Complete Genome Sequencing of the Divergent Guiana Dolphin Morbillivirus (GDMV), Brazil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material
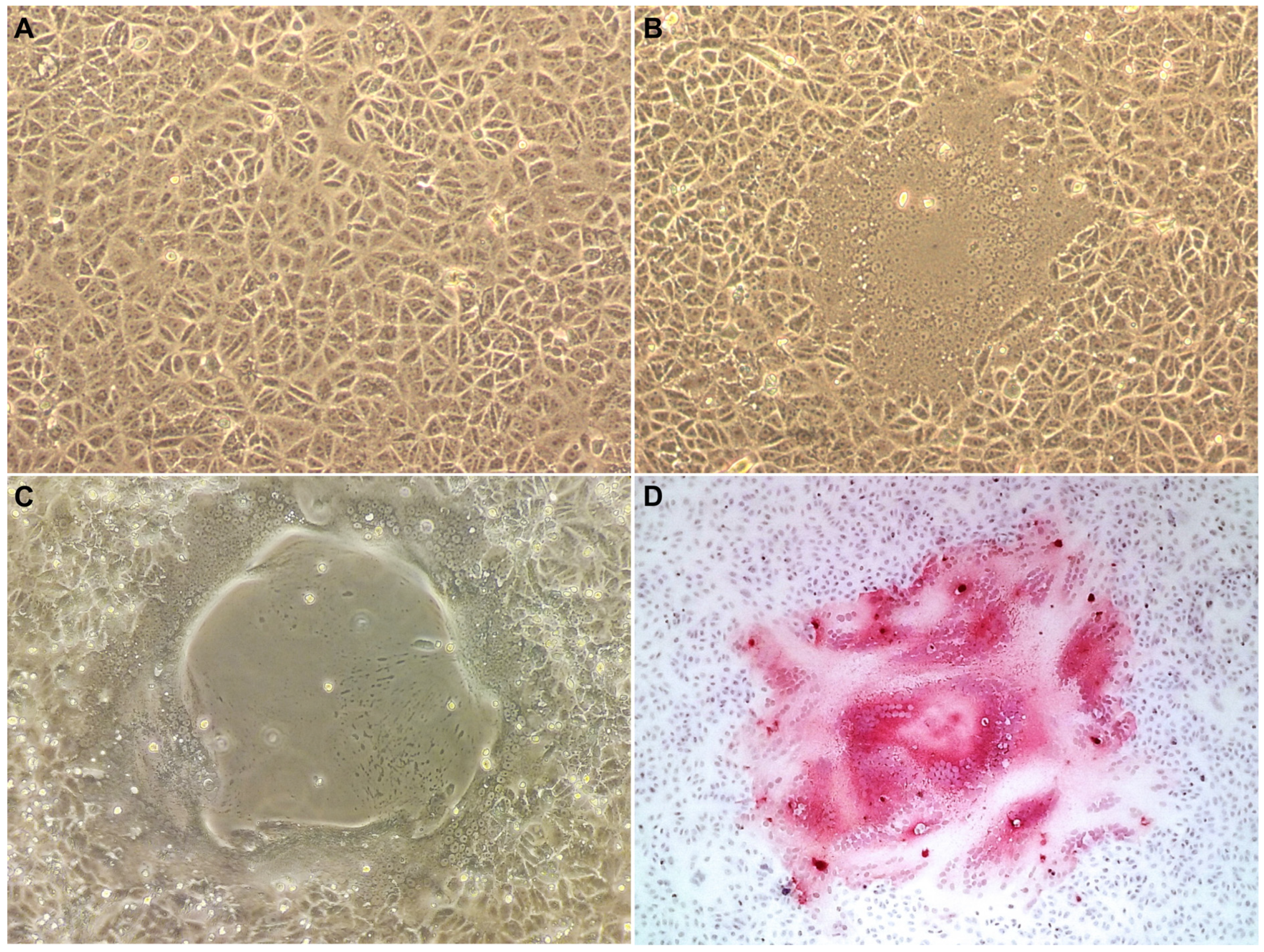
2.2. Viral Isolation
2.3. Immunohistochemical Analysis
2.4. RNA Extraction and Deep Sequencing Analysis
2.5. Primer Design for Genome Gaps Closure
2.6. Conventional RT-PCR
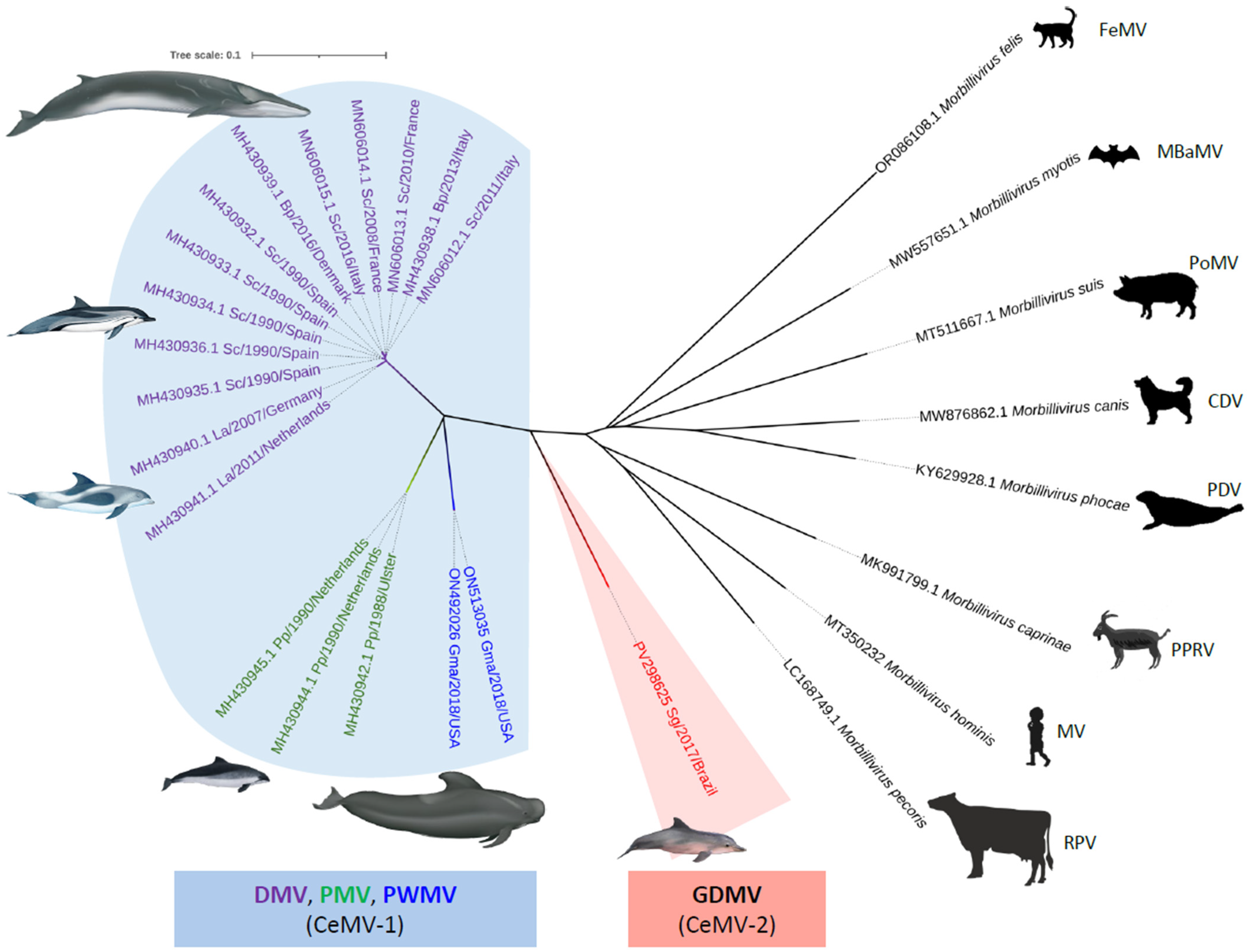
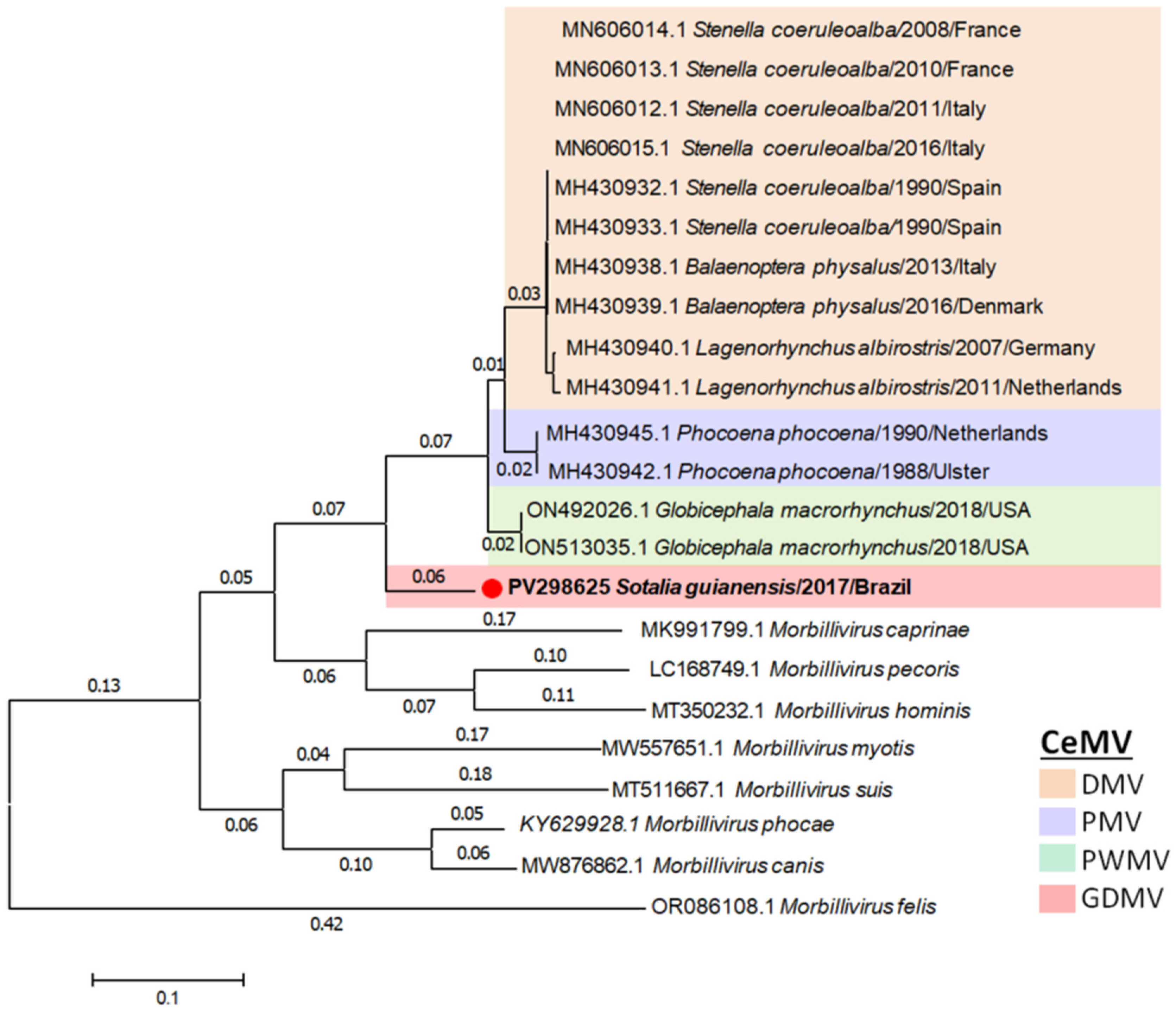
2.7. Phylogenetic Analysis
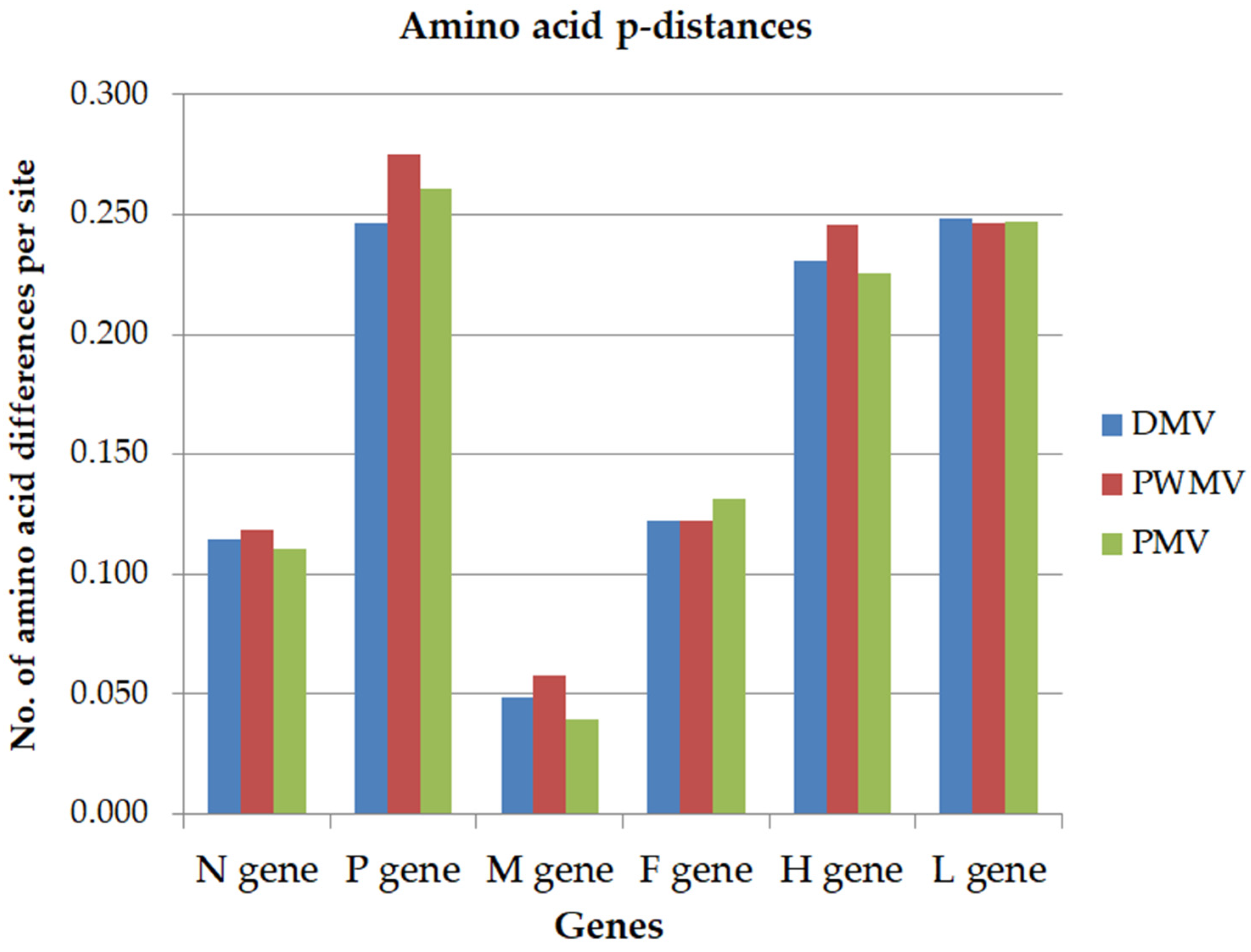
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plemper, R.K.; Lamb, R.A. Paramyxoviridae: The viruses and their replication. In Fields Virology: Emerging Viruses, 7th ed.; Howley, P.M., Knipe, D.M., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2021; Volume 1, pp. 504–558. [Google Scholar]
- de Vries, R.D.; Duprex, W.P.; de Swart, R.L. Morbillivirus infections: An introduction. Viruses 2015, 7, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Domingo, M.; Ferrer, L.; Pumarola, M.; Marco, A.; Plana, J.; Kennedy, S.; McAliskey, M.; Rima, B.K. Morbillivirus in dolphins. Nature 1990, 348, 21. [Google Scholar] [CrossRef]
- McCullough, S.J.; McNeilly, F.; Allan, G.M.; Kennedy, S.; Smyth, J.A.; Cosby, S.L.; McQuaid, S.; Rima, B.K. Isolation and characterisation of a porpoise morbillivirus. Arch. Virol. 1991, 118, 247–252. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Tsai, M.M.; Atkin, T.J.; Fanning, T.G.; Krafft, A.E.; Moeller, R.B.; Kodsi, S.E.; Mense, M.G.; Lipscomb, T.P. Molecular genetic evidence of a novel morbillivirus in a long-finned pilot whale (Globicephalus melas). Emerg. Infect. Dis. 2000, 6, 42–45. [Google Scholar] [CrossRef]
