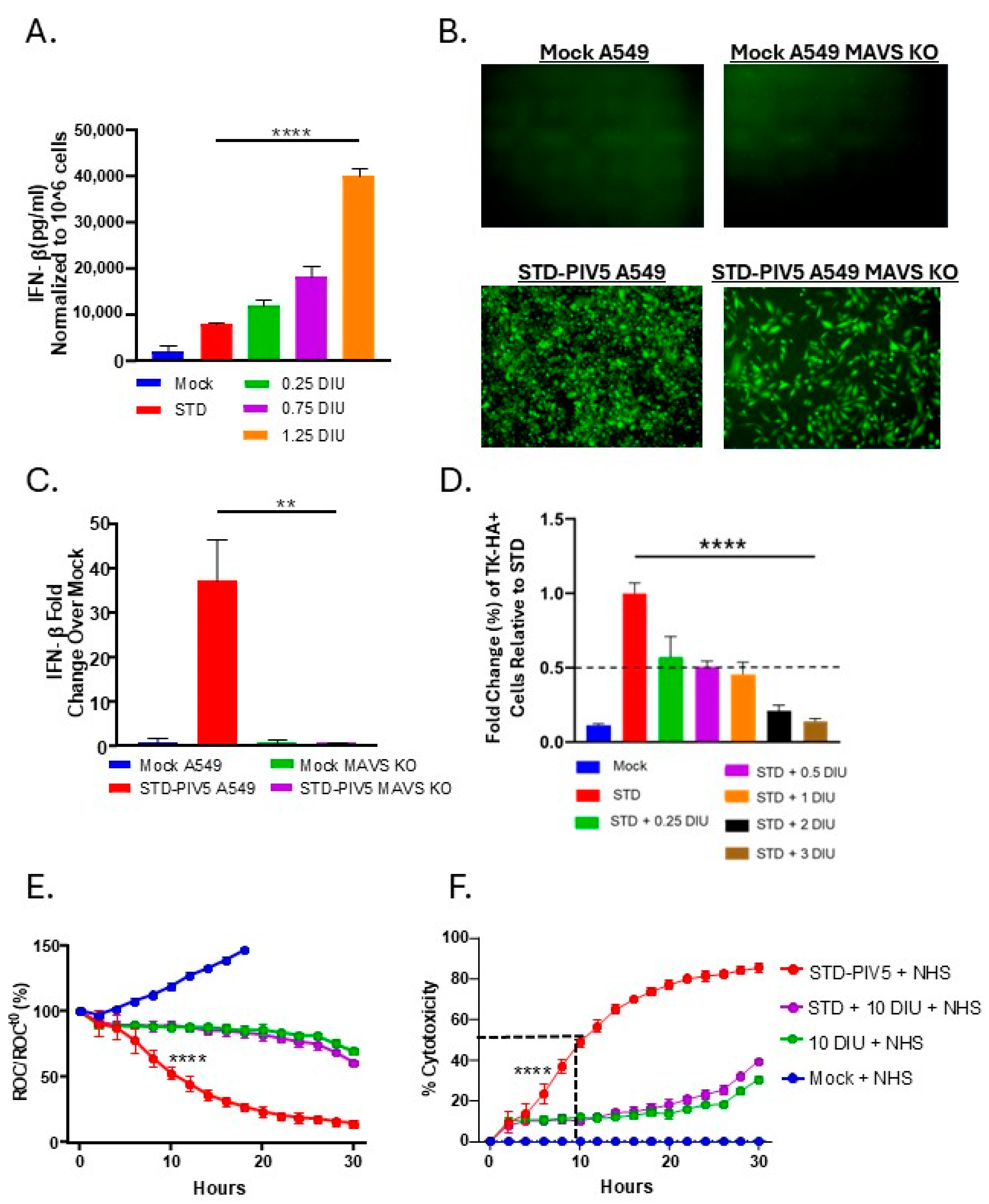
3.4. DIP Inhibition of C′-Mediated Killing of PIV5 Cells Is Dependent on DIP Replication but Independent of IFN-I Induction
3.5. Linear Relationship Between DIP-Mediated Changes in Cell Surface Glycoprotein Expression and C′-Mediated Killing
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Chakrabartty, I.; Khan, M.; Mahanta, S.; Chopra, H.; Dhawan, M.; Choudhary, O.P.; Bibi, S.; Mohanta, Y.K.; Emran, T.B. Comparative overview of emerging RNA viruses: Epidemiology, pathogenesis, diagnosis and current treatment. Ann. Med. Surg. 2022, 79, 103985. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.J.; Brierley, L. Epidemiological characteristics of human-infective RNA viruses. Sci. Data 2018, 5, 180017. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Blanco, M.A.; Ooi, E.E.; Sessions, O.M. RNA Viruses, Pandemics and Anticipatory Preparedness. Viruses 2022, 14, 2176. [Google Scholar] [CrossRef]
- Miranda, M.N.S.; Pingarilho, M.; Pimentel, V.; Torneri, A.; Seabra, S.G.; Libin, P.J.K.; Abecasis, A.B. A Tale of Three Recent Pandemics: Influenza, HIV and SARS-CoV-2. Front. Microbiol. 2022, 13, 889643. [Google Scholar] [CrossRef]
- Klein, J.; Wood, J.; Jaycox, J.R.; Dhodapkar, R.M.; Lu, P.; Gehlhausen, J.R.; Tabachnikova, A.; Greene, K.; Tabacof, L.; Malik, A.A.; et al. Distinguishing features of long COVID identified through immune profiling. Nature 2023, 623, 139–148. [Google Scholar] [CrossRef]
- Genoyer, E.; López, C.B. The Impact of Defective Viruses on Infection and Immunity. Annu. Rev. Virol. 2019, 6, 547–566. [Google Scholar] [CrossRef]
- Wu, M.; Zhou, E.; Sheng, R.; Fu, X.; Li, J.; Jiang, C.; Su, W. Defective Interfering Particles of Influenza Virus and Their Characteristics, Impacts, and Use in Vaccines and Antiviral Strategies: A Systematic Review. Viruses 2022, 14, 2273. [Google Scholar] [CrossRef]
- Li, D.; Lott, W.B.; Lowry, K.; Jones, A.; Thu, H.M.; Aaskov, J. Defective interfering viral particles in acute dengue infections. PLoS ONE 2011, 6, e19447. [Google Scholar] [CrossRef]
- Noppornpanth, S.; Smits, S.L.; Lien, T.X.; Poovorawan, Y.; Osterhaus, A.D.; Haagmans, B.L. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J. Virol. 2007, 81, 12496–12503. [Google Scholar] [CrossRef]
- Nüesch, J.P.; de Chastonay, J.; Siegl, G. Detection of defective genomes in hepatitis A virus particles present in clinical specimens. J. Gen. Virol. 1989, 70 Pt 12, 3475–3480. [Google Scholar] [CrossRef]
- Pesko, K.N.; Fitzpatrick, K.A.; Ryan, E.M.; Shi, P.-Y.; Zhang, B.; Lennon, N.J.; Newman, R.M.; Henn, M.R.; Ebel, G.D. Internally deleted WNV genomes isolated from exotic birds in New Mexico: Function in cells, mosquitoes, and mice. Virology 2012, 427, 10–17. [Google Scholar] [CrossRef]
- Saira, K.; Lin, X.; DePasse, J.V.; Halpin, R.; Twaddle, A.; Stockwell, T.; Angus, B.; Cozzi-Lepri, A.; Delfino, M.; Dugan, V.; et al. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J. Virol. 2013, 87, 8064–8074. [Google Scholar] [CrossRef]
