The Dynamics of HCF-1 Modulation of Herpes Simplex Virus Chromatin during Initiation of Infection
Abstract
:Abbreviations
| HCF-1 | host cell factor-1 |
| FHL2 | four and a half limb domain protein-2 |
| GABP | GA-binding protein |
| Oct-1 | octamer binding protein-1 |
| VZV | varicella zoster virus |
| ASF1 | anti-silencing function protein 1 |
| PCAF | histone acetyltransferase KAT2B |
| SWI/SNF | switch/sucrose non-fermentable |
| ISWI | Imitation SWI |
| TBP | tata-binding protein |
| Med | mediator |
| MLL | histone-lysine N-methyltransferase MLL1 |
| LSD1 | lysine-specific demethylase 1 |
| JMJD2 | Jumonji domain-containing protein 2 |
| PHF8 | PHD finger protein 8 |
| CHD8 | chromodomain helicase DNA binding protein 8 |
| NURF | nucleosome remodeling factor |
| THAP | thanatos-associated domain-containing apoptosis-associated protein |
| PGC | peroxisome proliferator-activated receptor gamma coactivator |
| PRC | peroxisome proliferator-activated receptor gamma coactivator PGC-1-related coactivator |
1. Introduction
2. Viral and Cellular Factors that Govern IE Gene Expression
3. The Essential HCF-1 Coactivator

4. HCF-1 and Multiple Chromatin Modulation Complexes
4.1. Chromatin Modulation of HSV Gene Expression
4.2. HCF-1 Couples Removal of Repressive Histone H3K9-Methylation with Deposition of Activating Histone H3K4-Methylation
4.3. Multiple Chromatin Modulation Components/Complexes Associated with the HCF-1 Coactivator

4.3.1. Histone Methyltransferases and Demethylases
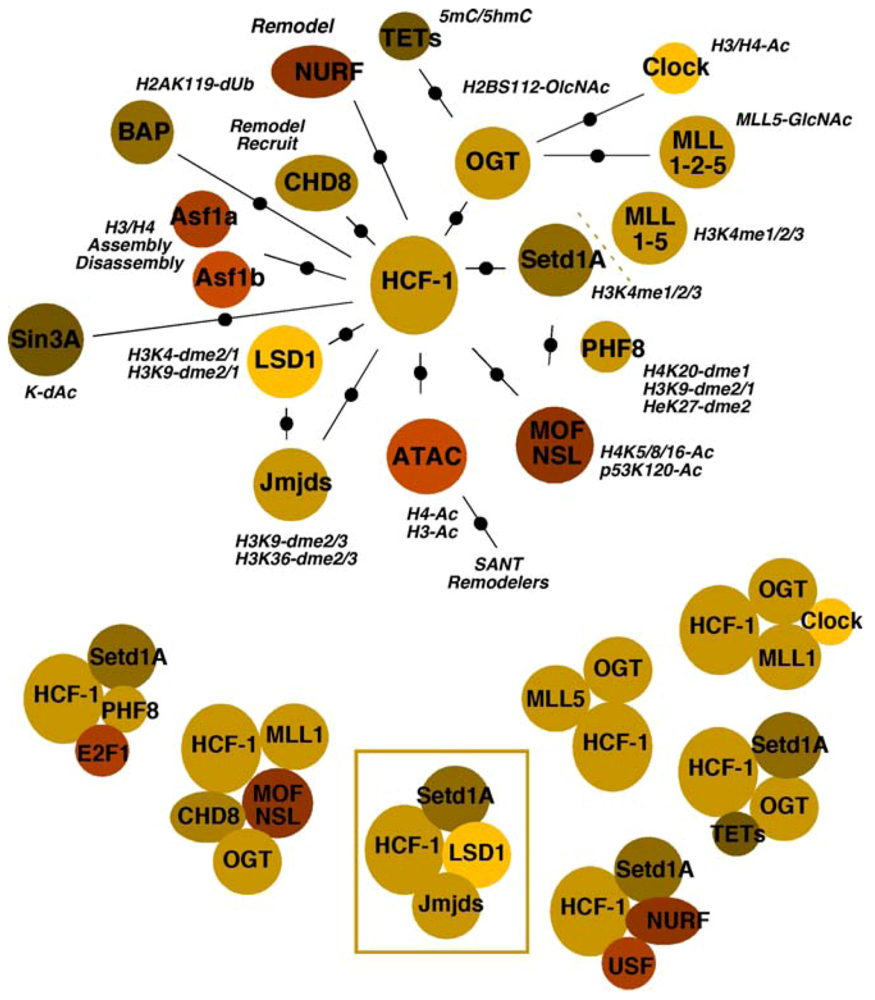
4.3.2. Acetyltransferases and Deacetylases
4.3.3. Chaperones
4.3.4. Remodelers
4.3.5. OGT
4.3.6. BAP1
4.3.7. The Potential for Multiple Roles of HCF-1 Complexes in Viral IE Gene Expression
6. HCF-1 Chromatin Modulation Complexes and HSV Latency-Reactivation
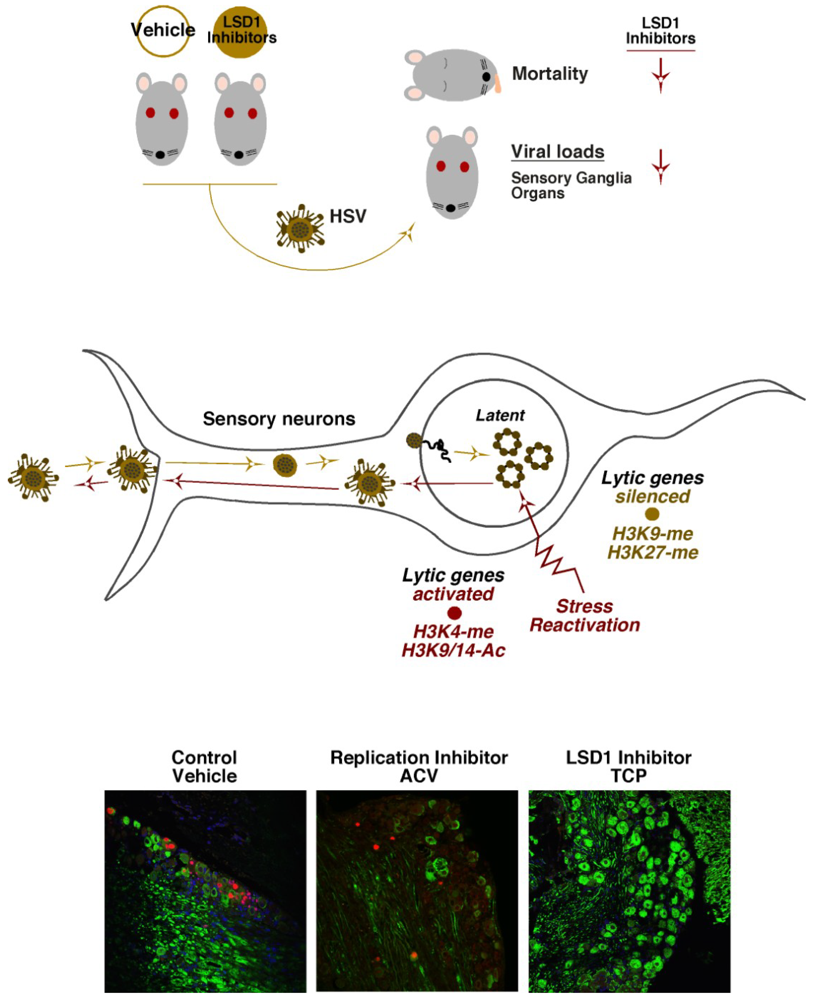
7. Concluding Remarks
Acknowledgments
Conflict of Interest
References and Notes
- Kristie, T.M. Early Events Pre-Initiation of Alphaherpes Viral Gene Expression. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Roizman, B.; Knipe, D.M.; Whitley, R.J. Herpes Simplex Viruses. In Fields Virology, 6th; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Kristie, T.M.; LeBowitz, J.H.; Sharp, P.A. The octamer-binding proteins form multi-protein--DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 1989, 8, 4229–4238. [Google Scholar]
- Kristie, T.M.; Pomerantz, J.L.; Twomey, T.C.; Parent, S.A.; Sharp, P.A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem. 1995, 270, 4387–4394. [Google Scholar]
- Kristie, T.M.; Sharp, P.A. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-trans-induction factor (VP16). J. Biol. Chem. 1993, 268, 6525–6534. [Google Scholar]
- Wilson, A.C.; LaMarco, K.; Peterson, M.G.; Herr, W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 1993, 74, 115–125. [Google Scholar] [CrossRef]
- Jones, K.A.; Tjian, R. Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro. Nature 1985, 317, 179–182. [Google Scholar] [CrossRef]
