Abstract
Gag proteins play an important role in many stages of the retroviral replication cycle. They orchestrate viral assembly, interact with numerous host cell proteins, engage in regulation of viral gene expression, and provide the main driving force for virus intracellular trafficking and budding. Foamy Viruses (FV), also known as spumaviruses, display a number of unique features among retroviruses. Many of these features can be attributed to their Gag proteins. FV Gag proteins lack characteristic orthoretroviral domains like membrane-binding domains (M domains), the major homology region (MHR), and the hallmark Cys-His motifs. In contrast, they contain several distinct domains such as the essential Gag-Env interaction domain and the glycine and arginine rich boxes (GR boxes). Furthermore, FV Gag only undergoes limited maturation and follows an unusual pathway for nuclear translocation. This review summarizes the known FV Gag domains and motifs and their functions. In particular, it provides an overview of the unique structural and functional properties that distinguish FV Gag proteins from orthoretroviral Gag proteins.
1. Introduction
While the retroviral Env mediates the important steps of receptor binding and membrane fusion, and Pol provides the key enzymatic functions required for viral replication, it is the Gag protein that is the major structural component of the viral particle. Gag functions are, however, not restricted to capsid formation; retroviral Gag proteins have numerous and very complex roles in the viral life cycle. Gag, (i) is the major component involved in intracellular trafficking processes; (ii) orchestrates viral assembly and disassembly; (iii) regulates viral gene expression; (iv) mediates correct encapsidation of Pol, the viral genome, and accessory proteins; (v) is involved in spatiotemporal regulation of the essential, viral, enzymatic reactions; and (vi) is essential for viral budding [reviewed in 1].
The family of Retroviridae (retroviruses) is divided into two subfamilies: the Orthoretrovirinae (orthoretroviruses) and the Spumaretrovirinae (spumaretroviruses). Whereas the subfamily of orthoretroviruses consists of six genera, the subfamily of spumaretroviruses consist of only one genus—the spumaviruses, also termed Foamy Viruses (FV). FV infection is endemic in many new- and old-world monkeys from which sporadic zoonotic transmissions to humans can occur. Transmission directly between humans has never been observed [reviewed in 2]. Other natural hosts include cats (feline FV, FFV), cattle (bovine FV, BFV) and horses (equine FV, EFV) [3]. Although FVs share the major characteristics of retroviruses in that they reverse-transcribe and integrate their genome, FV replication deviates in many aspects from that of orthoretroviruses [reviewed in 4,5]. In particular the FV Gag proteins have strikingly different domains and functions compared to orthoretroviral Gag, many of which are reminiscent of another reverse transcriptase encoding virus family—the hepadnaviruses [reviewed in 6].
This review summarizes the current knowledge of the different FV Gag domains and motifs and their functions, highlighting common features, but more importantly concentrating on the differences between foamyviral and orthoretroviral Gag proteins. The following parts will focus mainly on the features of prototype FV (PFV) Gag, as PFV is the best characterized isolate and represents the model for the FV family. PFV was isolated in the early 70s from a human nasopharyngal carcinoma [7], with the isolate initially named human FV (HFV), but was later renamed to PFV when large homologies to simian FV (SFV) of chimpanzees became apparent [8,9]. If amino acid positions are specified in the text these refer to the respective position in PFV Gag, however, statements are generalized if applicable.
2. Functional Domains of PFV Gag and Their Role in Viral Replication
Nascent FV Gag is translated as a precursor protein on free ribosomes in the cytoplasm from the unspliced, genomic RNA. In contrast to all other retroviruses, FV only express a Gag, and not a Gag-Pol fusion protein [10,11,12]. In general, the primate FV Gag proteins are slightly larger in size ranging from 572 aa (SFV of squirrel monkey) to 653 aa (SFV of chimpanzees) compared to 559 aa, 544 aa, and 489 aa for EFV, BFV and FFV Gag respectively. PFV Gag has a size of 648 aa. In contrast to orthoretroviruses, FV Gag is not processed into matrix (MA), capsid (CA) and nucleocapsid (NC). Instead, only a limited primary proteolysis by the FV protease (PR) occurs, which leads to the removal of a small C-terminal peptide [13,14]. In the case of PFV, cleavage of the 71 kDa precursor (p71Gag) results in a 68 kDa (p68Gag) and a 3 kDa (p3Gag) cleavage product. FV Gag processing is essential for viral infectivity [15,16,17]. In infectious PFV particles, p71Gag and p68Gag are present in a ratio between 1:1 and 1:4 [4,18]. Due to its small size it is unknown if p3Gag is present in released viral particles. It is also unknown if it has a specific function in the viral life cycle.
In addition to the primary Gag cleavage site, three secondary cleavage sites were identified in the central Gag region at positions 311/312, 339/340, and 352/353. Processing at these sites was shown for primate and non-primate FV isolates [14,19]. Secondary processing occurs with much lower efficiency and C-terminal p68Gag/p3Gag processing seems to be a prerequisite for further proteolysis [14]. The importance of secondary FV Gag processing has been debated. Mutations at any of the three secondary cleavage sites do not abrogate particle release, but lead to noninfectious viruses [14,20]. Interestingly, despite their close proximity, a mutation in one cannot be functionally substituted by the presence of the two other cleavage sites. This suggests that all three secondary cleavage sites have an important function. Initially it was thought that further Gag processing by Pol at secondary cleavage sites was required for capsid disassembly upon target cell entry [13,14,20]. However, we recently found that viral protease activity is fully dispensable during target cell entry [21]. Thus, the exact function of secondary Gag cleavage remains to be determined. Despite the lack of processing into mature subunits orthoretroviral-like MA, CA and NC domains were initially assigned to the N-terminal part, the central region, and the C-terminal part of FV Gag, respectively, because these different parts of FV Gag harbor similar motifs and often perform similar functionality as the orthoretroviral MA, CA, and NC subunits (Figure 1) [reviewed in 4].
Sequence homology between different retroviral Gag proteins is quite low. In line with that, Gag proteins of different foamyviral isolates have on average of less than 50% amino acid similarity. However, as all retroviral Gag proteins perform similar functions to achieve viral replication, similar functional domains are present in most retroviral Gag proteins (Figure 1).
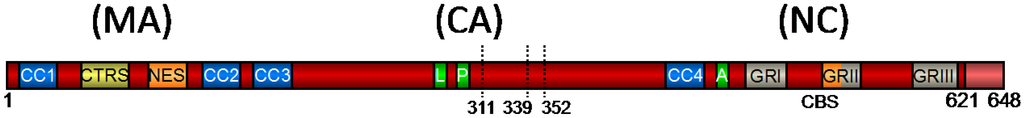
Figure 1.
