Phage ?Pan70, a Putative Temperate Phage, Controls Pseudomonas aeruginosa in Planktonic, Biofilm and Burn Mouse Model Assays
Abstract
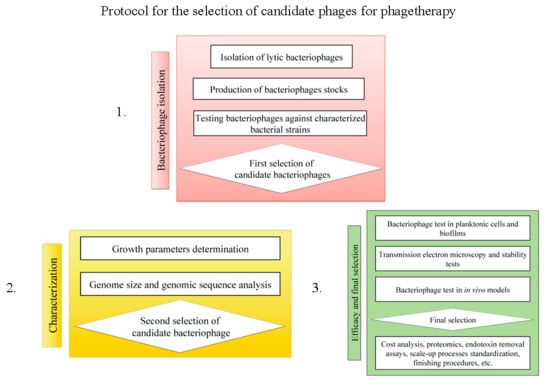
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Bacterial Culture Conditions
2.3. Bacteriophage Isolation and Propagation
2.4. Host Range, Plaque Morphology and Propagation of ΦPan70
2.5. Genome Size Estimation
2.6. Transmission Electron Microscopy (TEM)
2.7. Activity of ΦPan70 against Planktonic Cells
2.8. Activity of Bacteriophages on Biofilms
2.9. Activity of Bacteriophages in the Burned Mouse Model
2.10. Genome Sequencing, Assembly and Annotation
3. Results
3.1. ΦPan70 is a Member of the Myoviridae Family


3.2. ΦPan70 Reduces Planktonic Cells
3.3. ΦPan70 Reduces Biofilm Formation and Existing Biofilms
3.4. Phage-Therapy with ΦPan70 Improved Survival and Health in the Burned Mouse Model


3.5. Genome Sequencing, Assembly and Annotation

| ORF | Function | |
| gp001 | Small Subunit DNA Packaging Dimer (Pseudomonas Phage F10) | |
| gp002 | Large Subunit DNA Packaging Dimer (Pseudomonas Phage F10) | |
| gp004 | Portal Protein (Pseudomonas Phage F10) | |
| gp005 * | Putative ClpP Protease (Pseudomonas Phage F10) | |
| gp013 * | Putative Tail Length Tape Measure Protein (P. aeruginosa LESB58) | |
| gp017 | Tail Fiber Protein (P. aeruginosa PA21_ST175) | |
| gp028 | DNA Modification Protein (Desulfovibrio vulgaris str. 'Miyazaki F') | |
| gp029 | Site Specific Recombinase (P. aeruginosa ATCC 700888) | |
| gp043 | LuxR Family Transcriptional Regulator (P. aeruginosa LESB58) | |
| gp044 | Transcriptional Regulator (P. aeruginosa ATCC 25324) | |
| gp045 | Repressor (Pseudomonas syringae) | |
| gp052 | Rha Family Phage Regulatory Protein (P. aeruginosa LESB58) | |
| gp055 | DNA Replication protein, DnaC (P. aeruginosa LESB58) | |
| gp056 | Putative DnaB-like Replicative Helicase (P. aeruginosa LESB58) | |
| gp058 | Putative Metallophosphoesterase (Pseudomonas PhageF10) | |
| gp061 | Holin (P. aeruginosa LESB58) | |
| gp62 | Chitinase (P. aeruginosa LESB58) | |
4. Discussion
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Taylor, P. Multiple Drug Resistant Bacteria. Drug Discov. Today 2003, 8, 978–979. [Google Scholar] [CrossRef]
- Welte, T.; Pletz, M. Antimicrobial treatment of nosocomial methicillin-resistant Staphylococcus aureus (MRSA) pneumonia: Current and future options. Int. J. Antimicrob. Agents 2010, 36, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Kerr, K.; Senelling, A. Pseudomonas aeruginosa: A formidable and ever-present adversary. J. Hosp. Infect. 2009, 73, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Prada, G. Impacto de la Resistencia a los Antibióticos en el desarrollo de la Medicina Contemporánea. Rev. Fac. Med. 2007, 16, 9–11. [Google Scholar]
- Leal, A.; Álvarez, C.; Buitrago, G.; Méndez, M.; GREBO. Canales Endémicos y Marcadores de Resistencia Bacteriana, en Instituciones de Tercer Nivel de Bogota, Colombia. Rev. Salud Publica 2006, 8, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Lampert, R.; Boyce, F.; McFetridge, E. Bacteriophage therapy. A clinical study, with special reference to the technique of application. Am. J. Surg. 1935, 29, 436–443. [Google Scholar] [CrossRef]
- Parfitt, T. Georgia: An unlikely stronghold for bacteriophage therapy. Lancet 2005, 365, 2166–2167. [Google Scholar] [CrossRef]
- Carlson, K. Appendix: Working with Bacteriophages: Common techniques and methodological approaches. In Bacteriophages: Biology and Applications, 1st ed.; Sulakvelidze, A., Kutter, E., Eds.; CRC Press: Boca Raton, FL, USA, 2005; Volume 1, pp. 439–490. [Google Scholar]
- Brüssow, H. Bacteriophage Therapy: Potential and Problems. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; Volume 1, pp. 267–273. [Google Scholar]
- Kutateladze, M.; Adamia, R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 2010, 28, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.; Kuhl, S.; Blasdel, B.; Kutter, E. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.; Vives, M.; Gónzalez, A. Bioprospección de bacteriófagos activos contra Pseudomonas aeruginosa. In Proceedings of the XIX Congreso Latinoamericano de Microbiología, Quito, Ecuador, 15–18 October 2008.