- Triezenberg, S.J.; LaMarco, K.L.; McKnight, S.L. Evidence of DNA: Protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988, 2, 730–742. [Google Scholar] [CrossRef]
- Vogel, J.L.; Kristie, T.M. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000, 19, 683–690. [Google Scholar] [CrossRef]
- Vogel, J.L.; Kristie, T.M. Site-specific proteolysis of the transcriptional coactivator HCF-1 can regulate its interaction with protein cofactors. Proc. Natl. Acad. Sci. USA 2006, 103, 6817–6822. [Google Scholar] [CrossRef]
- Narayanan, A.; Nogueira, M.L.; Ruyechan, W.T.; Kristie, T.M. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J. Biol. Chem. 2005, 280, 1369–1375. [Google Scholar]
- Nogueira, M.L.; Wang, V.E.; Tantin, D.; Sharp, P.A.; Kristie, T.M. Herpes simplex virus infections are arrested in Oct-1-deficient cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1473–1478. [Google Scholar]
- Gunther, M.; Laithier, M.; Brison, O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol. Cell. Biochem. 2000, 210, 131–142. [Google Scholar] [CrossRef]
- Delehouzee, S.; Yoshikawa, T.; Sawa, C.; Sawada, J.; Ito, T.; Omori, M.; Wada, T.; Yamaguchi, Y.; Kabe, Y.; Handa, H. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells 2005, 10, 717–731. [Google Scholar] [CrossRef]
- Lu, R.; Misra, V. Potential role for luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency. J. Virol. 2000, 74, 934–943. [Google Scholar] [CrossRef]
- Lu, R.; Yang, P.; O'Hare, P.; Misra, V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 1997, 17, 5117–5126. [Google Scholar]
- Lu, R.; Yang, P.; Padmakumar, S.; Misra, V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J. Virol. 1998, 72, 6291–6297. [Google Scholar]
- Luciano, R.L.; Wilson, A.C. N-Terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif. Proc. Natl. Acad. Sci. USA 2000, 97, 10757–10762. [Google Scholar] [CrossRef]
- Luciano, R.L.; Wilson, A.C. HCF-1 functions as a coactivator for the zinc finger protein Krox20. J. Biol. Chem. 2003, 278, 51116–51124. [Google Scholar] [CrossRef]
- Knez, J.; Piluso, D.; Bilan, P.; Capone, J.P. Host cell factor-1 and E2F4 interact via multiple determinants in each protein. Mol. Cell. Biochem. 2006, 288, 79–90. [Google Scholar] [CrossRef]
- Liu, W.; Tanasa, B.; Tyurina, O.V.; Zhou, T.Y.; Gassmann, R.; Liu, W.T.; Ohgi, K.A.; Benner, C.; Garcia-Bassets, I.; Aggarwal, A.K.; et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010, 466, 508–512. [Google Scholar]
- Tyagi, S.; Chabes, A.L.; Wysocka, J.; Herr, W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 2007, 27, 107–119. [Google Scholar] [CrossRef]
- Dejosez, M.; Krumenacker, J.S.; Zitur, L.J.; Passeri, M.; Chu, L.F.; Songyang, Z.; Thomson, J.A.; Zwaka, T.P. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 2008, 133, 1162–1174. [Google Scholar] [CrossRef]
- Dejosez, M.; Levine, S.S.; Frampton, G.M.; Whyte, W.A.; Stratton, S.A.; Barton, M.C.; Gunaratne, P.H.; Young, R.A.; Zwaka, T.P. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010, 24, 1479–1484. [Google Scholar] [CrossRef]