Schematic Representation of the Prototype Foamy Virus (PFV) Gag Precursor Protein [adapted from 22]. Numbers indicate the amino acid position in PFV Gag. The primary cleavage site is indicated by its amino acid position (621). Secondary cleavage sites are indicated by dotted lines and their amino acid position. CC1 – CC4, predicted coiled-coil motifs 1–4; CTRS, cytoplasmic targeting and retention signal; NES, nuclear export signal; L, late assembly domain; P, Pro-Pro-Pro-Ile motif (PPPI motif); A, assembly domain; GRI–III, glycine-arginine boxes I–III; CBS, chromatin-binding site; MA, matrix domain; CA, capsid domain; NC, nucleocapsid domain.
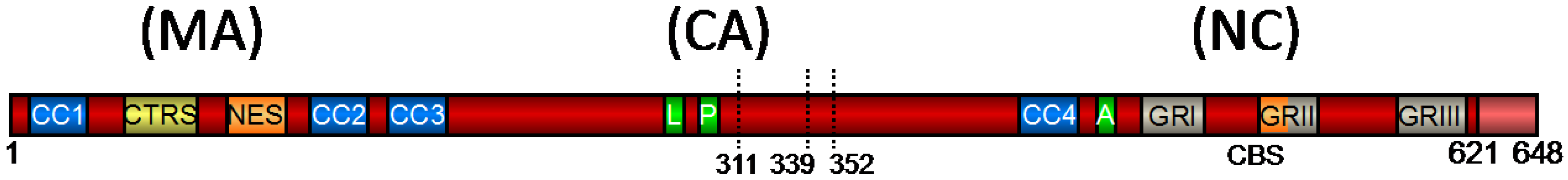
Figure 1.
Schematic Representation of the Prototype Foamy Virus (PFV) Gag Precursor Protein [adapted from 22]. Numbers indicate the amino acid position in PFV Gag. The primary cleavage site is indicated by its amino acid position (621). Secondary cleavage sites are indicated by dotted lines and their amino acid position. CC1 – CC4, predicted coiled-coil motifs 1–4; CTRS, cytoplasmic targeting and retention signal; NES, nuclear export signal; L, late assembly domain; P, Pro-Pro-Pro-Ile motif (PPPI motif); A, assembly domain; GRI–III, glycine-arginine boxes I–III; CBS, chromatin-binding site; MA, matrix domain; CA, capsid domain; NC, nucleocapsid domain.
2.1. FV Gag Domains Found Also in Orthoretroviruses
To begin our overview of the FV Gag domains we shall briefly review common Gag domains, which FV Gag shares with some of its orthoretroviral counterparts. These are the domains for incorporation of the envelope protein (Env), the cytoplasmic targeting and retention signal (CTRS) that directs Gag to the region where particle assembly occurs, interaction (I) domains that are essential for Gag-Gag interactions during capsid assembly, and late (L) domains that are required for particle release [23].
Several studies show that the N terminus of FV Gag is essential for the interaction of Gag with Env [18,24,25,26]. Thereby, the structural integrity of a predicted coiled-coil motif (CC1) at aa 4–19 seems to be important for the interaction [18]. Other retroviral Gag proteins, like HIV-1 Gag, contain similar domains required for efficient Env incorporation in the Gag N terminus [1]. However, FV are unique among retroviruses as FV Gag lacks a membrane-targeting domain (see below). Therefore, viral budding requires the co-expression and interaction of Gag and Env [27,28]. This feature is most closely resembled by Mason Pfizer Monkey Virus (MPMV), where although Env co-expression is not required, it was shown to enhance viral particle release by mediating capsid transport [29]. Similarly, it is thought that FV Env interacts with the preassembled capsids at a pericentriolar region, probably the trans-Golgi network, which then leads to membrane targeting of the fully assembled viral particles [30].
Like for B/D type orthoretroviruses, FV Gag preassembles into capsid structures in the cytoplasm at the centrosome [18,30,31]. Targeting of assembly is mediated by a CTRS at aa 43–60. The CTRS comprises of a [GXWGX3RX7L(Q/V)D] motif centered around an essential arginine residue, which is conserved among all known FV Gag proteins [32]. The presence of a CTRS in the Gag N terminus is reminiscent of MPMV, which also uses a B/D‑type egress strategy [33].
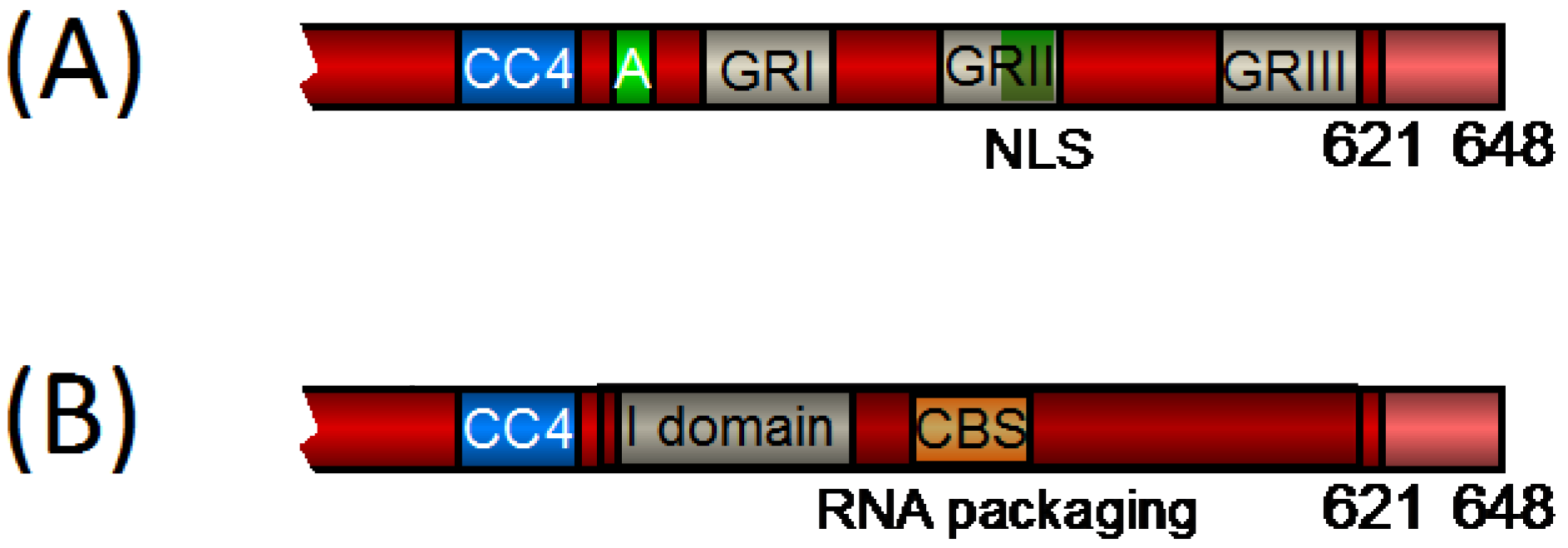
Viral I domains are essential for capsid formation, as they mediate Gag-Gag interaction and thereby Gag multimerization. They contain high amounts of basic amino acids and can be functionally substituted between different retroviruses. The I domains function to provide the proper density to viral particles. It is thought that they influence particle density by interacting with nucleic acids and using them as a scaffold to facilitate the subsequent Gag-Gag protein interactions [reviewed in 34]. Most orthoretroviral I domains are found in the C-terminal part of the CA subunit and in the NC subunit. Similarly, FV Gag contains an I domain in the very C-terminal part of the central region. The CC2, found at aa 130–150 was shown to be essential for Gag-Gag interactions [35]. Additional regions with putative I domain function can be found in the C-terminal part of FV Gag, the respective NC domain (also see Figure 1). At aa 464–469 a Tyr-X-X-Leu-Gly-Leu motif (YXXLGL motif) with important functions for capsid assembly was found. According to its function the YXXLGL motif was termed ‘assembly domain’ [35,36,37]. The assembly domain is well conserved among all FV species and was attributed to I domain function [37]. In close proximity to the assembly motif lies glycine and arginine rich box (GR box) I (GRI) (aa 485 – 512), which is also required for correct capsid assembly [38]. Notably, orthoretroviral I domains often overlap with the Cys-His motifs [36] and the FV GR boxes are the functional equivalent of the Cys-His motifs (also see below) [38,39]. While according to the original definition, GRI is not an I domain, because particles with mutations in GRI appear at the same buoyant density as wild-type particles [38], Cimarelli and Luban showed for HIV‑1 Gag that the determinants of the NC I domain and of virion density are genetically distinguishable [40]. Thus, it is possible that the PFV Gag region comprising the YXXLGL motif as well as GRI together form a larger I domain with functions in particle assembly and particle morphology, but not for particle density.