- Sidhu, M.S.; Crowley, J.; Lowenthal, A.; Karcher, D.; Menonna, J.; Cook, S.; Udem, S.; Dowling, P. Defective Measles Virus in Human Subacute Sclerosing Panencephalitis Brain. Virology 1994, 202, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jain, D.; Koziol-White, C.J.; Genoyer, E.; Gilbert, M.; Tapia, K.; Panettieri, R.A., Jr.; Hodinka, R.L.; López, C.B. Immunostimulatory Defective Viral Genomes from Respiratory Syncytial Virus Promote a Strong Innate Antiviral Response during Infection in Mice and Humans. PLoS Pathog. 2015, 11, e1005122. [Google Scholar] [CrossRef] [PubMed]
- Vasilijevic, J.; Zamarreño, N.; Oliveros, J.C.; Rodriguez-Frandsen, A.; Gómez, G.; Rodriguez, G.; Pérez-Ruiz, M.; Rey, S.; Barba, I.; Pozo, F.; et al. Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients. PLoS Pathog. 2017, 13, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Gilliam, N.J.; Li, S.; Spaudau, S.; Osborn, R.M.; Anderson, C.S.; Mariani, T.J.; Thakar, J.; Dewhurst, S.; Mathews, D.H.; et al. Generation and functional analysis of defective viral genomes during SARS-CoV-2 infection. bioRxiv 2022, 14, e0025023. [Google Scholar] [CrossRef]
- Baczko, K.; Liebert, U.G.; Billeter, M.; Cattaneo, R.; Budka, H.; ter Meulen, V. Expression of defective measles virus genes in brain tissues of patients with subacute sclerosing panencephalitis. J. Virol. 1986, 59, 472–478. [Google Scholar] [CrossRef]
- Yagi, S.; Mori, K.; Tanaka, E.; Matsumoto, A.; Sunaga, F.; Kiyosawa, K.; Yamaguchi, K. Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients. J. Med. Virol. 2005, 77, 399–413. [Google Scholar] [CrossRef]
- Viola, M.V.; Scott, C.; Duffy, P.D. Persistent measles virus infection in vitro and in man. Arthritis Rheum. 1978, 21 (Suppl. 5), S47–S51. [Google Scholar]
- Iwai, A.; Marusawa, H.; Takada, Y.; Egawa, H.; Ikeda, K.; Nabeshima, M.; Uemoto, S.; Chiba, T. Identification of novel defective HCV clones in liver transplant recipients with recurrent HCV infection. J. Viral Hepat. 2006, 13, 523–531. [Google Scholar] [CrossRef]
- Li, X.; Ye, Z.; Plant, E.P. 5′ copyback defective viral genomes are major component in clinical and non-clinical influenza samples. Virus Res. 2024, 339, 199274. [Google Scholar] [CrossRef] [PubMed]
- Bellocq, C.; Mottet, G.; Roux, L. Wide occurrence of measles virus subgenomic RNAs in attenuated live-virus vaccines. Biologicals 1990, 18, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Gould, P.S.; Easton, A.J.; Dimmock, N.J. Live Attenuated Influenza Vaccine contains Substantial and Unexpected Amounts of Defective Viral Genomic RNA. Viruses 2017, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gil, L.; Goff, P.H.; Hai, R.; García-Sastre, A.; Shaw, M.L.; Palese, P. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J. Virol. 2013, 87, 1290–1300. [Google Scholar] [CrossRef]
- McLaren, L.C.; Holland, J.J. Defective interfering particles from poliovirus vaccine and vaccine reference strains. Virology 1974, 60, 579–583. [Google Scholar] [CrossRef]
- Killip, M.J.; Young, D.F.; Gatherer, D.; Ross, C.S.; Short, J.A.; Davison, A.J.; Goodbourn, S.; Randall, R.E. Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J. Virol. 2013, 87, 4798–4807. [Google Scholar] [CrossRef]