- Groch, K.R.; Colosio, A.C.; Marcondes, M.C.C.; Zucca, D.; Diaz-Delgado, J.; Niemeyer, C.; Marigo, J.; Brandao, P.E.; Fernandez, A.; Catão-Dias, J.L. Novel cetacean morbillivirus in Guiana dolphin, Brazil. Emerg. Infect. Dis. 2014, 20, 511–513. [Google Scholar] [CrossRef]
- Stephens, N.; Duignan, P.J.; Wang, J.; Bingham, J.; Finn, H.; Bejder, L.; Patterson, A.P.; Holyoake, C. Cetacean morbillivirus in coastal Indo-Pacific bottlenose dolphins, Western Australia. Emerg. Infect. Dis. 2014, 20, 666–670. [Google Scholar] [CrossRef] [PubMed]
- West, K.L.; Silva-Krott, I.; Landrau-Giovannetti, N.; Rotstein, D.; Saliki, J.; Raverty, S.; Nielsen, O.; Popov, V.L.; Davis, N.; Walker, W.A.; et al. Novel cetacean morbillivirus in a rare Fraser’s dolphin (Lagenodelphis hosei) stranding from Maui, Hawai’i. Sci. Rep. 2021, 11, 15986. [Google Scholar]
- Rima, B.K.; Collin, A.M.; Earle, J.A. Completion of the sequence of a cetacean morbillivirus and comparative analysis of the complete genome sequences of four morbilliviruses. Virus Genes 2005, 30, 113–119. [Google Scholar] [CrossRef]
- Barrett, T.; Visser, I.K.G.; Mamaev, L.; Goatley, L.; Bressem, M.-F.V.; Osterhaus, A.D.M.E. Dolphin and porpoise morbilliviruses are genetically distinct from phocine distemper virus. Virology 1993, 193, 1010–1012. [Google Scholar] [CrossRef]
- Krafft, A.; Lichy, J.H.; Lipscomb, T.P.; Klaunberg, B.A.; Kennedy, S.; Taubenberger, J.K. Postmortem diagnosis of morbillivirus infection in bottlenose dolphins (Tursiops truncatus) in the Atlantic and Gulf of Mexico epizootics by polymerase chain reaction-based assay. J. Wildl. Dis. 1995, 31, 410–415. [Google Scholar] [CrossRef]