The FV Gag assembly motif was initially discovered together with two additional motifs, when sequences that show homologies to known L domains were analyzed [41,42]. L domains function during the last step of the viral replication cycle. They mediate interaction with components of the vacuolar protein sorting (VPS) machinery for particle release [reviewed in 43]. L domains are found in different domains of the retroviral Gag and some Gag proteins contain several L domains. While the L domains of lentiviruses are located mainly in the C terminus within Gag, those of oncoretroviruses and FVs are often found in the central region and the N-terminal half of the Gag precursor [reviewed in 43]. L domains act independently of their respective localization in Gag and can be functionally substituted between different retroviruses [reviewed in 44,45]. PFV Gag contains a Pro-Ser-Ala-Pro motif (PSAP motif) with L domain function at aa 284–287 [41,42]. All known SFV Gag proteins except SFV of orangutan (SFVora) Gag contain PSAP motifs in the central region. In addition, SFVora Gag and also SFV of spider monkeys contain a PSAP motif in the very C terminus [41,46]. The isolates of bovine, equine, and feline FV lack both PSAP and also Pro-Pro-X-Tyr sequence motifs. They do, however, contain several Leu-X-X-Leu motifs, but these are not well conserved [41]. As the conserved YXXLGL motif has no classical L domain function (see above) it is currently unknown if and how these FV isolates utilize the VPS for particle release.
An additional well-conserved motif in primate FV Gag that was initially thought to have L domain function is a Pro-Pro-Pro-Ile motif (PPPI motif) at aa 298–301. This motif clearly is not an L domain, as it was found to be dispensable for particle release. However, mutant particles have strongly diminished infectivity [37,41,42].
2.4. Many Ways Lead to the Nucleus—FV Gag Nuclear Trafficking
FV Gag nuclear localization has long been known as a characteristic feature of FV infection [70]. In fact, the nuclear accumulation of the FV Gag protein is a diagnostic feature of FV in cell culture and previously it was regarded as a unique feature of FVs compared to orthoretroviruses [61,70]. However, nucleo-cytoplasmic shuttling was later shown for the Gag proteins of several retroviruses including HIV-1, MLV, MMTV, and RSV [61,71,72,73,74].
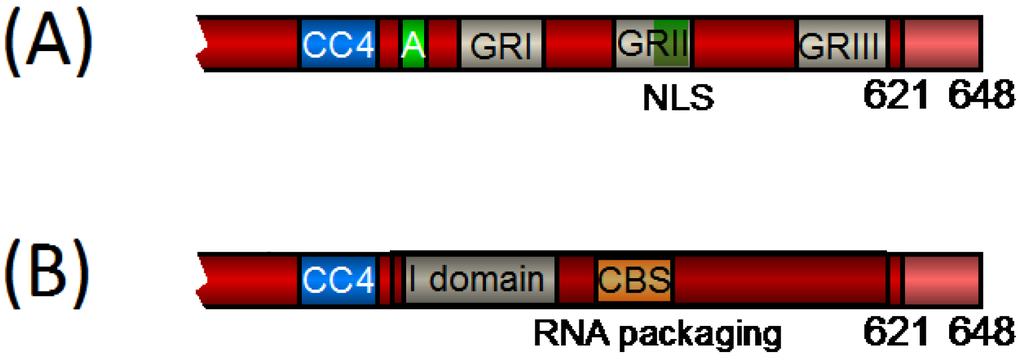
The Gag proteins of PFV, simian FV of macaques, EFV, and BFV were all shown to transiently traffic through the nucleus [3,31,62,75], while a lack of nuclear localization was reported only for FFV Gag [76]. As GRII, and especially its CBS subdomain, are better conserved in FFV Gag than in EFV or BFV Gag (Figure 2), the lack of nuclear localization of FFV Gag is most probably not caused by poor sequence conservation. While nuclear localization seems to be a common property of retroviral Gag proteins the mechanism of FV Gag nuclear translocation is unique. Initially, PFV GRII was reported to harbor a nuclear localization signal (NLS) in its C-terminal region responsible for transient nuclear targeting of PFV Gag during certain stages of the viral replication cycle [39,61]. However, recently, it has been highlighted that PFV Gag is not imported into the interphase cell nucleus and it does not contain a functional NLS. Gag nuclear translocation occurs by tethering of Gag via its CBS to accessible chromatin during mitosis [77,78]. This mechanism occurs for endogenously expressed Gag in the infected cell, as well as for Gag delivered by infecting viral particles [77,79]. Gag-chromatin association is mediated by binding of the PFV Gag CBS to H2A/H2B core histones [78].
Also very recently, a leptomycin B (LMB)-sensitive nuclear export sequence (NES) was identified in the N terminus of PFV Gag, at amino acid positions 95–112 [80]. An NES-defective Gag G110V mutant was retained in the cell nucleus and prevented viral particle release in a dominant-negative manner. Interestingly, while substitution of the PFV Gag NES with that of HIV-1 Rev led to nucleocytoplasmic redistribution of Gag, viral particle release and viral infectivity could not be restored [80]. Currently, it remains unclear how the CBS and the NES function are regulated during the viral life cycle.