- Mura, M.; Combredet, C.; Najburg, V.; Sanchez David, R.Y.; Tangy, F.; Komarova, A.V. Nonencapsidated 5′ Copy-Back Defective Interfering Genomes Produced by Recombinant Measles Viruses Are Recognized by RIG-I and LGP2 but Not MDA5. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Strahle, L.; Garcin, D.; Kolakofsky, D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 2006, 351, 101–111. [Google Scholar] [CrossRef]
- van den Hoogen, B.G.; van Boheemen, S.; de Rijck, J.; van Nieuwkoop, S.; Smith, D.J.; Laksono, B.; Gultyaev, A.; Osterhaus, A.; Fouchier, R.A.M. Excessive production and extreme editing of human metapneumovirus defective interfering RNA is associated with type I IFN induction. J. Gen. Virol. 2014, 95 Pt 8, 1625–1633. [Google Scholar] [CrossRef]
- Yoshida, A.; Kawabata, R.; Honda, T.; Sakai, K.; Ami, Y.; Sakaguchi, T.; Irie, T. A Single Amino Acid Substitution within the Paramyxovirus Sendai Virus Nucleoprotein Is a Critical Determinant for Production of Interferon-Beta-Inducing Copyback-Type Defective Interfering Genomes. J. Virol. 2018, 92, e02094-17. [Google Scholar] [CrossRef] [PubMed]
- Yount, J.S.; Gitlin, L.; Moran, T.M.; López, C.B. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai Virus defective interfering particles. J. Immunol. 2008, 180, 4910–4918. [Google Scholar] [CrossRef] [PubMed]
- Tapia, K.; Kim, W.K.; Sun, Y.; Mercado-López, X.; Dunay, E.; Wise, M.; Adu, M.; López, C.B. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog. 2013, 9, e1003703. [Google Scholar] [CrossRef] [PubMed]
- Marcus, P.I.; Sekellick, M.J. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 1977, 266, 815–819. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Xu, J.; Sun, Y.; Li, Y.; Ruthel, G.; Weiss, S.R.; Raj, A.; Beiting, D.; López, C.B. Replication defective viral genomes exploit a cellular pro-survival mechanism to establish paramyxovirus persistence. Nat. Commun. 2017, 8, 799. [Google Scholar] [CrossRef]
- Blue, C.E.; Spiller, O.B.; Blackbourn, D.J. The relevance of complement to virus biology. Virology 2004, 319, 176–184. [Google Scholar] [CrossRef]
- Gasque, P. Complement: A unique innate immune sensor for danger signals. Mol. Immunol. 2004, 41, 1089–1098. [Google Scholar] [CrossRef]
- Fox, C.R.; Parks, G.D. Complement Inhibitors Vitronectin and Clusterin Are Recruited from Human Serum to the Surface of Coronavirus OC43-Infected Lung Cells through Antibody-Dependent Mechanisms. Viruses 2021, 14, 29. [Google Scholar] [CrossRef]
- Fox, C.R.; Yousef, N.N.; Varudkar, N.; Shiffer, E.M.; Aquino, J.R.; Kedarinath, K.; Parks, G.D. Resistance to complement-mediated lysis of parainfluenza virus 5-infected cells is acquired after transition from acute to persistent infection. J. Virol. 2025, 99, e0189524. [Google Scholar] [CrossRef]
- Manuse, M.J.; Parks, G.D. Role for the paramyxovirus genomic promoter in limiting host cell antiviral responses and cell killing. J. Virol. 2009, 83, 9057–9067. [Google Scholar] [CrossRef] [PubMed]
