Estimating Hantavirus Risk in Southern Argentina: A GIS-Based Approach Combining Human Cases and Host Distribution
Abstract
:1. Introduction
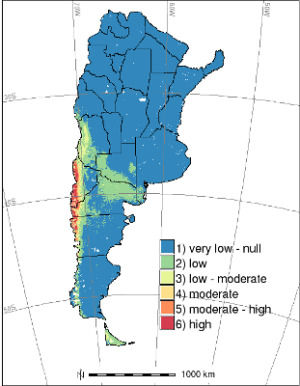
2. Results

| Variable | Description and units | HPS | Mean | SD | Median | K | B |
|---|---|---|---|---|---|---|---|
| ALT | Elevation above sea level (m) | 0 | 739.39 | 825.78 | 518.50 | 4,364.5 *** | 0.000018 ns |
| 1 | 749.03 | 358.73 | 726.00 | ||||
| BARE | Percentage of bare soil cover (%) | 0 | 34.26 | 33.94 | 26.00 | 8,506 *** | −0.0447 *** |
| 1 | 7.51 | 14.46 | 0.00 | ||||
| HERB | Percentage of grass cover (%) | 0 | 56.13 | 29.12 | 61.00 | 5,991.5 ns | −0.0024 ns |
| 1 | 54.28 | 23.18 | 59.00 | ||||
| TREE | Percentage of tree cover (%) | 0 | 9.61 | 15.65 | 3.00 | 1,909.5 *** | 0.05215 *** |
| 1 | 38.21 | 28.50 | 32.00 | ||||
| BIO1 | Annual mean temperature (x * 10, °C) | 0 | 131.02 | 53.56 | 135.50 | 8,256 *** | −0.0217 *** |
| 1 | 89.00 | 15.34 | 89.00 | ||||
| BIO2 | Mean diurnal range (Mean month (max-min), in °C) | 0 | 135.33 | 19.65 | 136.50 | 7,854 *** | −0.0308 *** |
| 1 | 125.31 | 12.61 | 123.00 | ||||
| BIO3 | Isothermality ((BIO2/BIO7) * 100) | 0 | 49.91 | 3.82 | 49.00 | 3,203.5 *** | 0.13419 ** |
| 1 | 51.66 | 1.87 | 52.00 | ||||
| BIO4 | Temperature seasonality (SD * 100) | 0 | 4,817.7 | 691.50 | 4,823.0 | 8,748 *** | −0.0015 *** |
| 1 | 4,284.7 | 317.77 | 4,248.0 | ||||
| BIO5 | Maximum temp of the warmest month (x * 10, °C) | 0 | 273.58 | 60.68 | 296.00 | 8,334.5 *** | −0.0153 *** |
| 1 | 228.61 | 23.10 | 224.00 | ||||
| BIO6 | Minimum temp of the coldest month (x * 10, °C) | 0 | 5.10 | 43.74 | 1.50 | 6,935.5 *** | −0.0124 ** |
| 1 | −11.46 | 10.41 | -11.00 | ||||
| BIO7 | Temperature annual range (BIO5-BIO6) (x * 10, °C) | 0 | 268.48 | 35.24 | 270.00 | 8,749 *** | −0.0302 *** |
| 1 | 240.07 | 18.52 | 237.00 | ||||
| BIO8 | Mean temp of the wettest quarter (x * 10, °C) | 0 | 141.24 | 95.40 | 152.00 | 8,886.5 *** | −0.0207 *** |
| 1 | 39.57 | 15.67 | 41.00 | ||||
| BIO9 | Mean temp of the driest quarter (x * 10, °C) | 0 | 112.47 | 39.82 | 109.50 | 2,965 *** | 0.0273 *** |
| 1 | 143.26 | 18.64 | 142.00 | ||||
| BIO10 | Mean temp of the warmest quarter (x * 10, °C) | 0 | 191.63 | 56.51 | 203.00 | 8,519.5 *** | −0.0201 *** |
| 1 | 144.44 | 18.17 | 143.00 | ||||
| BIO11 | Mean temp of the coldest quarter (x * 10, °C) | 0 | 68.54 | 51.69 | 69.00 | 7,706.5 *** | −0.0201 *** |
| 1 | 34.49 | 12.78 | 35.00 | ||||
| BIO12 | Annual precipitation (mm) | 0 | 494.92 | 338.62 | 402.50 | 1,705 *** | 0.00386 *** |
| 1 | 976.97 | 309.86 | 1,011.0 | ||||
| BIO13 | Precipitation of the wettest month (mm) | 0 | 75.13 | 48.68 | 63.50 | 1,182.5 *** | 0.0351 *** |
| 1 | 168.69 | 52.52 | 170.00 | ||||
| BIO14 | Precipitation of the driest month (mm) | 0 | 15.34 | 14.54 | 11.00 | 1,835.5 *** | 0.0623 *** |
| 1 | 28.67 | 11.41 | 26.00 | ||||
| BIO15 | Precipitation seasonality (variation coefficient) | 0 | 48.94 | 23.73 | 45.50 | 3,915 *** | 0.0200 ** |