What is the functional relevance of transient Gag nuclear targeting? PFV Gag nuclear localization is thought to occur at least twice during the viral replication cycle, once early as part of the incoming pre-integration complex (PIC), and once late when nascent Gag is expressed and viral capsids are assembled. Therefore, there are two basic hypotheses, which are not mutually exclusive—either Gag nuclear targeting is required during virus entry, or during virus assembly (also see Table 1).
In the case of virus entry it has been speculated that the CBS in GRII has a role in tethering the viral genome to the host cell chromatin in order to facilitate integration [78]. Hence, the dependency on cell division for Gag to enter the nucleus might explain why the FV genome is only integrated in dividing cells [81]. However, recent studies suggest that the differential cell cycle dependence of retroviral infection is not determined by nuclear targeting signals of the viral proteins and that nuclear import is not the limiting step for infection of non-dividing cells [reviewed in 82]. Interestingly, the primate FV genome can undergo intracellular retrotransposition, while FFV does not display efficient intracellular retrotransposition [83,84]. As FFV Gag is also the only foamyviral Gag protein that was not found to enter the nucleus, Gag nuclear translocation might be linked to efficient intracellular retrotransposition.
There are several potential roles for Gag nuclear targeting during late stages of the viral life cycle. For RSV, initial Gag-Gag dimerization occurs in the cell nucleus [85]. RSV Gag nuclear trafficking is important for viral RNA export from the nucleus and for efficient incorporation of vRNA [reviewed in 86]. Thus, RSV Gag oligomerization is linked to Gag-RNA binding, both occurring in the cell nucleus. Morozov and colleagues suggested a similar model for FVs, where initial complex formation of Gag, Pol, and RNA would take place in the nucleus [87]. In line with this, Renault et al. recently reported PFV Gag proteins to interact in the nucleus and proposed a model where Gag would be required for nuclear export of the unspliced viral RNA [80]. However, we found that Gag nuclear localization is not essential for genomic RNA encapsidation [38]. Further contradicting the proposed model, Bodem et al. recently reported that FV RNA is exported from the nucleus independently of viral proteins [88]. Deletion of the entire GRII significantly reduces Pol packaging [38,64], but detailed analysis of GRII showed that Pol packaging could be distinguished from Gag nuclear localization [77]. Also for Gag-Gag interactions the picture is unclear. While PFV Gag-Gag interactions were demonstrated in the nucleus [35,80], Gag mutants lacking nuclear localization do not display severe defects in viral assembly [38]. Thus, the concept of Gag nuclear translocation being required for the formation of a Gag-Pol-RNA complex remains highly debated. Other functions for retroviral Gag proteins in the cell nucleus could include transcriptional regulation of viral or cellular genes.

Table 1.
Overview of Potential Functions of Retroviral Gag Proteins in the Cell Nucleus. A question mark indicates that a function has been proposed, but not conclusively shown for the respective Gag protein, or that there are contradicting reports on the subject.
| Viral entry | Nuclear translocation of the PIC | HIV‑1 (?), PFV (?) |
| Integration target site selection | MLV, HIV‑1, PFV (?) | |
| Chromatin tethering | PFV (?) | |
| Transcriptional regulation | Transcriptional enhancement of early gene expression | HIV‑1 |
| Regulation of cellular gene expression | (?) | |
| Viral assembly | Gag dimerization | RSV, PFV (?) |
| RNA binding/export | MLV, FIV, RSV, PFV (?) | |
| Pol binding/packaging | PFV (?) | |
| Other | Binding to translation machinery | MMTV |
| Intracellular retrotransposition | PFV (?) | |
| Regulation of splicing | MLV |
In summary, while several retroviral Gag proteins translocate to the cell nucleus, the exact functions for most of them remain unknown. Similarly, the function of FV Gag nuclear trafficking is controversially discussed. However, it is tempting to speculate that the unique mechanism of FV Gag nuclear translocation might also suggest a different functionality compared to nuclear localization of other retroviral Gag proteins.
3. Interaction Between FV Gag and Host Cell Proteins
As illustrated above, the Gag proteins have numerous functions in the retroviral replication cycle. To accomplish these functions, retroviral Gag proteins are highly reliant on host cell proteins. Interactions between Gag and host cell proteins are required to employ the cellular machinery for viral replication. Consequently, numerous protein interaction partners are reported for orthoretroviral Gag proteins, in particular for HIV-1 Gag [89,90].
Unlike the long list of HIV-1 Gag interacting partners, very little is known about FV Gag intracellular interactions. Given the many unconventional features of their replication, it is expected that FV Gag interaction partners await identification. However, so far only few cellular proteins have been reported to interact with foamyviral Gag proteins (Table 2).

Table 2.
Described Interactors of FV Gag Proteins. The table contains both, confirmed and predicted/putative interactors. Functions are indicated as proposed in the original research articles. ‘Assay’ indicates the method by which an interaction was found. The column indicates ‘not shown’ if the interaction is not confirmed. Y2H, yeast two-hybrid.
| Interactor | Function | Interaction Domain | Assay |
|---|---|---|---|
| Actin | Regulation of secondary Gag cleavage | Unknown | Co-immunoprecipitation |
| DyneinLC8 | Incoming capsid transport | CC3 | Co-immunoprecipitation, domain mutation |
| H2A/H2B | Chromatin tethering | CBS | Co-immunoprecipitation, domain mutation |
| CRM1 | Nuclear export | NES | Not shown |
| DDX6 | RNA encapsidation | Unknown | Co-localization |
| TSG101 | Viral budding | PSAP L domain | Y2H |
An ‘interaction partner’ that FV Gag proteins share with orthoretroviral Gag proteins is Trim5α. Trim5α is a restriction factor of the innate immune system, acting on the CA domain of Gag [reviewed in 91]. Trim5α proteins can arrest replication of several retroviruses, including equine infectious anemia virus (EIAV), MLV, HIV-1, and FV [reviewed in 92,93]. Apart from the innate immune response, an early report showed reciprocal immunoprecipitation of PFV proteins, in particular Gag, and actin. The interaction is thought to occur during viral entry. Furthermore, the authors found indications that association of Gag with actin prevents recognition of the secondary Gag cleavage sites [13]. Also during viral entry, PFV Gag interacts with the dynein light chain 8 (LC8). Petit and colleagues showed that the interaction requires the integrity of the third coiled-coil motif (CC3) at aa 160–180. Helix-breaking point mutations in this region abolish the trafficking of incoming viral capsids to the centrosome [94]. Interestingly, trafficking of different incoming retroviruses involves a centriolar step [reviewed in 95].
