High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Virus
2.2. Preparation of Antibody and Fabs
2.3. Cryo-Electron Microscopy
2.4. Image Processing
2.5. Model Building
2.6. Magnitude Ratio Calculation
2.7. Competition Capture ELISAs
3. Results
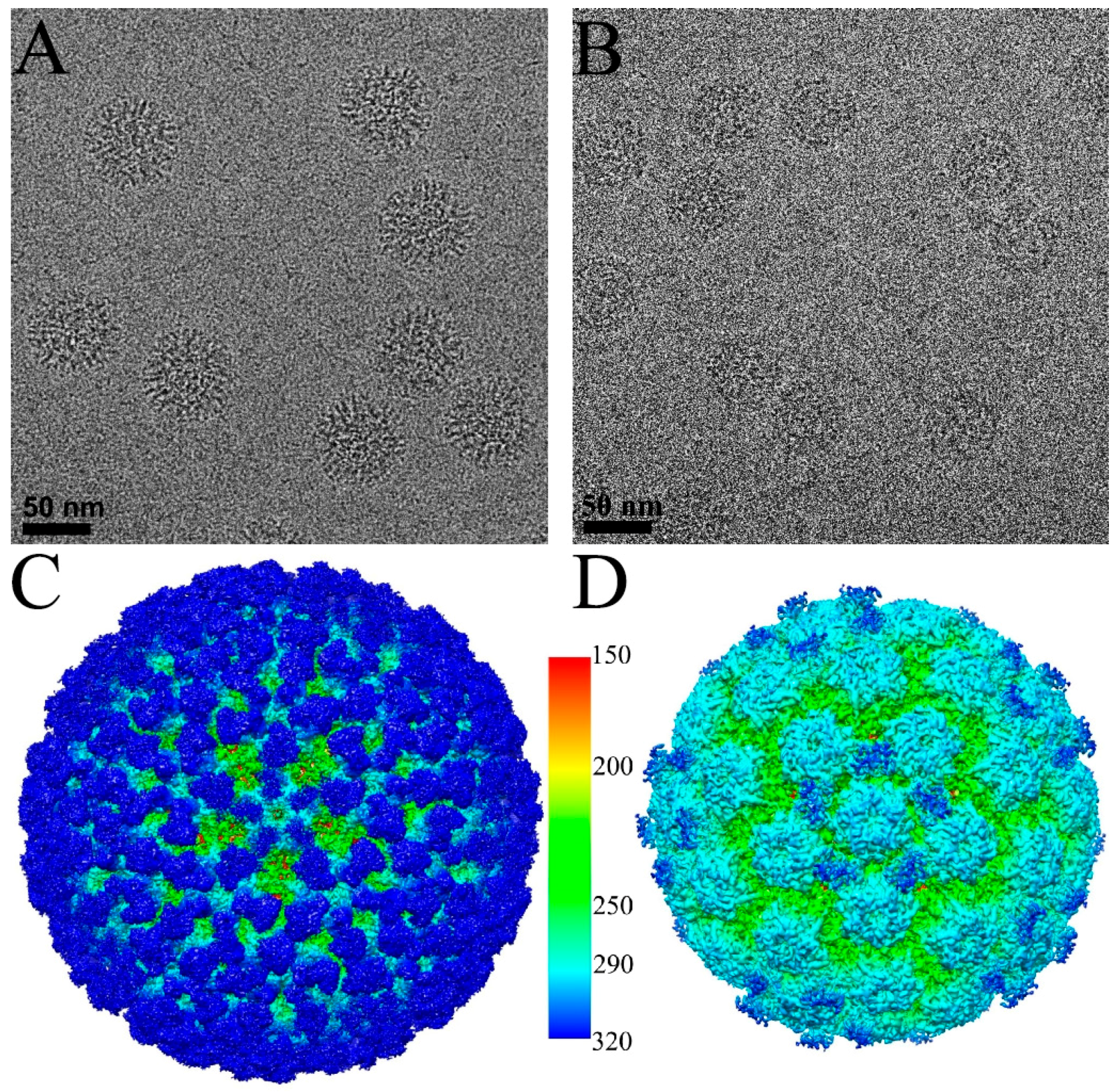
3.1. Cryo-EM Reconstructions of HPV-V5 Fab and HPV-U4 Fab Complexes
3.2. Building the V5 Fab and L1 Protein Structures into the High-Resolution Cryo-EM Maps
3.3. V5 Binding Induces Order to Capsomer Loops and Improves Resolution Significantly
3.4. U4 Fab Binding Suggests MAbs Bind Monovalently
3.5. Assessing Capsid Conformational Changes Initiated by U4 Binding
3.6. V5 and U4 Bind to Different Sites
3.7. V5/U4 mAb Competition Is Sequentially Dependent
4. Discussion
4.1. The Near-Atomic Resolution Map Allowed Building of the 50 kDa Fab Structure
4.2. V5 Neutralizes by Stabilization
4.3. V5 Preferentially Interacts with the Hexavalent Capsomer
4.4. U4 Blocks the Accessibility of Pentavalent Capsomer Sites
4.5. The Conformational Epitopes of V5 and U4 Were Identified Using the High-Resolution Maps
4.6. The V5/U4 Conformational Epitopes Are Non-Overlapping but V5 Outcompetes U4
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pullos, A.N.; Castilho, R.M.; Squarize, C.H. HPV infection of the head and neck region and its stem cells. J. Dent. Res. 2015, 94, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association: Human papillomavirus. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.Y.; Bracken, K. Update on the new 9-valent vaccine for human papillomavirus prevention. Can. Fam. Physician Méd. Fam. Can. 2016, 62, 399–402. [Google Scholar]
- Hammer, A.; Rositch, A.; Qeadan, F.; Gravitt, P.E.; Blaakaer, J. Age-specific prevalence of HPV16/18 genotypes in cervical cancer: A systematic review and meta-analysis: Age-specific HPV in cervical cancer. Int. J. Cancer 2016, 138, 2795–2803. [Google Scholar] [CrossRef] [PubMed]
- Dickson, E.L.; Vogel, R.I.; Luo, X.; Downs, L.S. Recent trends in type-specific HPV infection rates in the United States. Epidemiol. Infect. 2015, 143, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.M. HPV: The global burden. Nature 2012, 488, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Viens, L.J.; Henley, S.J.; Watson, M.; Markowitz, L.E.; Thomas, C.C.; Thompson, T.D.; Razzaghi, H.; Saraiya, M. Human papillomavirus–Associated cancers—United States, 2008–2012. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Nicol, A.F.; de Andrade, C.V.; Russomano, F.B.; Rodrigues, L.S.; Oliveira, N.S.; Provance, D.W., Jr.; Nuovo, G.J. HPV vaccines: Their pathology-based discovery, benefits and adverse effects. Ann. Diagn. Pathol. 