- Wignall-Fleming, E.B.; Vasou, A.; Young, D.; Short, J.A.L.; Hughes, D.J.; Goodbourn, S.; Randall, R.E. Innate Intracellular Antiviral Responses Restrict the Amplification of Defective Virus Genomes of Parainfluenza Virus 5. J. Virol. 2020, 94, 13. [Google Scholar] [CrossRef]
- Fox, C.R.; Parks, G.D. Histone Deacetylase Inhibitors Enhance Cell Killing and Block Interferon-Beta Synthesis Elicited by Infection with an Oncolytic Parainfluenza Virus. Viruses 2019, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Wansley, E.K.; Parks, G.D. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 2002, 76, 10109–10121. [Google Scholar] [CrossRef]
- Parks, G.D.; Young, V.A.; Koumenis, C.; Wansley, E.K.; Layer, J.L.; Cooke, K.M. Controlled cell killing by a recombinant nonsegmented negative-strand RNA virus. Virology 2002, 293, 192–203. [Google Scholar] [CrossRef][Green Version]
- Murphy, S.K.; Parks, G.D. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology 1997, 232, 145–157. [Google Scholar] [CrossRef]
- Robinson, R.; Dowdle, W.; Lennette, E.; Schmidt, H. Diagnostic Procedures for Viral and Rickettsial Infections; Lennette, E.H., Schmidt, N.J., Eds.; American Public Health Association: New York, NY, USA, 1969; p. 414. [Google Scholar]
- Capraro, G.A.; Johnson, J.B.; Kock, N.D.; Parks, G.D. Virus growth and antibody responses following respiratory tract infection of ferrets and mice with WT and P/V mutants of the paramyxovirus Simian Virus 5. Virology 2008, 376, 416–428. [Google Scholar] [CrossRef]
- Parks, G.D.; Ward, K.R.; Rassa, J.C. Increased readthrough transcription across the simian virus 5 M-F gene junction leads to growth defects and a global inhibition of viral mRNA synthesis. J. Virol. 2001, 75, 2213–2223. [Google Scholar] [CrossRef]
- Fox, C.R.; Parks, G.D. Parainfluenza Virus Infection Sensitizes Cancer Cells to DNA-Damaging Agents: Implications for Oncolytic Virus Therapy. J. Virol. 2018, 92, 7. [Google Scholar] [CrossRef]
- Von Magnus, P. Incomplete forms of influenza virus. Adv. Virus Res. 1954, 2, 59–79. [Google Scholar] [CrossRef]
- Devaux, P.; Christiansen, D.; Plumet, S.; Gerlier, D. Cell surface activation of the alternative complement pathway by the fusion protein of measles virus. J. Gen. Virol. 2004, 85 Pt 6, 1665–1673. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.; Wolinsky, J.; Winkelstein, J. Activation of the alternative complement pathway by mumps infected cells: Relationship to viral neuraminidase activity. Arch. Virol. 1986, 87, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.B.; Capraro, G.A.; Parks, G.D. Differential mechanisms of complement-mediated neutralization of the closely related paramyxoviruses simian virus 5 and mumps virus. Virology 2008, 376, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Mc Sharry, J.J.; Pickering, R.J.; Caliguiri, L.A. Activation of the alternative complement pathway by enveloped viruses containing limited amounts of sialic acid. Virology 1981, 114, 507–515. [Google Scholar] [CrossRef]
- Johnson John, B.; Schmitt Anthony, P.; Parks Griffith, D. Point Mutations in the Paramyxovirus F Protein That Enhance Fusion Activity Shift the Mechanism of Complement-Mediated Virus Neutralization. J. Virol. 2013, 87, 9250–9259. [Google Scholar] [CrossRef]