FV Gag from incoming capsids, as well as newly synthesized Gag translocates to the cell nucleus by interaction of the CBS with H2A/H2B core histones (see above) [77,78]. Consequently, H2A/H2B readily co-immunoprecipitates with Gag [78]. While FV Gag enters the cell nucleus by interaction with core histones, nuclear export of Gag is reported to rely on a leucine-rich, LMB-sensitive NES [80]. Leucine-rich NES are recognized by exportin 1, also named CRM1 [reviewed in 96]. Interestingly, Bodem et al. found that nuclear export of unspliced FV mRNA is CRM1 dependent [88]. However, FV Gag is not required for the nuclear export of FV mRNA (see above) [88] and direct interaction between Gag and CRM1 has not yet been demonstrated. Recently, the RNA helicase DDX6 was found to relocate and co-localize with viral RNA and Gag at the viral assembly site. DDX6 knockdown led to decreased RNA packaging. However, co-immunoprecipitation experiments failed to confirm a stable interaction between PFV Gag and DDX6 [97]. Finally, primate FV Gag contains classical PSAP L domains, which mediate viral budding through binding to components of the ESCRT-I complex (endosomal sorting complex required for transport), namely Tsg101 (tumor-susceptibility gene product 101) (see above) [41,42].
Based on the analogy with HIV-1 Gag, which reportedly binds to more than 50 different human proteins [89,90], it can be expected that the list of known FV Gag interaction partners is far from complete. New interaction partners and the respective interaction domains will likely be identified in the future. One possible candidate might be the fourth predicted coiled-coil motif (CC4) at aa 433–454. As CC1 is important for Gag-Env interaction, CC2 mediates Gag-Gag interactions, and the CC3 interacts with LC8, it is tempting to speculate that also CC4 might be a protein interaction domain.
4. Conclusions
In his review on HIV-1 Gag in 1998 Eric Freed concluded with: “A large number of studies […] have contributed significantly to our knowledge of how retroviral Gag proteins function in virus replication. Despite this wealth of knowledge, many questions remain unanswered.” [1]. Now, highlighting the similarities, but more importantly the unique features that distinguish FV Gag proteins from orthoretroviral Gag proteins leads to a very similar conclusion. Despite apparent progress on FV Gag biology, central questions are far from answered. That is, where do Gag and Env interact? How is RNA encapsidation and Pol packaging accomplished? Why does Gag localize to the cell nucleus? What is the functional role of p3Gag? And which host cell factors interact with Gag? Some of these questions are on the brink of being answered; some will stick around for some time to come, and new ones are certain to arise along the way. But most importantly, these are not just interesting scientific questions. The lack of an associated pathogenicity and the preferable integration pattern has led to a recent success of FV vectors in gene therapy [reviewed in 22,98,99]. However, it is clear, in order to further develop FV-derived vectors as a tool for modern molecular medicine more work is required to understand the basic molecular biology of these unconventional retroviruses.
Acknowledgments
This manuscript is dedicated to past and present members of the Molecular Retrovirology lab in Dresden, Germany. Thanks for the constant support. I thank Helena Silva Cascales, Dirk Lindemann, Richard Mills, and Christopher Bot for critically reading the manuscript. Furthermore, I thank Viruses for waiving the publishing fees.
Conflict of Interest
The author declares no conflict of interest.
References and Notes
- Freed, E.O. Hiv–1 gag proteins: Diverse functions in the virus life cycle. Virology 1998, 251, 1–15. [Google Scholar]
- Meiering, C.D.; Linial, M.L. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 2001, 14, 165–176. [Google Scholar]
- Saib, A. Non–primate foamy viruses. Curr. Top. Microbiol. Immunol. 2003, 277, 197–211. [Google Scholar]
- Linial, M.L. Foamy viruses are unconventional retroviruses. J. Virol. 1999, 73, 1747–1755. [Google Scholar]
- Rethwilm, A. Foamy virus vectors: An awaited alternative to gammaretro– and lentiviral vectors. Curr. Gene Ther. 2007, 7, 261–271. [Google Scholar] [CrossRef]
- Bruss, V. Hepatitis b virus morphogenesis. World J. Gastroenterol. 2007, 13, 65–73. [Google Scholar]
- Achong, B.G.; Mansell, P.W.; Epstein, M.A.; Clifford, P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 1971, 46, 299–307. [Google Scholar]
- Herchenroder, O.; Renne, R.; Loncar, D.; Cobb, E.K.; Murthy, K.K.; Schneider, J.; Mergia, A.; Luciw, P.A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (sfvcpz): High homology to human foamy virus (hfv). Virology 1994, 201, 187–199. [Google Scholar] [CrossRef]
- Schweizer, M.; Turek, R.; Hahn, H.; Schliephake, A.; Netzer, K.O.; Eder, G.; Reinhardt, M.; Rethwilm, A.; Neumann–Haefelin, D. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: Appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res. Hum. Retroviruses 1995, 11, 161–170. [Google Scholar]
- Enssle, J.; Jordan, I.; Mauer, B.; Rethwilm, A. Foamy virus reverse transcriptase is expressed independently from the gag protein. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 4137–4141. [Google Scholar]
- Lochelt, M.; Flugel, R.M. The human foamy virus pol gene is expressed as a pro–pol polyprotein and not as a gag–pol fusion protein. J. Virol. 1996, 70, 1033–1040. [Google Scholar]
- Yu, S.F.; Baldwin, D.N.; Gwynn, S.R.; Yendapalli, S.; Linial, M.L. Human foamy virus replication: A pathway distinct from that of retroviruses and hepadnaviruses. Science 1996, 271, 1579–1582. [Google Scholar]
- Giron, M.L.; Colas, S.; Wybier, J.; Rozain, F.; Emanoil–Ravier, R. Expression and maturation of human foamy virus gag precursor polypeptides. J. Virol. 1997, 71, 1635–1639. [Google Scholar]
- Pfrepper, K.I.; Lochelt, M.; Rackwitz, H.R.; Schnolzer, M.; Heid, H.; Flugel, R.M. Molecular characterization of proteolytic processing of the gag proteins of human spumavirus. J. Virol. 1999, 73, 7907–7911. [Google Scholar]
- Enssle, J.; Fischer, N.; Moebes, A.; Mauer, B.; Smola, U.; Rethwilm, A. Carboxy–terminal cleavage of the human foamy virus gag precursor molecule is an essential step in the viral life cycle. J. Virol. 1997, 71, 7312–7317. [Google Scholar]
- Zemba, M.; Wilk, T.; Rutten, T.; Wagner, A.; Flugel, R.M.; Lochelt, M. The carboxy–terminal p3gag domain of the human foamy virus gag precursor is required for efficient virus infectivity. Virology 1998, 247, 7–13. [Google Scholar] [CrossRef]
- Konvalinka, J.; Lochelt, M.; Zentgraf, H.; Flugel, R.M.; Krausslich, H.G. Active foamy virus proteinase is essential for virus infectivity but not for formation of a pol polyprotein. J. Virol. 1995, 69, 7264–7268. [Google Scholar]
- Cartellieri, M.; Herchenroder, O.; Rudolph, W.; Heinkelein, M.; Lindemann, D.; Zentgraf, H.; Rethwilm, A. N–terminal gag domain required for foamy virus particle assembly and export. J. Virol. 2005, 79, 12464–12476. [Google Scholar] [CrossRef]
- Wang, J.; Guo, H.Y.; Jia, R.; Xu, X.; Tan, J.; Geng, Y.Q.; Qiao, W.T. Preparation of bfv gag antiserum and preliminary study on cellular distribution of bfv. Virol. Sin. 2010, 25, 115–122. [Google Scholar] [CrossRef]
- Lehmann–Che, J.; Renault, N.; Giron, M.L.; Roingeard, P.; Clave, E.; Tobaly –Tapiero, J.; Bittoun, P.; Toubert, A.; de The, H.; Saib, A. Centrosomal latency of incoming foamy viruses in resting cells. PLoS Pathog. 2007, 3, e74. [Google Scholar] [CrossRef]
- Hutter, S.; Mullers, E.; Stanke, N.; Reh, J.; Lindemann, D. Prototype foamy virus protease activity is essential for intra–particle reverse transcription initiation but not absolutely required for uncoating upon host cell entry. J. Virol. 2013.