2015, 19, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Culp, T.D.; Spatz, C.M.; Reed, C.A.; Christensen, N.D. Binding and neutralization efficiencies of monoclonal antibodies, Fab fragments and scFv specific for L1 epitopes on the capsid of infectious HPV particles. Virology 2007, 361, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Modis, Y.; High, K.; Towne, V.; Meng, Y.; Wang, Y.; Alexandroff, J.; Brown, M.; Carragher, B.; Potter, C.S.; et al. Disassembly and reassembly of human papillomavirus virus-like particles produces more virion-like antibody reactivity. Virol. J. 2012, 9. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Bywaters, S.M.; Brendle, S.A.; Lee, H.; Ashley, R.E.; Makhov, A.M.; Conway, J.F.; Christensen, N.D.; Hafenstein, S. Structural comparison of four different antibodies interacting with human papillomavirus 16 and mechanisms of neutralization. Virology 2015, 483, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Bywaters, S.M.; Brendle, S.A.; Lee, H.; Ashley, R.E.; Christensen, N.D.; Hafenstein, S. The U4 antibody epitope on human papillomavirus 16 identified by cryo-electron microscopy. J. Virol. 2015, 89, 12108–12117. [Google Scholar] [CrossRef] [PubMed]
- Seyferth, E.R.; Bratic, J.S.; Bocchini, J.A. Human papillomavirus epidemiology and vaccine recommendations: Selected review of the recent literature. Curr. Opin. Pediatr. 2016, 28, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Trimble, C.L.; Frazer, I.H. Development of therapeutic HPV vaccines. Lancet Oncol. 2009, 10, 975–980. [Google Scholar] [CrossRef]
- Markowitz, L.E.; Tsu, V.; Deeks, S.L.; Cubie, H.; Wang, S.A.; Vicari, A.S.; Brotherton, J. Human Papillomavirus vaccine introduction—The first five years. Vaccine 2012, 30, F139–F148. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.S.; Newcomb, W.W.; Olson, N.H.; Cowsert, L.M.; Olson, C.; Brown, J.C. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys. J. 1991, 60, 1445–1456. [Google Scholar] [CrossRef]
- Trus, B.L.; Roden, R.B.; Greenstone, H.L.; Vrhel, M.; Schiller, J.T.; Booy, F.P. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat. Struct. Biol. 1997, 4, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Modis, Y. Atomic model of the papillomavirus capsid. EMBO J. 2002, 21, 4754–4762. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Cheng, N.; Thompson, C.D.; Lowy, D.R.; Steven, A.C.; Schiller, J.T.; Trus, B.L. Arrangement of L2 within the papillomavirus capsid. J. Virol. 2008, 82, 5190–5197. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.J.; Wipf, G.C.; Benki, S.F.; Christensen, N.D.; Galloway, D.A. Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1. J. Virol. 2003, 77, 11625–11632. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G.; Johnson, J.E. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989, 58, 533–569. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Garcea, R.L.; Grigorieff, N.; Harrison, S.C. Subunit interactions in bovine papillomavirus. Proc. Natl. Acad. Sci. USA 2010, 107, 6298–6303. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Thompson, C.D.; Pang, Y.-S.; Lowy, D.R.; Schiller, J.T. Maturation of papillomavirus capsids. J. Virol. 2005, 79, 2839–2846. [Google Scholar] [CrossRef] [PubMed]