- Mazars, R.; Gonzalez-de-Peredo, A.; Cayrol, C.; Lavigne, A.C.; Vogel, J.L.; Ortega, N.; Lacroix, C.; Gautier, V.; Huet, G.; Ray, A.; et al. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: A link between DYT6 and DYT3 dystonias. J. Biol. Chem. 2010, 285, 13364–13371. [Google Scholar]
- Lin, J.; Puigserver, P.; Donovan, J.; Tarr, P.; Spiegelman, B.M. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002, 277, 1645–1648. [Google Scholar]
- Vercauteren, K.; Gleyzer, N.; Scarpulla, R.C. PGC-1-related coactivator complexes with HCF-1 and NRF-2beta in mediating NRF-2(GABP)-dependent respiratory gene expression. J. Biol. Chem. 2008, 283, 12102–12111. [Google Scholar] [CrossRef]
- Herrera, F.J.; Triezenberg, S.J. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 2004, 78, 9689–9696. [Google Scholar]
- Dechassa, M.L.; Sabri, A.; Pondugula, S.; Kassabov, S.R.; Chatterjee, N.; Kladde, M.P.; Bartholomew, B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 2010, 38, 590–602. [Google Scholar] [CrossRef]
- Hirai, H.; Tani, T.; Kikyo, N. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. Biol. 2010, 54, 1589–1596. [Google Scholar] [CrossRef]
- Mittler, G.; Stuhler, T.; Santolin, L.; Uhlmann, T.; Kremmer, E.; Lottspeich, F.; Berti, L.; Meisterernst, M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003, 22, 6494–6504. [Google Scholar] [CrossRef]
- Rafalska-Metcalf, I.U.; Powers, S.L.; Joo, L.M.; LeRoy, G.; Janicki, S.M. Single cell analysis of transcriptional activation dynamics. PLoS One 2010, 5, e10272. [Google Scholar]
- Sikorski, T.W.; Joo, Y.J.; Ficarro, S.B.; Askenazi, M.; Buratowski, S.; Marto, J.A. Proteomic analysis demonstrates activator- and chromatin-specific recruitment to promoters. J. Biol. Chem. 2012, 287, 35397–35408. [Google Scholar]
- Uhlmann, T.; Boeing, S.; Lehmbacher, M.; Meisterernst, M. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J. Biol. Chem. 2007, 282, 2163–2173. [Google Scholar]
- Lacasse, J.J.; Schang, L.M. During lytic infections, herpes simplex virus type 1 DNA is in complexes with the properties of unstable nucleosomes. J. Virol. 2010, 84, 1920–1933. [Google Scholar] [CrossRef]
- Lacasse, J.J.; Schang, L.M. Herpes simplex virus 1 DNA is in unstable nucleosomes throughout the lytic infection cycle, and the instability of the nucleosomes is independent of DNA replication. J. Virol. 2012, 86, 11287–11300. [Google Scholar] [CrossRef]
- Oh, J.; Fraser, N.W. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 2008, 82, 3530–3537. [Google Scholar] [CrossRef]
- Placek, B.J.; Huang, J.; Kent, J.R.; Dorsey, J.; Rice, L.; Fraser, N.W.; Berger, S.L. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 2009, 83, 1416–1421. [Google Scholar] [CrossRef]
- Peng, H.; Nogueira, M.L.; Vogel, J.L.; Kristie, T.M. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc. Natl. Acad. Sci. USA 2010, 107, 2461–2466. [Google Scholar]
- Cliffe, A.R.; Knipe, D.M. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 2008, 82, 12030–12038. [Google Scholar] [CrossRef]