- Cruz, M.A.; Parks, G.D. La Crosse Virus Infection of Human Keratinocytes Leads to Interferon-Dependent Apoptosis of Bystander Non-Infected Cells In Vitro. Viruses 2020, 12, 253. [Google Scholar] [CrossRef]
- Ho, T.-H.; Kew, C.; Lui, P.-Y.; Chan, C.-P.; Satoh, T.; Akira, S.; Jin, D.-Y.; Kok, K.-H. PACT- and RIG-I-Dependent Activation of Type I Interferon Production by a Defective Interfering RNA Derived from Measles Virus Vaccine. J. Virol. 2016, 90, 1557–1568. [Google Scholar] [CrossRef]
- Shivakoti, R.; Siwek, M.; Hauer, D.; Schultz, K.L.; Griffin, D.E. Induction of dendritic cell production of type I and type III interferons by wild-type and vaccine strains of measles virus: Role of defective interfering RNAs. J. Virol. 2013, 87, 7816–7827. [Google Scholar] [CrossRef]
- Xiao, Y.; Lidsky, P.V.; Shirogane, Y.; Aviner, R.; Wu, C.T.; Li, W.; Zheng, W.; Talbot, D.; Catching, A.; Doitsh, G.; et al. A defective viral genome strategy elicits broad protective immunity against respiratory viruses. Cell 2021, 184, 6037–6051.e14. [Google Scholar] [CrossRef]
- Brennan, J.W.; Sun, Y. Defective viral genomes: Advances in understanding their generation, function, and impact on infection outcomes. mBio 2024, 15, e0069224. [Google Scholar] [CrossRef]
- Andzhaparidze, O.G.; Boriskin Yu, S.; Bogomolova, N.N.; Drynov, I.D. Mumps virus-persistently infected cell cultures release defective interfering virus particles. J. Gen. Virol. 1982, 63, 499–503. [Google Scholar] [CrossRef]
- Fodor, E.; Mingay, L.J.; Crow, M.; Deng, T.; Brownlee, G.G. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase promotes the generation of defective interfering RNAs. J. Virol. 2003, 77, 5017–5020. [Google Scholar] [CrossRef] [PubMed]
- Girgis, S.; Xu, Z.; Oikonomopoulos, S.; Fedorova, A.D.; Tchesnokov, E.P.; Gordon, C.J.; Schmeing, T.M.; Götte, M.; Sonenberg, N.; Baranov, P.V.; et al. Evolution of naturally arising SARS-CoV-2 defective interfering particles. Commun. Biol. 2022, 5, 1140. [Google Scholar] [CrossRef] [PubMed]
- Hillung, J.; Olmo-Uceda, M.J.; Muñoz-Sánchez, J.C.; Elena, S.F. Accumulation dynamics of defective genomes during experimental evolution of two betacoronaviruses. bioRxiv 2024, 16, 644. [Google Scholar] [CrossRef]
- Odagiri, T.; Tobita, K. Mutation in NS2, a nonstructural protein of influenza A virus, extragenically causes aberrant replication and expression of the PA gene and leads to generation of defective interfering particles. Proc. Natl. Acad. Sci. USA 1990, 87, 5988–5992. [Google Scholar] [CrossRef]
- Pfaller, C.K.; Mastorakos, G.M.; Matchett, W.E.; Ma, X.; Samuel, C.E.; Cattaneo, R. Measles Virus Defective Interfering RNAs Are Generated Frequently and Early in the Absence of C Protein and Can Be Destabilized by Adenosine Deaminase Acting on RNA-1-Like Hypermutations. J. Virol. 2015, 89, 7735–7747. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Mounce, B.C.; Rozen-Gagnon, K.; Hooikaas, P.J.; Stapleford, K.A.; Moratorio, G.; Vignuzzi, M. Low-Fidelity Polymerases of Alphaviruses Recombine at Higher Rates To Overproduce Defective Interfering Particles. J. Virol. 2015, 90, 2446–2454. [Google Scholar] [CrossRef]