- Kirnbauer, R.; Booy, F.; Cheng, N.; Lowy, D.R.; Schiller, J.T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 1992, 89, 12180–12184. [Google Scholar] [CrossRef] [PubMed]
- Kirnbauer, R.; Taub, J.; Greenstone, H.; Roden, R.; Dürst, M.; Gissmann, L.; Lowy, D.R.; Schiller, J.T. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 1993, 67, 6929–6936. [Google Scholar] [PubMed]
- Hernandez, B.Y.; Ton, T.; Shvetsov, Y.B.; Goodman, M.T.; Zhu, X. Human Papillomavirus (HPV) L1 and L1-L2 virus-like particle-based multiplex assays for measurement of HPV virion antibodies. Clin. Vaccine Immunol. 2012, 19, 1348–1352. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Pastrana, D.V.; Lowy, D.R.; Schiller, J.T. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 2004, 78, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Christensen, N.D. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir. Chem. Chemother. 2005, 16, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Christensen, N.D.; Dillner, J.; Eklund, C.; Carter, J.J.; Wipf, G.C.; Reed, C.A.; Cladel, N.M.; Galloway, D.A. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 1996, 223, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Christensen, N.; Schiller, J.T.; Dillner, J. A monoclonal antibody against intact human papillomavirus type 16 capsids blocks the serological reactivity of most human sera. J. Gen. Virol. 1997, 78 Pt 9, 2209–2215. [Google Scholar] [CrossRef] [PubMed]
- White, W.I.; Wilson, S.D.; Palmer-Hill, F.J.; Woods, R.M.; Ghim, S.; Hewitt, L.A.; Goldman, D.M.; Burke, S.J.; Jenson, A.B.; Koenig, S.; et al. Characterization of a major neutralizing epitope on human papillomavirus type 16 L1. J. Virol. 1999, 73, 4882–4889. [Google Scholar] [PubMed]
- Christensen, N.D.; Cladel, N.M.; Reed, C.A.; Budgeon, L.R.; Embers, M.E.; Skulsky, D.M.; McClements, W.L.; Ludmerer, S.W.; Jansen, K.U. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 2001, 291, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Day, P.M.; Thompson, C.D.; Buck, C.B.; Pang, Y.-S.; Lowy, D.R.; Schiller, J.T. Neutralization of human papillomavirus with monoclonal antibodies reveals different mechanisms of inhibition. J. Virol. 2007, 81, 8784–8792. [Google Scholar] [CrossRef] [PubMed]