- Lindemann, D.; Rethwilm, A. Foamy virus biology and its application for vector development. Viruses 2011, 3, 561–585. [Google Scholar] [CrossRef]
- Swanstrom, R.; Wills, J.W. Synthesis, assembly, and processing of viral proteins. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor (NY), 1997. [Google Scholar]
- Geiselhart, V.; Schwantes, A.; Bastone, P.; Frech, M.; Lochelt, M. Features of the env leader protein and the n–terminal gag domain of feline foamy virus important for virus morphogenesis. Virology 2003, 310, 235–244. [Google Scholar]
- Life, R.B.; Lee, E.G.; Eastman, S.W.; Linial, M.L. Mutations in the amino terminus of foamy virus gag disrupt morphology and infectivity but do not target assembly. J. Virol. 2008, 82, 6109–6119. [Google Scholar] [CrossRef]
- Wilk, T.; Geiselhart, V.; Frech, M.; Fuller, S.D.; Flugel, R.M.; Lochelt, M. Specific interaction of a novel foamy virus env leader protein with the n–terminal gag domain. J. Virol. 2001, 75, 7995–8007. [Google Scholar] [CrossRef]
- Pietschmann, T.; Heinkelein, M.; Heldmann, M.; Zentgraf, H.; Rethwilm, A.; Lindemann, D. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 1999, 73, 2613–2621. [Google Scholar]
- Lindemann, D.; Pietschmann, T.; Picard–Maureau, M.; Berg, A.; Heinkelein, M.; Thurow, J.; Knaus, P.; Zentgraf, H.; Rethwilm, A. A particle–associated glycoprotein signal peptide essential for virus maturation and infectivity. 2001, 75, 5762–5771. [Google Scholar]
- Sfakianos, J.N.; Hunter, E. M–pmv capsid transport is mediated by env/gag interactions at the pericentriolar recycling endosome. Traffic 2003, 4, 671–680. [Google Scholar] [CrossRef]
- Yu, S.F.; Eastman, S.W.; Linial, M.L. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in gag. Traffic 2006, 7, 966–977. [Google Scholar] [CrossRef]
- Linial, M.L.; Eastman, S.W. Particle assembly and genome packaging. Curr. Top. Microbiol. Immunol. 2003, 277, 89–110. [Google Scholar]
- Eastman, S.W.; Linial, M.L. Identification of a conserved residue of foamy virus gag required for intracellular capsid assembly. J. Virol. 2001, 75, 6857–6864. [Google Scholar] [CrossRef]
- Choi, G.; Park, S.; Choi, B.; Hong, S.; Lee, J.; Hunter, E.; Rhee, S.S. Identification of a cytoplasmic targeting/retention signal in a retroviral gag polyprotein. J. Virol. 1999, 73, 5431–5437. [Google Scholar]
- Craven, R.C.; Parent, L.J. Dynamic interactions of the gag polyprotein. Curr. Top. Microbiol. Immunol. 1996, 214, 65–94. [Google Scholar]
- Tobaly–Tapiero, J.; Bittoun, P.; Giron, M.L.; Neves, M.; Koken, M.; Saib, A.; de The, H. Human foamy virus capsid formation requires an interaction domain in the n terminus of gag. J. Virol. 2001, 75, 4367–4375. [Google Scholar]
- Bowzard, J.B.; Bennett, R.P.; Krishna, N.K.; Ernst, S.M.; Rein, A.; Wills, J.W. Importance of basic residues in the nucleocapsid sequence for retrovirus gag assembly and complementation rescue. J. Virol. 1998, 72, 9034–9044. [Google Scholar]
- Mannigel, I.; Stange, A.; Zentgraf, H.; Lindemann, D. Correct capsid assembly mediated by a conserved yxxlgl motif in prototype foamy virus gag is essential for infectivity and reverse transcription of the viral genome. J. Virol. 2007, 81, 3317–3326. [Google Scholar] [CrossRef]
- Mullers, E.; Uhlig, T.; Stirnnagel, K.; Fiebig, U.; Zentgraf, H.; Lindemann, D. Novel functions of prototype foamy virus gag glycine– arginine–rich boxes in reverse transcription and particle morphogenesis. J. Virol. 2011, 85, 1452–1463. [Google Scholar] [CrossRef]
- Yu, S.F.; Edelmann, K.; Strong, R.K.; Moebes, A.; Rethwilm, A.; Linial, M.L. The carboxyl terminus of the human foamy virus gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 1996, 70, 8255–8262. [Google Scholar]
- Cimarelli, A.; Luban, J. Human immunodeficiency virus type 1 virion density is not determined by nucleocapsid basic residues. J. Virol. 2000, 74, 6734–6740. [Google Scholar] [CrossRef]
- Stange, A.; Mannigel, I.; Peters, K.; Heinkelein, M.; Stanke, N.; Cartellieri, M.; Gottlinger, H.; Rethwilm, A.; Zentgraf, H.; Lindemann, D. Characterization of prototype foamy virus gag late assembly domain motifs and their role in particle egress and infectivity. J. Virol. 2005, 79, 5466–5476. [Google Scholar] [CrossRef]
- Patton, G.S.; Morris, S.A.; Chung, W.; Bieniasz, P.D.; McClure, M.O. Identification of domains in gag important for prototypic foamy virus egress. J. Virol. 2005, 79, 6392–6399. [Google Scholar] [CrossRef]
- Pincetic, A.; Leis, J. The mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009, 2009, 6239691–6239699. [Google Scholar]
- Pornillos, O.; Garrus, J.E.; Sundquist, W.I. Mechanisms of enveloped rna virus budding. Trends Cell. Biol. 2002, 12, 569–579. [Google Scholar] [CrossRef]
- Morita, E.; Sundquist, W.I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004, 20, 395–425. [Google Scholar] [CrossRef]
- Thumer, L.; Rethwilm, A.; Holmes, E.C.; Bodem, J. The complete nucleotide sequence of a new world simian foamy virus. Virology 2007, 369, 191–197. [Google Scholar] [CrossRef]
- Maurer, B.; Flugel, R.M. Genomic organization of the human spumaretrovirus and its relatedness to aids and other retroviruses. AIDS Res. Hum. 1988, 4, 467–473. [Google Scholar] [CrossRef]