- Merabishvili, M.; Pirnay, J.P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; van Parys, L.; et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS ONE 2009, 4, 4944–4949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clavijo, V. Efecto de bacteriófagos sobre biopelículas de Pseudomonas aeruginosa resistentes a múltiples drogas (MDR). Master Thesis, Universidad de los Andes, Bogotá, Colombia, 2010. [Google Scholar]
- Recipe. SM buffer with gelatin. Cold Spring Harb. Protoc. 2006. [CrossRef]
- Ackermann, H. Phage Classification and Characterization. In Bacteriophages methods and protocols, 1st ed.; Clokie, M., Kropinski, A., Eds.; Humana: New York, NY, USA, 2009; Volume 1, pp. 127–141. [Google Scholar]
- Frampton, R.; Taylor, C.; Holguín, A.; Visnovsky, S.; Petty, N.; Pitman, A.; Fineran, P. Identification of bacteriophages for biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv. actinidiae. Appl. Environ. Microbiol. 2014, 80, 2216–2228. [Google Scholar] [CrossRef] [PubMed]
- Ardila, C.; Castro, J.; Holguin, A.; Clavijo, V.; Vives, M.; González, A. Determination of the Optimal Bacteriophage Dose to Control Pseudomonas aeruginosa using evolutionary programming and stochastic kinetics. Unpublished data.
- Burton, E.; Yakandawala, N.; LoVetri, K.; Madhyastha, M. A microplate spectrofluorometric assay for bacterial biofilms. J. Ind. Microbiol. Biotechnol. 2007, 34, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Recipe. Phosphate-buffered saline (PBS). Cold Spring Harb. Protoc. 2006. [CrossRef]
- Colombia Ministerio de Salud. Resolución No. 008430 de 1993. Available online: https://www.invima.gov.co/index.php?option=com_content&view=article&id=2977:resolucion-no-8430-del-4-de-octubre-de-1993&catid=147:resoluciones-medicamentos-&Itemid=203 (accessed on 20 July 2013).
- National Research Council (Ed.) Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011.
- Heo, Y.; Lee, Y.; Jung, H.; Lee, J.; Ko, G.; Cho, Y. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob. Agents Chemother. 2009, 53, 2469–2474. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.; Eddy, S. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lukashin, A.; Borodovsky, M. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 1998, 26, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Glimmer Microbial Gene-Finding System. Available online: https://ccb.jhu.edu/software/glimmer/ (accessed on 9 July 2013).
- CD-HIT Representative sequences. Available online: http://weizhongli-lab.org/cd-hit/ (accessed on 10 July 2013).
- Aziz, R.; Bartels, D.; Best, A.; de Jongh, M.; Disz, T.; Edwards, R.; Formsma, K.; Gerdes, S.; Glass, E.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, e75. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.; Mau, B.; Blattner, F.; Perna, N. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- VISTA Tools for Comparative Genomics. Available online: http://genome.lbl.gov/vista/aboutus.shtml (accessed on 20 July 2013).