| 1 | 58.02 | 7.70 | 58.00 | ||||
| BIO16 | Precipitation of the wettest quarter (mm) | 0 | 201.12 | 131.03 | 177.50 | 1,081 *** | 0.01396 *** |
| 1 | 455.36 | 131.42 | 476.00 | ||||
| BIO17 | Precipitation of the driest quarter (mm) | 0 | 54.09 | 49.44 | 40.00 | 1,728.5 *** | 0.02017 *** |
| 1 | 105.52 | 40.98 | 101.00 | ||||
| BIO18 | Precipitation of the warmest quarter (mm) | 0 | 151.59 | 128.00 | 88.00 | 5,584 ns | −0.00410 ** |
| 1 | 105.84 | 41.44 | 101.00 | ||||
| BIO19 | Precipitation of the coldest quarter (mm) | 0 | 94.67 | 98.46 | 56.00 | 438 *** | 0.0159 *** |
| 1 | 431.13 | 129.22 | 451.00 |
| Model | Variables | AIC | ΔAIC |
|---|---|---|---|
| m1 | BARE + BIO3 + BIO6 + BIO18 + BIO12 | 76.07 | 0.00 |
| m2 | BARE + BIO3 + BIO6 + BIO18 + BIO19 | 76.76 | 0.69 |
| m3 | BARE + BIO3 + BIO6 + BIO18 + BIO19 + TREE | 78.32 | 2.25 |
| m4 | BIO3 + BIO6 + BIO12 + BIO18 | 79.79 | 3.71 |
| m5 | BARE + BIO3 + BIO4 + BIO6 + BIO18 + BIO19 + TREE | 79.86 | 3.78 |
| m6 | BIO3 + BIO4 + BIO6 + BIO12 + BIO18 | 80.69 | 4.62 |
| m7 | BIO3 + BIO6 + BIO18 + BIO19 + TREE | 80.71 | 4.63 |
| m8 | HERB + BIO3 + BIO6 + BIO18 + BIO12 | 81.45 | 5.38 |
| m9 | BARE + BIO3 + BIO4 + BIO6 + BIO15 + BIO18 + BIO19 + TREE | 81.86 | 5.78 |
| m10 | BIO9 + BIO19 | 88.12 | 12.04 |
| m11 | BIO12 + BIO19 | 98.20 | 22.13 |
| m12 | BIO4 + BIO19 | 100.42 | 24.35 |

| Criteria | Threshold | Sensitivity | Specificity | False positive rate | False negative rate | Positive predictive value | Negative predictive value | K |
|---|---|---|---|---|---|---|---|---|
| GLM | ||||||||
| Min occurrence prediction | 0.019 | 1.000 | 0.742 | 0.258 | 0.000 | 0.560 | 1.00 | 0.59 |
| Mean occurrence prediction | 0.869 | 0.754 | 0.989 | 0.011 | 0.246 | 0.958 | 0.925 | 0.80 |
| 10% omission | 0.550 | 0.902 | 0.962 | 0.038 | 0.098 | 0.887 | 0.968 | 0.86 |
| Sens = Specif, Max Sens + Specif, Max prop correct, Max K, Min ROC plot distance | 0.430 | 0.951 | 0.952 | 0.048 | 0.049 | 0.866 | 0.983 | 0.87 |
| MaxEnt | ||||||||
| Min occurrence prediction | 0.093 | 1.000 | 0.505 | 0.495 | 0.000 | 0.399 | 1.000 | 0.33 |
| Mean occurrence prediction | 0.799 | 0.738 | 0.989 | 0.011 | 0.262 | 0.957 | 0.920 | 0.79 |
| 10% omission | 0.730 | 0.902 | 0.968 | 0.032 | 0.098 | 0.902 | 0.968 | 0.87 |
| Sens = Specif | 0.570 | 0.951 | 0.952 | 0.048 | 0.049 | 0.866 | 0.983 | 0.87 |
| Max Sens+Specif, Max prop correct, Max K, Min ROC plot distance | 0.650 | 0.951 | 0.968 | 0.032 | 0.049 | 0.906 | 0.984 | 0.90 |


3. Discussion
4. Materials and Methods
4.1. Hantavirus Pulmonary Syndrome Data
4.2. Environmental Data
4.3. Spatial and Statistical Modeling
| Performance measure | Definition | Formula |
|---|---|---|
| Sensitivity (True positive rate) | Proportion true presences correctly predicted | TP/P |
| Specificity (True negative rate) | Proportion true absences correctly predicted | TN/N |