- Huang, J.; Kent, J.R.; Placek, B.; Whelan, K.A.; Hollow, C.M.; Zeng, P.Y.; Fraser, N.W.; Berger, S.L. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 2006, 80, 5740–5746. [Google Scholar] [CrossRef]
- Kent, J.R.; Zeng, P.Y.; Atanasiu, D.; Gardner, J.; Fraser, N.W.; Berger, S.L. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 2004, 78, 10178–10186. [Google Scholar] [CrossRef]
- Kutluay, S.B.; DeVos, S.L.; Klomp, J.E.; Triezenberg, S.J. Transcriptional coactivators are not required for herpes simplex virus type 1 immediate-early gene expression in vitro. J. Virol. 2009, 83, 3436–3449. [Google Scholar] [CrossRef]
- Bryant, K.F.; Colgrove, R.C.; Knipe, D.M. Cellular SNF2H chromatin-remodeling factor promotes herpes simplex virus 1 immediate-early gene expression and replication. MBio 2011, 2, e00330-10. [Google Scholar]
- Kutluay, S.B.; Triezenberg, S.J. Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection. J. Virol. 2009, 83, 5835–5845. [Google Scholar] [CrossRef]
- Kalamvoki, M.; Roizman, B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 17721–17726. [Google Scholar] [CrossRef]
- Kalamvoki, M.; Roizman, B. The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J. Virol. 2011, 85, 9472–9477. [Google Scholar] [CrossRef]
- Ferenczy, M.W.; DeLuca, N.A. Reversal of heterochromatic silencing of quiescent herpes simplex virus type 1 by ICP0. J. Virol. 2011, 85, 3424–3435. [Google Scholar] [CrossRef]
- Ferenczy, M.W.; Ranayhossaini, D.J.; Deluca, N.A. Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J. Virol. 2011, 85, 4993–5002. [Google Scholar] [CrossRef]
- Gu, H.; Roizman, B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. USA 2007, 104, 17134–17139. [Google Scholar] [CrossRef]
- Liang, Y.; Vogel, J.L.; Narayanan, A.; Peng, H.; Kristie, T.M. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 2009, 15, 1312–1317. [Google Scholar] [CrossRef]
- Narayanan, A.; Ruyechan, W.T.; Kristie, T.M. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. USA 2007, 104, 10835–10840. [Google Scholar] [CrossRef]
- Silva, L.; Cliffe, A.; Chang, L.; Knipe, D.M. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 2008, 4, e1000071. [Google Scholar] [CrossRef]
- Revenko, A.S.; Kalashnikova, E.V.; Gemo, A.T.; Zou, J.X.; Chen, H.W. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell. Biol. 2010, 30, 5260–5272. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- van Nuland, R.; Smits, A.H.; Pallaki, P.; Jansen, P.W.; Vermeulen, M.; Timmers, H.T. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell. Biol. 2013, 33, 2067–2077. [Google Scholar] [CrossRef]
- Wysocka, J.; Myers, M.P.; Laherty, C.D.; Eisenman, R.N.; Herr, W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003, 17, 896–911. [Google Scholar] [CrossRef]
- Yokoyama, A.; Wang, Z.; Wysocka, J.; Sanyal, M.; Aufiero, D.J.; Kitabayashi, I.; Herr, W.; Cleary, M.L. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 2004, 24, 5639–5649. [Google Scholar] [CrossRef]