- Vignuzzi, M.; López, C.B. Defective viral genomes are key drivers of the virus-host interaction. Nat. Microbiol. 2019, 4, 1075–1087. [Google Scholar] [CrossRef]
- Lebel, M.; Langlois, M.P.; Daudelin, J.F.; Tarrab, E.; Savard, P.; Leclerc, D.; Lamarre, A. Complement Component 3 Regulates IFN-α Production by Plasmacytoid Dendritic Cells following TLR7 Activation by a Plant Virus-like Nanoparticle. J. Immunol. 2017, 198, 292–299. [Google Scholar] [CrossRef]
- Jodele, S.; Medvedovic, M.; Luebbering, N.; Chen, J.; Dandoy, C.E.; Laskin, B.L.; Davies, S.M. Interferon-complement loop in transplant-associated thrombotic microangiopathy. Blood Adv. 2020, 4, 1166–1177. [Google Scholar] [CrossRef]
- Li, T.; Pattnaik, A.K. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: Identification of a sequence element that enhances DI RNA replication. Virology 1997, 232, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Calain, P.; Roux, L. Functional characterisation of the genomic and antigenomic promoters of Sendai virus. Virology 1995, 212, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.S.; Baltimore, D. Defective Viral Particles and Viral Disease Processes. Nature 1970, 226, 325–327. [Google Scholar] [CrossRef]
- Genoyer, E.; López, C.B. Defective Viral Genomes Alter How Sendai Virus Interacts with Cellular Trafficking Machinery, Leading to Heterogeneity in the Production of Viral Particles among Infected Cells. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef]
- Carroll, M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004, 5, 981–986. [Google Scholar] [CrossRef]
- Kemper, C.; Atkinson, J.P. T-cell regulation: With complements from innate immunity. Nat. Rev. Immunol. 2007, 7, 9–18. [Google Scholar] [CrossRef]



| Name | F′ Primer | R′ Primer |
|---|---|---|
| GAPDH | 5′-TTAAAAGCAGCCCTGGTGAC-3′ | 5′-CTCTGCTCCTGTTCGAC-3′ |
| F | 5′-ACGTGTTATGGTGACTGGCA-3′ | 5′-GAACAGCACGAATCGAGTGA-3′ |
| HN | 5′-TGACCAACCCTTCGTCTACC-3′ | 5′-CTTGACCGCTTGATCCAAAT-3′ |
| NP | 5′-TGACCAGTCACCAGAAGCTG-3′ | 5′-CGGAATCAACGAAAGGTGTT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aquino, J.R.; Fox, C.R.; Parks, G.D. Role of Defective Interfering Particles in Complement-Mediated Lysis of Parainfluenza Virus-Infected Cells. Viruses 2025, 17, 488. https://doi.org/10.3390/v17040488
Aquino JR, Fox CR, Parks GD. Role of Defective Interfering Particles in Complement-Mediated Lysis of Parainfluenza Virus-Infected Cells. Viruses. 2025; 17(4):488. https://doi.org/10.3390/v17040488
Chicago/Turabian StyleAquino, Jenna R., Candace R. Fox, and Griffith D. Parks. 2025. "Role of Defective Interfering Particles in Complement-Mediated Lysis of Parainfluenza Virus-Infected Cells" Viruses 17, no. 4: 488. https://doi.org/10.3390/v17040488
APA StyleAquino, J. R., Fox, C. R., & Parks, G. D. (2025). Role of Defective Interfering Particles in Complement-Mediated Lysis of Parainfluenza Virus-Infected Cells. Viruses, 17(4), 488. https://doi.org/10.3390/v17040488