- Winstanley, C.; Langille, M.; Fothergill, J.; Kukavica-Ibrulj, I.; Paradis-Bleau, C.; Sanschagrin, F.; Thomson, N.; Winsor, G.; Quail, M.; Lennard, N.; et al. Newly Introduced Genomic Prophage Islands Are Critical Determinants of in Vivo Competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2013, 19, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Buyck, J.; Tulkens, P.; Bambeke, V. Pharmacodynamic evaluation of the intracellular activity of antibiotics towards Pseudomonas aeruginosa PAO1 in a model of THP-1 human monocytes. Antimicrob. Agents Chemother. 2013, 57, 2310–2318. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.; Erwin, T.; McLean, R.; Aron, G. Bacteriophage Ecology in Escherichia coli and Pseudomonas aeruginosa Mixed-Biofilm Communities. Appl. Environ. Microbiol. 2011, 77, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Sillankorva, S.; Oliveira, R.; Vieira, M.; Sutherland, I.; Azeredo, J. Bacteriophage Φ S1 Infection of Pseudomonas fluorescens Planktonic Cells vs. Biofilms. Biofouling J. Bioadhesion Biofilm Res. 2004, 20, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Phee, A.; Bondy-Denomy, J.; Kishen, A.; Basrani, B.; Azarpazhooh, A.; Maxwell, K. Efficacy of Bacteriophage Treatment on Pseudomonas aeruginosa Biofilms. J. Endod. 2013, 39, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Zimecki, M.; Artym, J.; Kocieba, M.; Weber-Dabrowska, B.; Borysowski, J.; Gorski, A. Effects of Prophylactic Administration of Bacteriophages to Immunosuppressed Mice Infected with Staphylococcus aureus. BMC Microbiol. 2009, 9, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Miernikiewicz, P.; Dąbrowska, K.; Piotrowicz, A.; Owczarek, B.; Wojas-Turek, J.; Kicielińska, J.; Rossowska, J.; Pajtasz-Piasecka, E.; Hodyra, K.; Macegoniuk, K.; et al. T4 Phage and Its Head Surface Proteins do not Stimulate Inflammatory Mediator Production. PLoS ONE 2013, 8, e71036. [Google Scholar]
- Lu, T.; Koeris, M. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 2011, 14, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Harjai, K.; Chhibber, S. Bacteriophage Treatment of Burn Wound Infection Caused by Pseudomonas aeruginosa PAO in BALB/c Mice. Am. J. Biomed. Sci. 2009, 1, 385–394. [Google Scholar] [CrossRef]
- Siebenhaar, F.; Syska, W.; Weller, K.; Magerl, M.; Zuberbier, T.; Metz, M.; Maurer, M. Control of Pseudomonas aeruginosa Skin Infections in Mice Is Mast Cell-Dependent. Am. J. Pathol. 2007, 170, 1910–1915. [Google Scholar] [CrossRef] [PubMed]
- Golkar, Z.; Bagasra, O.; Jamil, N. Experimental phage therapy on multiple drug resistant Pseudomonas aeruginosa infection in mice. J. Antivir. Antiretrovir. 2007, 51, 1934–1938. [Google Scholar]
- Tony, K.; Liu, J.; DuBow, M.; Gros, P.; Pelletier, J. Comparative Genomic Analysis of 18 Pseudomonas aeruginosa Bacteriophages. J. Bacteriol. 2006, 188, 1184–1187. [Google Scholar]
- The Félix d´Hérelle Reference Center for Bacterial Viruses. Available online: http://www.phage.ulaval.ca/?pageDemandee=phage&noPhage=11&id=41&L=1 (accessed on 6 August 2013).
- Hendrix, R.; Smith, M.; Burns, R.; Ford, M.; Hatfull, G. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. 1999, 96, 2192–2197. [Google Scholar] [CrossRef] [PubMed]
- PHACTS. Available online: http://edwards.sdsu.edu/PHACTS/ (accessed on 1 March 2015).
- McNair, K.; Bailey, B.; Edwards, R. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res 2007, 171, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Sim, N.; Cho, Y. Antibacterial Efficacy of Temperate Phage-Mediated Inhibition of Bacterial Group Motilities Antimicrobial. Agents Chemother. 2012, 56, 5612–5617. [Google Scholar] [CrossRef] [PubMed]
- Krylov, V.; Shaburova, O.; Krylov, S.; Pleteneva, E. A Genetic Approach to the Development of New Therapeutic Phages to Fight Pseudomonas aeruginosa in Wound Infections. J. Viruses 2012, 5, 15–53. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holguín, A.V.; Rangel, G.; Clavijo, V.; Prada, C.; Mantilla, M.; Gomez, M.C.; Kutter, E.; Taylor, C.; Fineran, P.C.; Barrios, A.F.G.; et al. Phage ?Pan70, a Putative Temperate Phage, Controls Pseudomonas aeruginosa in Planktonic, Biofilm and Burn Mouse Model Assays. Viruses 2015, 7, 4602-4623. https://doi.org/10.3390/v7082835
Holguín AV, Rangel G, Clavijo V, Prada C, Mantilla M, Gomez MC, Kutter E, Taylor C, Fineran PC, Barrios AFG, et al. Phage ?Pan70, a Putative Temperate Phage, Controls Pseudomonas aeruginosa in Planktonic, Biofilm and Burn Mouse Model Assays. Viruses. 2015; 7(8):4602-4623. https://doi.org/10.3390/v7082835
Chicago/Turabian StyleHolguín, Angela V., Guillermo Rangel, Viviana Clavijo, Catalina Prada, Marcela Mantilla, María Catalina Gomez, Elizabeth Kutter, Corinda Taylor, Peter C. Fineran, Andrés Fernando González Barrios, and et al. 2015. "Phage ?Pan70, a Putative Temperate Phage, Controls Pseudomonas aeruginosa in Planktonic, Biofilm and Burn Mouse Model Assays" Viruses 7, no. 8: 4602-4623. https://doi.org/10.3390/v7082835
APA StyleHolguín, A. V., Rangel, G., Clavijo, V., Prada, C., Mantilla, M., Gomez, M. C., Kutter, E., Taylor, C., Fineran, P. C., Barrios, A. F. G., & Vives, M. J. (2015). Phage ?Pan70, a Putative Temperate Phage, Controls Pseudomonas aeruginosa in Planktonic, Biofilm and Burn Mouse Model Assays. Viruses, 7(8), 4602-4623. https://doi.org/10.3390/v7082835