- Whetstine, J.R.; Nottke, A.; Lan, F.; Huarte, M.; Smolikov, S.; Chen, Z.; Spooner, E.; Li, E.; Zhang, G.; Colaiacovo, M.; et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006, 125, 467–481. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Liang, Y.; Quenelle, D.; Vogel, J.L.; Mascaro, C.; Ortega, A.; Kristie, T.M. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. MBio 2013, 4, e00558-12. [Google Scholar]
- Liang, Y.; Vogel, J.L.; Arbuckle, J.H.; Rai, G.; Jadhav, A.; Simeonov, A.; Maloney, D.J.; Kristie, T.M. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci. Transl. Med. 2013, 5, 167ra165. [Google Scholar]
- Fujiki, R.; Chikanishi, T.; Hashiba, W.; Ito, H.; Takada, I.; Roeder, R.G.; Kitagawa, H.; Kato, S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 2009, 459, 455–459. [Google Scholar] [CrossRef]
- Mishra, B.P.; Ansari, K.I.; Mandal, S.S. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. FEBS J. 2009, 276, 1629–1640. [Google Scholar] [CrossRef]
- Cai, Y.; Jin, J.; Swanson, S.K.; Cole, M.D.; Choi, S.H.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 2010, 285, 4268–4272. [Google Scholar] [CrossRef]
- Dou, Y.; Milne, T.A.; Tackett, A.J.; Smith, E.R.; Fukuda, A.; Wysocka, J.; Allis, C.D.; Chait, B.T.; Hess, J.L.; Roeder, R.G. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 2005, 121, 873–885. [Google Scholar] [CrossRef]
- Li, X.; Wu, L.; Corsa, C.A.; Kunkel, S.; Dou, Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol. Cell 2009, 36, 290–301. [Google Scholar] [CrossRef]
- Smith, E.R.; Cayrou, C.; Huang, R.; Lane, W.S.; Cote, J.; Lucchesi, J.C. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 2005, 25, 9175–9188. [Google Scholar] [CrossRef]
- Guelman, S.; Kozuka, K.; Mao, Y.; Pham, V.; Solloway, M.J.; Wang, J.; Wu, J.; Lill, J.R.; Zha, J. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol. Cell. Biol. 2009, 29, 1176–1188. [Google Scholar] [CrossRef]
- Guelman, S.; Suganuma, T.; Florens, L.; Swanson, S.K.; Kiesecker, C.L.; Kusch, T.; Anderson, S.; Yates, J.R., III; Washburn, M.P.; Abmayr, S.M.; et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol. Cell. Biol. 2006, 26, 871–882. [Google Scholar] [CrossRef]
- Krebs, A.R.; Karmodiya, K.; Lindahl-Allen, M.; Struhl, K.; Tora, L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol. Cell 2011, 44, 410–423. [Google Scholar] [CrossRef]
- Suganuma, T.; Gutierrez, J.L.; Li, B.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Abmayr, S.M.; Workman, J.L. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008, 15, 364–372. [Google Scholar] [CrossRef]
- Wang, Y.L.; Faiola, F.; Xu, M.; Pan, S.; Martinez, E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 2008, 283, 33808–33815. [Google Scholar] [CrossRef]
- Doi, M.; Hirayama, J.; Sassone-Corsi, P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006, 125, 497–508. [Google Scholar] [CrossRef]
- Katada, S.; Sassone-Corsi, P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 2010, 17, 1414–1421. [Google Scholar] [CrossRef]