- Matthes, D.; Wiktorowicz, T.; Zahn, J.; Bodem, J.; Stanke, N.; Lindemann, D.; Rethwilm, A. Basic residues in the foamy virus gag protein. J. Virol. 2011, 85, 3986–3995. [Google Scholar] [CrossRef]
- Fischer, N.; Heinkelein, M.; Lindemann, D.; Enssle, J.; Baum, C.; Werder, E.; Zentgraf, H.; Muller, J.G.; Rethwilm, A. Foamy virus particle formation. J. Virol. 1998, 72, 1610–1615. [Google Scholar]
- Lindemann, D.; Goepfert, P.A. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 2003, 277, 111–129. [Google Scholar]
- Zhadina, M.; McClure, M.O.; Johnson, M.C.; Bieniasz, P.D. Ubiquitin–dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. USA 2007, 104, 20031–20036. [Google Scholar]
- Liu, Y.; Kim, Y.B.; Lochelt, M. N–terminally myristoylated feline foamy virus gag allows env–independent budding of sub–viral particles. Viruses 2011, 3, 2223–2237. [Google Scholar] [CrossRef]
- Swiersy, A.; Wiek, C.; Zentgraf, H.; Lindemann, D. Characterization and manipulation of foamy virus membrane interactions. Cell. Microbiol. 2012.
- Wills, J.W.; Craven, R.C. Form, function, and use of retroviral gag proteins. Aids 1991, 5, 639–654. [Google Scholar] [CrossRef]
- Dorfman, T.; Luban, J.; Goff, S.P.; Haseltine, W.A.; Gottlinger, H.G. Mapping of functionally important residues of a cysteine–histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993, 67, 6159–6169. [Google Scholar]
- Aldovini, A.; Young, R.A. Mutations of rna and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990, 64, 1920–1926. [Google Scholar]
- D'Souza, V.; Summers, M.F. How retroviruses select their genomes. Nature Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef]
- Schwartz, M.D.; Fiore, D.; Panganiban, A.T. Distinct functions and requirements for the cys–his boxes of the human immunodeficiency virus type 1 nucleocapsid protein during rna encapsidation and replication. J. Virol. 1997, 71, 9295–9305. [Google Scholar]
- Winkler, I.; Bodem, J.; Haas, L.; Zemba, M.; Delius, H.; Flower, R.; Flugel, R.M.; Lochelt, M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J. Virol. 1997, 71, 6727–6741. [Google Scholar]
- Gabus, C.; Ivanyi–Nagy, R.; Depollier, J.; Bucheton, A.; Pelisson, A.; Darlix, J.L. Characterization of a nucleocapsid–like region and of two distinct primer trnalys,2 binding sites in the endogenous retrovirus gypsy. Nucleic Acids Res. 2006, 34, 5764–5777. [Google Scholar] [CrossRef]
- Schliephake, A.W.; Rethwilm, A. Nuclear localization of foamy virus gag precursor protein. J. Virol. 1994, 68, 4946–4954. [Google Scholar]
- Tobaly–Tapiero, J.; Bittoun, P.; Neves, M.; Guillemin, M.C.; Lecellier, C.H.; Puvion–Dutilleul, F.; Gicquel, B.; Zientara, S.; Giron, M.L.; de The, H.; et al. Isolation and characterization of an equine foamy virus. J. Virol. 2000, 74, 4064–4073. [Google Scholar] [CrossRef]
- Hatton, T.; Zhou, S.; Standring, D.N. Rna– and DNA–binding activities in hepatitis b virus capsid protein: A model for their roles in viral replication. J. Virol. 1992, 66, 5232–5241. [Google Scholar]
- Stenbak, C.R.; Linial, M.L. Role of the c terminus of foamy virus gag in rna packaging and pol expression. J. Virol. 2004, 78, 9423–9430. [Google Scholar] [CrossRef]
- Heinkelein, M.; Leurs, C.; Rammling, M.; Peters, K.; Hanenberg, H.; Rethwilm, A. Pregenomic rna is required for efficient incorporation of pol polyprotein into foamy virus capsids. J. Virol. 2002, 76, 10069–10073. [Google Scholar] [CrossRef]
- Peters, K.; Wiktorowicz, T.; Heinkelein, M.; Rethwilm, A. Rna and protein requirements for incorporation of the pol protein into foamy virus particles. J. Virol. 2005, 79, 7005–7013. [Google Scholar] [CrossRef]
- Lee, E.G.; Linial, M.L. The c terminus of foamy retrovirus gag contains determinants for encapsidation of pol protein into virions. J. Virol. 2008, 82, 10803–10810. [Google Scholar] [CrossRef]
- Lee, E.G.; Linial, M.L. Deletion of a cys–his motif from the alpharetrovirus nucleocapsid domain reveals late domain mutant–like budding defects. Virology 2006, 347, 226–233. [Google Scholar] [CrossRef]
- Muriaux, D.; Costes, S.; Nagashima, K.; Mirro, J.; Cho, E.; Lockett, S.; Rein, A. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 2004, 78, 12378–12385. [Google Scholar] [CrossRef]
- Hooks, J.J.; Gibbs, C.J., Jr. The foamy viruses. Bacteriol. Rev. 1975, 39, 169–185. [Google Scholar]
- Dupont, S.; Sharova, N.; DeHoratius, C.; Virbasius, C.M.; Zhu, X.; Bukrinskaya, A.G.; Stevenson, M.; Green, M.R. A novel nuclear export activity in hiv–1 matrix protein required for viral replication. Nature 1999, 402, 681–685. [Google Scholar]
- Nash, M.A.; Meyer, M.K.; Decker, G.L.; Arlinghaus, R.B. A subset of pr65gag is nucleus associated in murine leukemia virus–infected cells. J. Virol. 1993, 67, 1350–1356. [Google Scholar]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and crm1–dependent nuclear export of the rous sarcoma virus gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar]
- Beyer, A.R.; Bann, D.V.; Rice, B.; Pultz, I.S.; Kane, M.; Goff, S.P.; Golovkina, T.V.; Parent, L.J. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein l9. J. Virol. 2013, 87, 1069–1082. [Google Scholar] [CrossRef]