- Saliki, J.T.; Cooper, E.J.; Gustavson, J.P. Emerging morbillivirus infections of marine mammals—Development of two diagnostic approaches. Ann. N. Y. Acad. Sci. 2002, 969, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Tsai, M.; Krafft, A.E.; Lichy, J.H.; Reid, A.H.; Schulman, Y.; Lipscomb, T.P. Two morbilliviruses implicated in bottlenose dolphin epizootics. Emerg. Infect. Dis. 1996, 2, 213–216. [Google Scholar]
- Groch, K.R.; Taniwaki, S.A.; Favero, C.M.; Brandao, P.E.; Diaz-Delgado, J.; Fernandez, A.; Catao-Dias, J.L.; Sierra, E. A novel real-time PCR to detect Cetacean morbillivirus in Atlantic cetaceans. J. Virol. Methods 2020, 285, 113964. [Google Scholar] [CrossRef]
- Van Bressem, M.F.; Visser, I.K.; Van de Bildt, M.W.; Teppema, J.S.; Raga, J.A.; Osterhaus, A.D. Morbillivirus infection in Mediterranean striped dolphins (Stenella coeruleoalba). Vet. Rec. 1991, 129, 471–472. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, O.; Smith, G.; Weingartl, H.; Lair, S.; Measures, L. Use of a SLAM transfected Vero cell line to isolate and characterize marine mammal morbilliviruses using an experimental ferret model. J. Wildl. Dis. 2008, 44, 600–611. [Google Scholar] [CrossRef]
- Jo, W.K.; Kruppa, J.; Habierski, A.; van de Bildt, M.; Mazzariol, S.; Di Guardo, G.; Siebert, U.; Kuiken, T.; Jung, K.; Osterhaus, A.; et al. Evolutionary evidence for multi-host transmission of cetacean morbillivirus. Emerg. Microbes Infect. 2018, 7, 201. [Google Scholar] [CrossRef] [PubMed]
- Peletto, S.; Caruso, C.; Cerutti, F.; Modesto, P.; Biolatti, C.; Pautasso, A.; Grattarola, C.; Giorda, F.; Mazzariol, S.; Mignone, W. Efficient isolation on Vero. DogSLAMtag cells and full genome characterization of Dolphin Morbillivirus (DMV) by next generation sequencing. Sci. Rep. 2018, 8, 860. [Google Scholar] [CrossRef]