- Parker, J.B.; Palchaudhuri, S.; Yin, H.; Wei, J.; Chakravarti, D. A transcriptional regulatory role of the THAP11-HCF-1 complex in colon cancer cell function. Mol. Cell. Biol. 2012, 32, 1654–1670. [Google Scholar] [CrossRef]
- Mousson, F.; Ochsenbein, F.; Mann, C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma 2007, 116, 79–93. [Google Scholar] [CrossRef]
- Schulz, L.L.; Tyler, J.K. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J. 2006, 20, 488–490. [Google Scholar]
- Oh, J.; Ruskoski, N.; Fraser, N.W. Chromatin assembly on herpes simplex virus 1 DNA early during a lytic infection is Asf1a dependent. J. Virol. 2012, 86, 12313–12321. [Google Scholar] [CrossRef]
- Nishiyama, M.; Skoultchi, A.I.; Nakayama, K.I. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-beta-catenin signaling pathway. Mol. Cell. Biol. 2012, 32, 501–512. [Google Scholar] [CrossRef]
- Yuan, C.C.; Zhao, X.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Hernandez, N. CHD8 associates with human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol. Cell. Biol. 2007, 27, 8729–8738. [Google Scholar] [CrossRef]
- Huang, S.; Li, X.; Yusufzai, T.M.; Qiu, Y.; Felsenfeld, G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol. Cell. Biol. 2007, 27, 7991–8002. [Google Scholar] [CrossRef]
- Li, X.; Wang, S.; Li, Y.; Deng, C.; Steiner, L.A.; Xiao, H.; Wu, C.; Bungert, J.; Gallagher, P.G.; Felsenfeld, G.; et al. Chromatin boundaries require functional collaboration between the hSET1 and NURF complexes. Blood 2011, 118, 1386–1394. [Google Scholar] [CrossRef]
- Li, M.D.; Ruan, H.B.; Hughes, M.E.; Lee, J.S.; Singh, J.P.; Jones, S.P.; Nitabach, M.N.; Yang, X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013, 17, 303–310. [Google Scholar] [CrossRef]
- Fujiki, R.; Hashiba, W.; Sekine, H.; Yokoyama, A.; Chikanishi, T.; Ito, S.; Imai, Y.; Kim, J.; He, H.H.; Igarashi, K.; et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 2011, 480, 557–560. [Google Scholar]
- Sakabe, K.; Wang, Z.; Hart, G.W. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. USA 2010, 107, 19915–19920. [Google Scholar] [CrossRef]
- Balasubramani, A.; Rao, A. O-GlcNAcylation and 5-methylcytosine oxidation: An unexpected association between OGT and TETs. Mol. Cell 2013, 49, 618–619. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, Y.; Bian, C.; Fujiki, R.; Yu, X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 2013, 493, 561–564. [Google Scholar]
- Deplus, R.; Delatte, B.; Schwinn, M.K.; Defrance, M.; Mendez, J.; Murphy, N.; Dawson, M.A.; Volkmar, M.; Putmans, P.; Calonne, E.; et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013, 32, 645–655. [Google Scholar] [CrossRef]
- Vella, P.; Scelfo, A.; Jammula, S.; Chiacchiera, F.; Williams, K.; Cuomo, A.; Roberto, A.; Christensen, J.; Bonaldi, T.; Helin, K.; et al. Tet proteins connect the O-linked N-acetylglucosamine transferase ogt to chromatin in embryonic stem cells. Mol. Cell 2013, 49, 645–656. [Google Scholar] [CrossRef]