| False positive rate | FP/N | |
| False negative rate | FN/P | |
| Positive predictive value (Precision) | Percentage of predicted presences that were real | TP/(TP + FP) |
| Negative predictive value | Percentage of predicted absences that were real | TN/(TN + FN) |
4.4. HPS Risk Mapping: Integration with Previous Potential Distribution Map of Host

5. Conclusions
Acknowledgments
Conflicts of Interest
References and Notes
- Enria, D.A.M.; Levis, S.C. Zoonosis virales emergentes: Las infecciones por Hantavirus. Introducción e Historia. Rev. Sci. Tech. Off. Int. Epiz. 2004, 23, 595–611. (in Spanish). [Google Scholar]
- Hjelle, B.; Torres-Pérez, F. Hantaviruses in the Americas and their role as emerging pathogens. Viruses 2010, 2, 2559–2586. [Google Scholar] [CrossRef]
- Kang, H.J.; Bennett, S.N.; Hope, A.G.; Cook, J.A.; Yanagihara, R. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J. Virol. 2011, 85, 7496–7503. [Google Scholar] [CrossRef]
- Song, J.W.; Baek, L.J.; Schmaljohn, C.S.; Yanagihara, R. Thottapalayam virus, a prototype shrewborne hantavirus. Emerg. Infect. Dis. 2007, 13, 980–985. [Google Scholar] [CrossRef]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef]
- Jonsson, C.B.; Tadeu, L.; Figueiredo, M.; Vapalahti, O. A global perspective on hantavirus: Ecology, epidemiology and disease. Clin. Microbiol. Rev. 2010, 23, 412–441. [Google Scholar] [CrossRef]
- Glass, G.E.; Childs, J.E.; Korch, G.W.; LeDuc, J.W. Association of intraspecific wounding with hantaviral infection in wild rats (Rattus norvegicus). Epidemiol. Infect. 1988, 101, 459–472. [Google Scholar] [CrossRef]
- Childs, J.E.; Ksiazek, T.G.; Spiropoulou, C.F.; Krebs, J.W.; Morzunov, S.; Maupin, G.O.; Rollin, P.E.; Sarisky, J.; Enscore, R.E. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J. Infect. Dis. 1994, 169, 1271–1280. [Google Scholar] [CrossRef]
- Botten, J.; Mirowsky, K.; Ye, C.Y.; Gottlieb, K.; Saavedra, M.; Ponce, L.; Hjelle, B. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J. Virol. 2002, 76, 7587–7594. [Google Scholar] [CrossRef]
- Padula, P.; Figueroa, R.; Navarrete, M.; Pizarro, E.; Cadiz, R.; Bellomo, C.; Jofre, C.; Zaror, L.; Rodriguez, E.; Murua, R. Transmission study of Andes hantavirus infection in wild sigmodontine rodents. J. Virol. 2004, 78, 11972–11979. [Google Scholar] [CrossRef]
- Mills, J.; Childs, J.E. Ecologic studies of rodent reservoirs: Their relevance for human health. Emerg. Infect. Dis. 1998, 4, 529–537. [Google Scholar] [CrossRef]
- Mills, J. The role of rodents in emerging human disease: Examples from the hantaviruses and arenaviruses. In Ecologically-Based Rodent Management; Singleton, G.R., Hinds, L.A., Leirs, H., Zhang, Z., Eds.; Arrawang Communication Group: Canberra, Australia, 1999; pp. 134–160. [Google Scholar]
- Yates, T.; Mills, J.; Parmenter, C.; Ksiazek, T.; Parmenter, R.; Vande Castle, J.; Calisher, C.; Nichol, S.; Abbott, K.; et al. The ecology and evolutionary history of an emergent disease: Hantavirus Pulmonary Syndrome. Bioscience 2002, 52, 989–998. [Google Scholar] [CrossRef]