- Groch, K.R.; Santos-Neto, E.B.; Díaz-Delgado, J.; Ikeda, J.M.P.; Carvalho, R.R.; Oliveira, R.B.; Guari, E.B.; Bisi, T.L.; Azevedo, A.F.; Lailson-Brito, J.; et al. Guiana dolphin unusual mortality event and link to cetacean morbillivirus, Brazil. Emerg. Infect. Dis. 2018, 24, 1349–1354. [Google Scholar] [CrossRef]
- Groch, K.R.; Diaz-Delgado, J.; Santos-Neto, E.B.; Ikeda, J.M.P.; Carvalho, R.R.; Oliveira, R.B.; Guari, E.B.; Flach, L.; Sierra, E.; Godinho, A.I.; et al. The pathology of cetacean morbillivirus infection and comorbidities in Guiana dolphins during an unusual mortality event (Brazil, 2017–2018). Vet. Pathol. 2020, 57, 845–857. [Google Scholar] [CrossRef]
- Cunha, H.A.; Santos-Neto, E.B.; Carvalho, R.R.; Ikeda, J.M.; Groch, K.R.; Díaz-Delgado, J.; Guari, E.B.; Brião, J.A.; Oliveira, R.B.; Flach, L. Epidemiological features of the first Unusual Mortality Event linked to cetacean morbillivirus in the South Atlantic (Brazil, 2017–2018). Mar. Mamm. Sci. 2021, 37, 1375–1390. [Google Scholar] [CrossRef]
- Groch, K.R.; Kolesnikovas, C.K.M.; de Castilho, P.V.; Moreira, L.M.P.; Barros, C.; Morais, C.R.; Renault-Braga, E.P.; Sierra, E.; Fernandez, A.; Catao-Dias, J.L.; et al. Cetacean morbillivirus in Southern right whales, Brazil. Transbound. Emerg. Dis. 2019, 66, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Groch, K.R.; Blazquez, D.N.H.; Marcondes, M.C.C.; Santos, J.; Colosio, A.; Diaz Delgado, J.; Catao-Dias, J.L. Cetacean morbillivirus in Humpback whales’ exhaled breath. Transbound. Emerg. Dis. 2021, 68, 1736–1743. [Google Scholar] [CrossRef] [PubMed]
- de Amorim, D.B.; de Camargo, L.J.; Ribeiro, P.R.; Budaszewski, R.D.F.; Menegatt, J.C.O.; Paz, M.C.; de Castro, L.T.; Almeida, P.R.; Olegario, J.C.; Canal, C.W.; et al. Characterization of Cetacean Morbillivirus in Humpback Whales, Brazil. Emerg. Infect. Dis. 2024, 30, 1296–1298. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, S.; Sacristan, C.; Duarte-Benvenuto, A.; Ewbank, A.C.; Soares, R.M.; Carvalho, V.L.; P, V.C.; Cremer, M.J.; Vieira, J.V.; Lemos, G.G.; et al. Morbillivirus and coronavirus survey in stranded cetaceans, Brazil. PLoS ONE 2025, 20, e0316050. [Google Scholar] [CrossRef]
- Groch, K.R.; Jerdy, H.; Marcondes, M.C.; Barbosa, L.A.; Ramos, H.G.; Pavanelli, L.; Fornells, L.A.M.; Silva, M.B.; Souza, G.S.; Kanashiro, M.M.; et al. Cetacean morbillivirus infection in a killer whale (Orcinus orca) from Brazil. J. Comp. Pathol. 2020, 181, 26–32. [Google Scholar] [CrossRef]
- Díaz-Delgado, J.; Groch, K.R.; Ressio, R.; Riskallah, I.P.; Sierra, E.; Sacchini, S.; Quesada-Canales, Ó.; Arbelo, M.; Fernández, A.; Santos-Neto, E. Comparative immunopathology of Cetacean morbillivirus infection in free-ranging dolphins from Western Mediterranean, Northeast-Central, and Southwestern Atlantic. Front. Immunol. 2019, 10, 485. [Google Scholar] [CrossRef]
- Díaz-Delgado, J.; Groch, K.R.; Sierra, E.; Sacchini, S.; Zucca, D.; Quesada-Canales, Ó.; Arbelo, M.; Fernández, A.; Santos, E.; Ikeda, J. Comparative histopathologic and viral immunohistochemical studies on CeMV infection among Western Mediterranean, Northeast-Central, and Southwestern Atlantic cetaceans. PLoS ONE 2019, 14, e0213363. [Google Scholar] [CrossRef]
- Bellière, E.N.; Esperón, F.; Sánchez-Vizcaíno, J.M. Genetic comparison among dolphin morbillivirus in the 1990–1992 and 2006–2008 Mediterranean outbreaks. Infect. Genet. Evol. 2011, 11, 1913–1920. [Google Scholar] [CrossRef]