- Vissers, J.H.; Nicassio, F.; van Lohuizen, M.; di Fiore, P.P.; Citterio, E. The many faces of ubiquitinated histone H2A: Insights from the DUBs. Cell Div. 2008, 3, e8. [Google Scholar] [CrossRef]
- Weake, V.M.; Workman, J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Wu, X.; Gong, Y.; Yue, J.; Qiang, B.; Yuan, J.; Peng, X. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008, 36, 3590–3599. [Google Scholar] [CrossRef]
- Machida, Y.J.; Machida, Y.; Vashisht, A.A.; Wohlschlegel, J.A.; Dutta, A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 2009, 284, 34179–34188. [Google Scholar]
- Misaghi, S.; Ottosen, S.; Izrael-Tomasevic, A.; Arnott, D.; Lamkanfi, M.; Lee, J.; Liu, J.; O'Rourke, K.; Dixit, V.M.; Wilson, A.C. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 2009, 29, 2181–2192. [Google Scholar] [CrossRef]
- Yu, H.; Mashtalir, N.; Daou, S.; Hammond-Martel, I.; Ross, J.; Sui, G.; Hart, G.W.; Rauscher, F.J., III; Drobetsky, E.; Milot, E.; et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 2010, 30, 5071–5085. [Google Scholar]
- Knipe, D.M.; Cliffe, A. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 2008, 6, 211–221. [Google Scholar] [CrossRef]
- Knipe, D.M.; Lieberman, P.M.; Jung, J.U.; McBride, A.A.; Morris, K.V.; Ott, M.; Margolis, D.; Nieto, A.; Nevels, M.; Parks, R.J.; et al. Snapshots: Chromatin control of viral infection. Virology 2013, 435, 141–156. [Google Scholar] [CrossRef]
- Kristie, T.M.; Liang, Y.; Vogel, J.L. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim. Biophys. Acta 2010, 1799, 257–265. [Google Scholar] [CrossRef]
- Placek, B.J.; Berger, S.L. Chromatin dynamics during herpes simplex virus-1 lytic infection. Biochim. Biophys. Acta 2010, 1799, 223–227. [Google Scholar]
- Best, J.D.; Carey, N. Epigenetic opportunities and challenges in cancer. Drug Discov. Today 2010, 15, 65–70. [Google Scholar] [CrossRef]
- Carey, N.; Marques, C.J.; Reik, W. DNA demethylases: A new epigenetic frontier in drug discovery. Drug Discov. Today 2011, 16, 683–690. [Google Scholar] [CrossRef]
- Copeland, R.A.; Olhava, E.J.; Scott, M.P. Targeting epigenetic enzymes for drug discovery. Curr. Opin. Chem. Biol. 2010, 14, 505–510. [Google Scholar] [CrossRef]
- Grant, S. Targeting histone demethylases in cancer therapy. Clin. Cancer Res. 2009, 15, 7111–7113. [Google Scholar] [CrossRef]
- Kelly, T.K.; de Carvalho, D.D.; Jones, P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef]
- Simon, J.A. Stopping a chromatin enzyme. Nat. Chem. Biol. 2012, 8, 875–876. [Google Scholar] [CrossRef]
- Kristie, T.M. The rise of epigenetic targets for the development of novel antivirals. Expert Rev. Anti-Infect. Ther. 2012, 10, 1359–1361. [Google Scholar] [CrossRef]
- Culhane, J.C.; Szewczuk, L.M.; Liu, X.; Da, G.; Marmorstein, R.; Cole, P.A. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 2006, 128, 4536–4537. [Google Scholar]
- Lee, M.G.; Wynder, C.; Schmidt, D.M.; McCafferty, D.G.; Shiekhattar, R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006, 13, 563–567. [Google Scholar] [CrossRef]