- Parmenter, R.; Yadav, E.; Parmenter, C.; Ettestad, P.; Gage, K. Incidence of plague associated with increased winter-spring precipitation in New Mexico. Am. J. Trop. Med. Hyg. 1999, 61, 814–821. [Google Scholar]
- Linard, C.; Lamarque, P.; Heyman, P.; Ducoffre, G.; Luyasu, V.; Tersago, K.; Vanwambeke, S.O.; Lambin, E.F. Determinants of the geographic distribution of Puumala virus and Lyme borreliosis infections in Belgium. Int. J. Health Geogr. 2007, 2, 6–15. [Google Scholar]
- Vanwambeke, S.O.; Šumilo, D.; Bormane, A.; Lambin, E.F.; Randolph, S.E. Landscape predictors of tick-borne encephalitis in Latvia: Land cover, land use, and land ownership. Vector Borne Zoonotic Dis. 2010, 10, 497–506. [Google Scholar] [CrossRef]
- Glass, G.; Cheek, J.; Patz, J.; Shields, T.; Doyle, T.; Thoroughman, D.; Hunt, D.; Enscore, R.; Gage, K.; Irland, C.; et al. Using remotely sensed data to identify areas at risk for Hantavirus Pulmonary Syndrome. Emerg. Infect. Dis. 2000, 6, 238–247. [Google Scholar] [CrossRef]
- Goodin, D.G.; Koch, D.E.; Owen, R.D.; Chu, Y.; Hutchinson, J.M.S.; Jonsson, C.B. Land cover associated with hantavirus presence in Paraguay. Glob. Ecol. Biogeogr. 2006, 15, 519–527. [Google Scholar]
- Andreo, V.; Glass, G.; Shields, T.; Provensal, C.; Polop, J. Modeling potential distribution of Oligoryzomys longicaudatus, the Andes Virus (Genus: Hantavirus) reservoir, in Argentina. EcoHealth 2011, 3, 332–348. [Google Scholar]
- Pearson, R.G.; Dawson, T.P. Predicting the impacts of climate change on the distribution of species: Are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudik, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species' distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species Distribution Models: Ecological explanation and prediction across space and time. Ann. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Franklin, J. Mapping Species Distributions. Spatial Inference and Prediction; Cambridge University Press: New York, NY, USA, 2010; p. 320. [Google Scholar]
- Sosa-Estani, S.; Salomón, O.D.; Gómez, A.O.; Esquivel, M.L.; Segura, E.L. Diferencias regionales y Síndrome Pulmonar por Hantavirus (enfermedad emergente y tropical en Argentina). Cad. Saude Publica 2001, 17, 47–57. (in Spanish). [Google Scholar]
- Padula, P.J.; Martinez, V.P.; Bellomo, C.; Maidana, S.; San Juan, J.; Tagliaferri, P.; Bargardi, S.; Vazquez, C.; Colucci, N.; Estevez, J.; et al. Pathogenic hantaviruses, northeastern Argentina and eastern Paraguay. Emerg. Infect. Dis. 2007, 13, 1211–1214. [Google Scholar] [CrossRef]
- Martinez, V.P.; Bellomo, C.M.; Cacace, M.L.; Suárez, P.; Bogni, L.; Padula, P.J. Hantavirus pulmonary syndrome in Argentina, 1995–2008. Emerg. Infect. Dis. 2010, 16, 1853–1860. [Google Scholar] [CrossRef]