- Budaszewski, R.F.; Pinto, L.D.; Weber, M.N.; Caldart, E.T.; Alves, C.D.; Martella, V.; Ikuta, N.; Lunge, V.R.; Canal, C.W. Genotyping of canine distemper virus strains circulating in Brazil from 2008 to 2012. Virus Res. 2014, 180, 76–83. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Van Bressem, M.-F.; Duignan, P.J.; Banyard, A.; Barbieri, M.; Colgrove, K.M.; De Guise, S.; Di Guardo, G.; Dobson, A.; Domingo, M.; Fauquier, D.; et al. Cetacean morbillivirus: Current knowledge and future directions. Viruses 2014, 6, 5145–5181. [Google Scholar] [CrossRef] [PubMed]
- Bolt, G.; Blixenkrone-Møller, M.; Gottschalck, E.; Wishaupt, R.G.; Welsh, M.J.; Earle, J.A.P.; Rima, B.K. Nucleotide and deduced amino acid sequences of the matrix (M) and fusion (F) protein genes of cetacean morbilliviruses isolated from a porpoise and a dolphin. Virus Res. 1994, 34, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Rima, B.K.; Earle, J.A.; Baczko, K.; ter Meulen, V.; Liebert, U.G.; Carstens, C.; Carabana, J.; Caballero, M.; Celma, M.L.; Fernandez-Munoz, R. Sequence divergence of measles virus haemagglutinin during natural evolution and adaptation to cell culture. J. Gen. Virol. 1997, 78 Pt 1, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Tatsuo, H.; Ono, N.; Tanaka, K.; Yanagi, Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature 2000, 406, 893–897. [Google Scholar] [CrossRef]
- Noyce, R.S.; Bondre, D.G.; Ha, M.N.; Lin, L.-T.; Sisson, G.; Tsao, M.-S.; Richardson, C.D. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011, 7, e1002240. [Google Scholar] [CrossRef]
- Mühlebach, M.D.; Mateo, M.; Sinn, P.L.; Prüfer, S.; Uhlig, K.M.; Leonard, V.H.; Navaratnarajah, C.K.; Frenzke, M.; Wong, X.X.; Sawatsky, B. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 2011, 480, 530–533. [Google Scholar] [CrossRef]