- Kong, X.H.; Yu, H.; Xuan, C.H.; Wang, J.Z.; Chen, Q.M.; Geng, Y.Q. The requirements and mechanism for capsid assembly and budding of bovine foamy virus. Arch. Virol. 2005, 150, 1677–1684. [Google Scholar] [CrossRef]
- Bodem, J.; Zemba, M.; Flugel, R.M. Nuclear localization of the functional bel 1 transactivator but not of the gag proteins of the feline foamy virus. Virology 1998, 251, 22–27. [Google Scholar] [CrossRef]
- Mullers, E.; Stirnnagel, K.; Kaulfuss, S.; Lindemann, D. Prototype foamy virus gag nuclear localization: A novel pathway among retroviruses. J. Virol. 2011, 85, 9276–9285. [Google Scholar] [CrossRef]
- Tobaly–Tapiero, J.; Bittoun, P.; Lehmann–Che, J.; Delelis, O.; Giron, M.L.; de The, H.; Saib, A. Chromatin tethering of incoming foamy virus by the structural gag protein. Traffic 2008, 9, 1717–1727. [Google Scholar]
- Stirnnagel, K.; Schupp, D.; Dupont, A.; Kudryavtsev, V.; Reh, J.; Mullers, E.; Lamb, D.C.; Lindemann, D. Differential ph–dependent cellular uptake pathways among foamy viruses elucidated using dual–colored fluorescent particles. Retrovirology 2012, 9, 71. [Google Scholar] [CrossRef]
- Renault, N.; Tobaly–Tapiero, J.; Paris, J.; Giron, M.L.; Coiffic, A.; Roingeard, P.; Saib, A. A nuclear export signal within the structural gag protein is required for prototype foamy virus replication. Retrovirology 2011, 8, 6. [Google Scholar] [CrossRef]
- Lo, Y.T.; Tian, T.; Nadeau, P.E.; Park, J.; Mergia, A. The foamy virus genome remains unintegrated in the nuclei of g1/s phase–arrested cells, and integrase is critical for preintegration complex transport into the nucleus. J. Virol. 2010, 84, 2832–2842. [Google Scholar] [CrossRef]
- Yamashita, M.; Emerman, M. Retroviral infection of non–dividing cells: Old and new perspectives. Virology 2006, 344, 88–93. [Google Scholar] [CrossRef]
- Heinkelein, M.; Pietschmann, T.; Jarmy, G.; Dressler, M.; Imrich, H.; Thurow, J.; Lindemann, D.; Bock, M.; Moebes, A.; Roy, J.; et al. Efficient intracellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 2000, 19, 3436–3445. [Google Scholar] [CrossRef]
- Roy, J.; Rudolph, W.; Juretzek, T.; Gartner, K.; Bock, M.; Herchenroder, O.; Lindemann, D.; Heinkelein, M.; Rethwilm, A. Feline foamy virus genome and replication strategy. J. Virol. 2003, 77, 11324–11331. [Google Scholar] [CrossRef]
- Kenney, S.P.; Lochmann, T.L.; Schmid, C.L.; Parent, L.J. Intermolecular interactions between retroviral gag proteins in the nucleus. J. Virol. 2008, 82, 683–691. [Google Scholar] [CrossRef]
- Parent, L.J. New insights into the nuclear localization of retroviral gag proteins. Nucleus 2011, 2, 92–97. [Google Scholar] [CrossRef]
- Morozov, V.A.; Copeland, T.D.; Nagashima, K.; Gonda, M.A.; Oroszlan, S. Protein composition and morphology of human foamy virus intracellular cores and extracellular particles. Virology 1997, 228, 307–317. [Google Scholar] [CrossRef]
- Bodem, J.; Schied, T.; Gabriel, R.; Rammling, M.; Rethwilm, A. Foamy virus nuclear rna export is distinct from that of other retroviruses. J. Virol. 2011, 85, 2333–2341. [Google Scholar]
- Pinney, J.W.; Dickerson, J.E.; Fu, W.; Sanders–Beer, B.E.; Ptak, R.G.; Robertson, D.L. Hiv–host interactions: A map of viral perturbation of the host system. Aids 2009, 23, 549–554. [Google Scholar]
- Ptak, R.G.; Fu, W.; Sanders–Beer, B.E.; Dickerson, J.E.; Pinney, J.W.; Robertson, D.L.; Rozanov, M.N.; Katz, K.S.; Maglott, D.R.; Pruitt, K.D.; et al. Cataloguing the hiv type 1 human protein interaction network. AIDS Res. Hum. Retroviruses 2008, 24, 1497–1502. [Google Scholar]
- Sastri, J.; Campbell, E.M. Recent insights into the mechanism and consequences of trim5alpha retroviral restriction. AIDS Res. Hum. Retroviruses 2011, 27, 231–238. [Google Scholar] [CrossRef]
- Bieniasz, P.D. Intrinsic immunity: A front–line defense against viral attack. Nat. Immunol. 2004, 5, 1109–1115. [Google Scholar] [CrossRef]
- Yap, M.W.; Lindemann, D.; Stanke, N.; Reh, J.; Westphal, D.; Hanenberg, H.; Ohkura, S.; Stoye, J.P. Restriction of foamy viruses by primate trim5alpha. J. Virol. 2008, 82, 5429–5439. [Google Scholar]
- Petit, C.; Giron, M.L.; Tobaly–Tapiero, J.; Bittoun, P.; Real, E.; Jacob, Y.; Tordo, N.; De The, H.; Saib, A. Targeting of incoming retroviral gag to the centrosome involves a direct interaction with the dynein light chain 8. J. Cell Sci. 2003, 116, 3433–3442. [Google Scholar] [CrossRef]
- Afonso, P.V.; Zamborlini, A.; Saib, A.; Mahieux, R. Centrosome and retroviruses: The dangerous liaisons. Retrovirology 2007, 4, 27. [Google Scholar] [CrossRef]
- Kutay, U.; Guttinger, S. Leucine–rich nuclear–export signals: Born to be weak. Trends Cell Biol. 2005, 15, 121–124. [Google Scholar] [CrossRef]
- Yu, S.F.; Lujan, P.; Jackson, D.L.; Emerman, M.; Linial, M.L. The dead–box rna helicase ddx6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 2011, 7, e1002303. [Google Scholar] [CrossRef]
- Erlwein, O.; McClure, M. Gene delivery the key to gene therapy: The case for foamy viruses. Ther. Deliv. 2011, 2, 681–684. [Google Scholar] [CrossRef]
- Trobridge, G.D.; Horn, P.A.; Beard, B.C.; Kiem, H.P. Large animal models for foamy virus vector gene therapy. Viruses 2012, 4, 3572–3588. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).