- Lopez, N.; Padula, P.; Rossi, C.; Lazaro, M.E.; Franze-Fernandez, M.T. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology 1996, 219, 1–4. [Google Scholar] [CrossRef]
- Levis, S.; Morzunov, S.; Rowe, J.; Enria, D.; Pini, N.; Calderón, G.; Sabattini, M.; St Jeor, S. Genetic diversity and epidemiology of hantaviruses in Argentina. J. Infect. Dis. 1998, 177, 529–538. [Google Scholar]
- Murúa, R.; Gonzalez, L.A. Microhabitat selection in two Chilean cricetid rodents. Oecologia 1982, 52, 12–15. [Google Scholar] [CrossRef]
- Pearson, O.P. Characteristics of a mammalian fauna from forests in Patagonia, Southern Argentina. J. Mammal. 1983, 64, 476–492. [Google Scholar] [CrossRef]
- Larrieu, E.; Herrero, E.; García Cachau, M.; Labanchi, J.L.; Mancini, S.; Padula, P.; Cantoni, G.; Cavagion, L.; Alvarez, E.; Bruni, M.; et al. Seroprevalencia de hantavirus en roedores y casos humanos en el sur de Argentina. Rev. Bras. Epidemiol. 2003, 6, 68–75. [Google Scholar] [CrossRef]
- Piudo, L.; Monteverde, M.; Gonzalez Capria, S.; Padula, P.; Carmanchahi, P. Distribution and abundance of sigmodontine rodents in relation to hantavirus in Neuquén, Argentina. J. Vector Ecol. 2005, 30, 119–125. [Google Scholar]
- Polop, F.J.; Provensal, M.C.; Pini, N.; Levis, S.C.; Priotto, J.W.; Enría, D.; Calderón, G.E.; Costa, F.; Polop, J.J. Temporal and spatial host abundance and prevalence of Andes Hantavirus in Southern Argentina. EcoHealth 2010, 7, 176–184. [Google Scholar] [CrossRef]
- Andreo, V.; Provensal, M.C.; Levis, S.C.; Pini, N.; Enria, D.; Polop, J. Summer-autumn distribution and abundance of the hantavirus host, Oligoryzomys longicaudatus, in northwestern Chubut, Argentina. J. Mammal. 2012, 93, 1559–1568. [Google Scholar] [CrossRef]
- Jaksic, F.M.; Lima, M. Myths and facts on ratadas: Bamboo blooms, rainfall peaks and rodent outbreaks in South America. Austral Ecol. 2003, 28, 237–251. [Google Scholar] [CrossRef]
- Sage, R.D.; Pearson, O.P.; Sanguinetti, J.; Pearson, A.K. Ratada 2001: A rodent outbreak following the flowering of bamboo (Chusquea culeou) in Southwestern Argentina. In The Quintessential Naturalist; Kelt, D., Kaspin, D., Eds.; University of California Press: Berkeley, CA, USA, 2007; pp. 177–224. [Google Scholar]
- Pearson, O.P. A perplexing outbreak of mice in Patagonia, Argentina. Stud. Neotrop. Fauna Environ. 2002, 37, 187–200. [Google Scholar] [CrossRef]
- Murua, R.; Gonzalez, L.A.; Lima, M. Population dynamics of rice rats (a Hantavirus reservoir) in southern Chile: Feedback structure and non-linear effects of climatic oscillations. Oikos 2003, 102, 137–145. [Google Scholar] [CrossRef]
- Cantoni, G.; Padula, P.; Calderón, G.; Mills, J.; Herrero, E.; Sandoval, P.; Martinez, V.; Pini, N.; Larrieu, E. Seasonal variation in prevalence of antibody to hantaviruses in rodents from southern Argentina. Trop. Med. Int. Health 2001, 6, 811–816. [Google Scholar] [CrossRef]