- Ohishi, K.; Maruyama, T.; Seki, F.; Takeda, M. Marine Morbilliviruses: Diversity and Interaction with Signaling Lymphocyte Activation Molecules. Viruses 2019, 11, 606. [Google Scholar] [CrossRef]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P. ICTV virus taxonomy profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef]
- McIlhatton, M.A.; Curran, M.D.; Rima, B.K. Nucleotide sequence analysis of the large (L) genes of phocine distemper virus and canine distemper virus (corrected sequence). J. Gen. Virol. 1997, 78 Pt 3, 571–576. [Google Scholar] [CrossRef]
| Primer No. | nt Gap (nt Position) * | Primer Name | Target Gene | 5′ → 3′ Sequence (Sense) | Tm | nt Position * | Template [Reference] |
|---|---|---|---|---|---|---|---|
| 1 | 985 (1–985) | DMV-1F | 3′ leading | ACCARACAAAGYTGGSTARGG (+) | 58 | 1–21 | DMV [29] |
| 2 | GDMV-1R | N | CCTTGGTTTATTCCCTGGTGT (−) | 58 | 835–855 | GDMV [20] | |
| 3 | DMV-2R | N | AATAGTCATCCGCCTCATCC (−) | 57 | 479–498 | DMV [this study] | |
| 4 | DMV-3F | N | ACCCAGATGTCAGCATCAGA (+) | 58 | 388–407 | DMV [this study] | |
| 5 | CDV-3F | N | ACAGRATTGCYGAGGACYTRT (+) | 59 | 851–871 | CDV [30] | |
| 6 | CDV-4R | N | CARRATAACCATGTAYGGTGC (−) | 58 | 1036–1056 | CDV [30] | |
| 7 | 52 (4137–4189) | GDMV-5F | M | CACAAAGGTTCAGGGTGGTC (+) | 59 | 3894–3913 | GDMV [this study] |
| 8 | GDMV-5R | M | TGAACTCCTGTGGGACTGAA (−) | 58 | 4364–4383 | GDMV [this study] | |
| 9 | 150 (5462–5612) | GDMV-7F | F | CAAATCCATTGGGGAAATCT (+) | 53 | 5352–5371 | GDMV [this study] |
| 10 | GDMV-7R | F | TTGTTAATATCTCCGCCAAGAG (−) | 56 | 5992–6013 | GDMV [this study] | |
| 11 | 524 (7387–7911) | GDMV-10F | H | CCAAGACCGAGAAATCATAGAG (+) | 56 | 7136–7157 | GDMV [this study] |
| 12 | GDMV-10R | H | AACGGTTCATCTTTGTAGCC (−) | 56 | 8015–8034 | GDMV [this study] | |
| 13 | 550 (13,375–13,924) | GDMV-13F | L | CCCCTATCATAGAAAAGGATG (+) | 43 | 13,246–13,266 | GDMV [this study] |