- Schmidt, D.M.; McCafferty, D.G. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 2007, 46, 4408–4416. [Google Scholar] [CrossRef]
- Amelio, A.L.; Giordani, N.V.; Kubat, N.J.; O'Neil, J.E.; Bloom, D.C. Deacetylation of the herpes simplex virus type 1 Latency-Associated Transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 2006, 80, 2063–2068. [Google Scholar] [CrossRef]
- Bloom, D.C.; Giordani, N.V.; Kwiatkowski, D.L. Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta 2010, 1799, 246–256. [Google Scholar]
- Cliffe, A.R.; Coen, D.M.; Knipe, D.M. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. MBio 2013, 4, e00590-12. [Google Scholar]
- Cliffe, A.R.; Garber, D.A.; Knipe, D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009, 83, 8182–8190. [Google Scholar] [CrossRef]
- Kubat, N.J.; Amelio, A.L.; Giordani, N.V.; Bloom, D.C. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 2004, 78, 12508–12518. [Google Scholar] [CrossRef]
- Kubat, N.J.; Tran, R.K.; McAnany, P.; Bloom, D.C. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 2004, 78, 1139–1149. [Google Scholar]
- Kwiatkowski, D.L.; Thompson, H.W.; Bloom, D.C. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 2009, 83, 8173–8181. [Google Scholar] [CrossRef]
- Neumann, D.M.; Bhattacharjee, P.S.; Giordani, N.V.; Bloom, D.C.; Hill, J.M. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J. Virol. 2007, 81, 13248–13253. [Google Scholar]
- Wang, Q.Y.; Zhou, C.; Johnson, K.E.; Colgrove, R.C.; Coen, D.M.; Knipe, D.M. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 2005, 102, 16055–16059. [Google Scholar]
- Kim, J.Y.; Mandarino, A.; Chao, M.V.; Mohr, I.; Wilson, A.C. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons. PLoS Pathog. 2012, 8, e1002540. [Google Scholar] [CrossRef]
- Kolb, G.; Kristie, T.M. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J. Virol. 2008. [Google Scholar] [CrossRef]
- Kristie, T.M.; Vogel, J.L.; Sears, A.E. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc. Natl. Acad. Sci. USA 1999, 96, 1229–1233. [Google Scholar] [CrossRef]
- Whitlow, Z.; Kristie, T.M. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J. Virol. 2009, 83, 9591–9595. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vogel, J.L.; Kristie, T.M. The Dynamics of HCF-1 Modulation of Herpes Simplex Virus Chromatin during Initiation of Infection. Viruses 2013, 5, 1272-1291. https://doi.org/10.3390/v5051272
Vogel JL, Kristie TM. The Dynamics of HCF-1 Modulation of Herpes Simplex Virus Chromatin during Initiation of Infection. Viruses. 2013; 5(5):1272-1291. https://doi.org/10.3390/v5051272
Chicago/Turabian StyleVogel, Jodi L., and Thomas M. Kristie. 2013. "The Dynamics of HCF-1 Modulation of Herpes Simplex Virus Chromatin during Initiation of Infection" Viruses 5, no. 5: 1272-1291. https://doi.org/10.3390/v5051272
APA StyleVogel, J. L., & Kristie, T. M. (2013). The Dynamics of HCF-1 Modulation of Herpes Simplex Virus Chromatin during Initiation of Infection. Viruses, 5(5), 1272-1291. https://doi.org/10.3390/v5051272