- Yadón, Z. Epidemiología del síndrome pulmonar por hantavirus en la Argentina (1991–1997). Medicina 1998, 58, 25–26. (in Spanish). [Google Scholar]
- Wells, R.; Sosa, S.; Yadón, Z.; Enría, D.; Padula, P.; Pini, N.; Mills, J.N.; Peters, C.J.; Segura, E.L. An unusual hantavirus outbreak in south Argentina: Person-to-person transmission? Emerg. Infect. Dis. 1997, 3, 171–174. [Google Scholar] [CrossRef]
- Padula, P.; Edelstein, A.; Miguel, S.; Lopez, M.; Rossi, C.; Rabinovish, R. Hantavirus pulmonary syndrome outbreak in Argentina: Molecular evidence of person-to-person transmission of Andes virus. Virology 1998, 15, 323–330. [Google Scholar]
- Martinez, V.P.; Bellomo, C.; San Juan, J.; Pinna, D.; Forlenza, R.; Elder, M.; Padula, P.J. Person-to-person transmission of Andes virus. Emerg. Infect. Dis. 2005, 11, 1848–1853. [Google Scholar] [CrossRef]
- Porcasi, X.; Calderon, G.; Lamfri, M.; Gardenal, N.; Polop, J.; Sabattini, M.; Scavuzzo, M. The use of satellite in modeling population dynamics and prevalence of infection in the rodent reservoir of Junin virus. Ecol. Model. 2005, 185, 437–449. [Google Scholar] [CrossRef]
- Carbajo, A.E.; Pardiñas, U.F.J. Spatial distribution model of a hantavirus reservoir, the long-tailed colilargo (Oligoryzomys longicaudatus), in Argentina. J. Mammal. 2007, 88, 1555–1568. [Google Scholar] [CrossRef]
- Belmar-Lucero, S.; Godoy, P.; Ferres, M.; Vial, P.; Palma, R.E. Range expansion of Oligoryzomys longicaudatus (Rodentia: Sigmodontinae) in Patagonian Chile and first record of Hantavirus in the region. Rev. Chil. Hist. Nat. 2009, 82, 265–275. [Google Scholar]
- Haylock, M.R.; Peterson, T.; Sousa, J.R.A.D.; Alves, L.M.; Ambrizzi, T.; Baez, J.; Barbosa, J.I.; Barros, V.R.; Berlato, M.A.; Bidegain, M.; et al. Trends in total and extreme South American rainfall in 1960–2000 and links with sea surface temperature. J. Climate 2006, 19, 1490–1512. [Google Scholar] [CrossRef]
- Rusticucci, M.; Penalba, O. Interdecadal changes in the precipitation seasonal cycle over Southern South America and their relationship with surface temperature. Clin. Res. 2000, 16, 1–15. [Google Scholar]
- Carbajo, A.E.; Vera, C.; González, P.L.M. Hantavirus reservoir Oligoryzomys longicaudatus spatial distribution sensitivity to climate change scenarios in Argentine Patagonia. Int. J. Health Geogr. 2009, 8, 44. [Google Scholar] [CrossRef]
- Schmaljohn, C.; Hjelle, B. Hantaviruses: A global disease problem. Emerg. Infect. Dis. 1997, 3, 95–104. [Google Scholar] [CrossRef]
- Olsson, G.E.; Dalerum, F.; Hörnfeldt, B.; Elgh, F.; Palo, T.R.; Juto, P.; Ahlm, C. Human hantavirus infections, Sweden. Emerg. Infect. Dis. 2003, 9, 1395–1401. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística y Censos (INDEC). Censo nacional de población, hogares y viviendas 2010. Censo del Bicentenario: resultados definitivos. (in Spanish). Buenos Aires, Argentina, 2012; p. 378. Available online: http://www.indec.gov.ar/ (accessed on 23 December 2013).