| 14 | GDMV-13R | L | TTTAACTGCTCCTCTCCTGA (−) | 45 | 14,062–14,081 | GDMV [this study] |
| Target Gene | Primer Combinations a | Annealing Temperature | Consensus Obtained (bp) | nt Position in the Reference Genome b |
|---|---|---|---|---|
| 3′ leading and N | 1–2 followed by 1–3 | 55 | 854 | 108–787 |
| N | 1–2 followed by 4–2 | 55 | 388 | 434–801 |
| N | 1–2 followed by 4–6 | 55 | 334 | 555–888 |
| N | 5–6 | 59 | 122 | 867–989 |
| M | 7–8 | 55 | 405 | 3947–4351 |
| F | 9–10 | 52 | 512 | 5408–5919 |
| H | 11–12 | 55 | 717 | 7249–7965 |
| L | 13–14 | 52 | 728 | 13,301–14,030 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Groch, K.R.; Miyagi, S.A.T.; Díaz-Delgado, J.; Santos-Neto, E.B.; Lailson-Brito, J.; Brandão, P.E.; Catão-Dias, J.L. Complete Genome Sequencing of the Divergent Guiana Dolphin Morbillivirus (GDMV), Brazil. Viruses 2025, 17, 582. https://doi.org/10.3390/v17040582
Groch KR, Miyagi SAT, Díaz-Delgado J, Santos-Neto EB, Lailson-Brito J, Brandão PE, Catão-Dias JL. Complete Genome Sequencing of the Divergent Guiana Dolphin Morbillivirus (GDMV), Brazil. Viruses. 2025; 17(4):582. https://doi.org/10.3390/v17040582
Chicago/Turabian StyleGroch, Kátia Regina, Sueli Akemi Taniwaki Miyagi, Josué Díaz-Delgado, Elitieri B. Santos-Neto, José Lailson-Brito, Paulo Eduardo Brandão, and José Luiz Catão-Dias. 2025. "Complete Genome Sequencing of the Divergent Guiana Dolphin Morbillivirus (GDMV), Brazil" Viruses 17, no. 4: 582. https://doi.org/10.3390/v17040582
APA StyleGroch, K. R., Miyagi, S. A. T., Díaz-Delgado, J., Santos-Neto, E. B., Lailson-Brito, J., Brandão, P. E., & Catão-Dias, J. L. (2025). Complete Genome Sequencing of the Divergent Guiana Dolphin Morbillivirus (GDMV), Brazil. Viruses, 17(4), 582. https://doi.org/10.3390/v17040582