- Sbriller, A.; Sepúlveda, L. La Rosa Mosqueta, el colilargo patagónico y el Hantavirus. Desde la Patagonia 2007, 5, 18–22. (in Spanish). [Google Scholar]
- Calisher, C.H.; Mills, J.N.; Sweeney, W.P.; Choate, J.R.; Sharp, D.E.; Canestorp, K.M.; Beaty, B.J. Do unusual site-specific population dynamics of rodent reservoirs provide clues to the natural history of hantaviruses? J. Wildl. Dis. 2001, 37, 280–288. [Google Scholar] [CrossRef]
- Madhav, N.K.; Wagoner, K.D.; Douglass, R.J.; Mills, J.N. Delayed density-dependent prevalence of Sin Nombre virus antibody in Montana deer mice (Peromyscus maniculatus) and implications for human disease risk. Vector Borne Zoonotic Dis. 2007, 7, 353–364. [Google Scholar]
- Bellomo, C.; Nudelman, J.; Kwaszka, R.; Vazquez, G.; Cantoni, G.; Weinzettel, B.; Larrieu, E.; Padula, P. Expansión geográfica del Síndrome Pulmonar por Hantavirus en la Argentina: Informe del caso más austral. Medicina (Buenos Aires) 2009, 69, 647–650. (in Spanish). [Google Scholar]
- Carbajo, A.E.; Cardo, M.V.; Vezzani, D. Is temperature the main cause of dengue rise in non-endemic countries? The case of Argentina. Int. J. Health Geogr. 2012, 11, 26. [Google Scholar] [CrossRef]
- Peterson, A.T. Ecological niche modelling and understanding the geography of disease transmission. Vet. Ital. 2007, 43, 393–400. [Google Scholar]
- SIGN. Sistema de Información Geográfica del Instituto Geográfico Nacional de la República Argentina. Available online: http://sig.ign.gob.ar/ (accessed on 23 December 2013).
- Lobo, J.M.; Jimenez-Valverde, A.; Hortal, J. The uncertain nature of absences and their importance in Species Distribution Modelling. Ecography 2010, 33, 103–114. [Google Scholar] [CrossRef]
- Wisz, M.S.; Guisan, A. Do pseudo-absence selection strategies influence Species Distribution Models and their predictions? An information-theoretic approach based on simulated data. BMC Ecol. 2009, 13, 1–13. [Google Scholar]
- GRASS Development Team. Geographic Resources Analysis Support System (GRASS) Software, Version 7; Open Source Geospatial Foundation: San Michele all’Adige, Italy, 2013. Available online: http://grass.osgeo.org/ (accessed on 21 Dec 2013).
- Neteler, M.; Bowman, M.H.; Landa, M.; Metz, M. GRASS GIS: A multi-purpose open source GIS. Environ. Model. Softw. 2012, 31, 124–130. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Hansen, M.; de Fries, R.S.; Townshend, J.R.G.; Carroll, M.; Dimiceli, C.; Sohlberg, R.A. Global percent tree cover at a spatial resolution of 500 meters: First results of the MODIS vegetation continuous fields algorithm. Earth Interact. 2003, 7, 1–15. [Google Scholar]
- Friedl, M.A.; McIver, D.K.; Hodges, J.C.F.; Zhang, X.; Muchoney, D.; Strahler, A.H.; Woodcock, C.E.; Gopal, S.; Schnieder, A.; Cooper, A.; et al. Global land cover from MODIS: Algorithms and early results. Remote Sens. Environ. 2002, 83, 135–148. [Google Scholar] [CrossRef]
- Zuur, A.F.; Ieno, E.N.; Walker, N.; Saveliev, A.; Smith, G. Mixed Effects Models and Extensions in Ecology with R; Springer: New York, NY, USA, 2009; p. 574. [Google Scholar]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A. A practical guide to MaxEnt for modeling species’ distributions: What it does, and why inputs and settings matter. Ecography 2013, 36, 1–12. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria, 2013. Available online: http://www.R-project.org/ (accessed on 23 December 2013).
- Rosenberg, M.S.; Anderson, C.D. PASSaGE: Pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol. Evol. 2011, 2, 229–232. [Google Scholar] [CrossRef]
- Vaughan, I.P.; Ormerod, S.J. The continuing challenges of testing species distribution. J. Appl. Ecol. 2005, 42, 720–730. [Google Scholar] [CrossRef]
- Manel, S. Evaluating presence–absence models in Ecology: The need to account for prevalence. J. Appl. Ecol. 2001, 38, 921–931. [Google Scholar] [CrossRef]
- Liu, C.; Berry, P.M.; Dawson, T.P.; Pearson, R.G. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 2005, 28, 385–393. [Google Scholar] [CrossRef]
- Freeman, E.A.; Moisen, G.G. A comparison of the performance of threshold criteria for binary classification in terms of predicted prevalence and kappa. Ecol. Model. 2008, 217, 48–58. [Google Scholar] [CrossRef]
- Liu, C.; White, M.; Newell, G. Selecting thresholds for the prediction of species occurrence with presence-only data. J. Biogeogr. 2013, 40, 778–789. [Google Scholar] [CrossRef]
- VanDerWal, J.; Falconi, L.; Januchowski, S.; Shoo, L.; Storlie, C. Species Distribution Modelling Tools: Tools for Processing Data Associated with Species Distribution Modelling Exercises. Package ‘SDMTools’ 2012. Available online: http://www.rforge.net/SDMTools/ (accessed on 23 December 2013).
- Anderson, R.P.; Lew, D.; Peterson, A.T. Evaluating predictive models of species’ distributions: Criteria for selecting optimal models. Ecol. Model. 2003, 162, 211–232. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Hanspach, J.; Kühn, I.; Schweiger, O.; Pompe, S.; Klotz, S. Geographical patterns in prediction errors of species distribution models. Glob. Ecol. Biogeogr. 2011, 20, 779–788. [Google Scholar] [CrossRef]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Andreo, V.; Neteler, M.; Rocchini, D.; Provensal, C.; Levis, S.; Porcasi, X.; Rizzoli, A.; Lanfri, M.; Scavuzzo, M.; Pini, N.; et al. Estimating Hantavirus Risk in Southern Argentina: A GIS-Based Approach Combining Human Cases and Host Distribution. Viruses 2014, 6, 201-222. https://doi.org/10.3390/v6010201
Andreo V, Neteler M, Rocchini D, Provensal C, Levis S, Porcasi X, Rizzoli A, Lanfri M, Scavuzzo M, Pini N, et al. Estimating Hantavirus Risk in Southern Argentina: A GIS-Based Approach Combining Human Cases and Host Distribution. Viruses. 2014; 6(1):201-222. https://doi.org/10.3390/v6010201
Chicago/Turabian StyleAndreo, Veronica, Markus Neteler, Duccio Rocchini, Cecilia Provensal, Silvana Levis, Ximena Porcasi, Annapaola Rizzoli, Mario Lanfri, Marcelo Scavuzzo, Noemi Pini, and et al. 2014. "Estimating Hantavirus Risk in Southern Argentina: A GIS-Based Approach Combining Human Cases and Host Distribution" Viruses 6, no. 1: 201-222. https://doi.org/10.3390/v6010201
APA StyleAndreo, V., Neteler, M., Rocchini, D., Provensal, C., Levis, S., Porcasi, X., Rizzoli, A., Lanfri, M., Scavuzzo, M., Pini, N., Enria, D., & Polop, J. (2014). Estimating Hantavirus Risk in Southern Argentina: A GIS-Based Approach Combining Human Cases and Host Distribution. Viruses, 6(1), 201-222. https://doi.org/10.3390/v6010201