- Ryding, J.; Dahlberg, L.; Wallen-Ohman, M.; Dillner, J. Deletion of a major neutralizing epitope of human papillomavirus type 16 virus-like particles. J. Gen. Virol. 2007, 88, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Brendle, S.A.; Bywaters, S.M.; Guan, J.; Ashley, R.E.; Yoder, J.D.; Makhov, A.M.; Conway, J.F.; Christensen, N.D.; Hafenstein, S. A CryoEM study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. J. Virol. 2015, 89, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Potter, C.S.; Carragher, B.; Lander, G.; Sworen, J.; Towne, V.; Abraham, D.; Duncan, P.; Washabaugh, M.W.; Sitrin, R.D. Characterization of virus-like particles in GARDASIL® by cryo transmission electron microscopy. Hum. Vaccines Immunother. 2014, 10, 734–739. [Google Scholar] [CrossRef]
- Guan, J.; Bywaters, S.M.; Brendle, S.A.; Ashley, R.E.; Makhov, A.M.; Conway, J.F.; Christensen, N.D.; Hafenstein, S. Cryoelectron microscopy maps of human papillomavirus 16 reveal L2 densities and heparin binding site. Struct. Lond. Engl. 2017, 25, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Organtini, L.J.; Makhov, A.M.; Conway, J.F.; Hafenstein, S.; Carson, S.D. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J. Virol. 2014, 88, 5755–5765. [Google Scholar] [CrossRef] [PubMed]
- Organtini, L.J.; Lee, H.; Iketani, S.; Huang, K.; Ashley, R.E.; Makhov, A.M.; Conway, J.F.; Parrish, C.R.; Hafenstein, S. Near-atomic resolution structure of a highly neutralizing fab bound to canine parvovirus. J. Virol. 2016, 90, 9733–9742. [Google Scholar] [CrossRef] [PubMed]
- Brendle, S.A.; Culp, T.D.; Broutian, T.R.; Christensen, N.D. Binding and neutralization characteristics of a panel of monoclonal antibodies to human papillomavirus 58. J. Gen. Virol. 2010, 91, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Mejia, A.F.; Culp, T.D.; Cladel, N.M.; Balogh, K.K.; Budgeon, L.R.; Buck, C.B.; Christensen, N.D. Preclinical model to test human papillomavirus virus (HPV) capsid vaccines in vivo using infectious HPV/cottontail rabbit papillomavirus chimeric papillomavirus particles. J. Virol. 2006, 80, 12393–12397. [Google Scholar] [CrossRef] [PubMed]
- Pyeon, D.; Lambert, P.F.; Ahlquist, P. Production of infectious human papillomavirus independently of viral replication and epithelial cell differentiation. Proc. Natl. Acad. Sci. USA 2005, 102, 9311–9316. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Pastrana, D.V.; Lowy, D.R.; Schiller, J.T. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 2005, 119, 445–462. [Google Scholar] [PubMed]
- Pastrana, D.V.; Buck, C.B.; Pang, Y.Y.; Thompson, C.D.; Castle, P.E.; FitzGerald, P.C.; Krüger Kjaer, S.; Lowy, D.R.; Schiller, J.T. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 2004, 321, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Christensen, N.D.; Kreider, J.W.; Cladel, N.M.; Patrick, S.D.; Welsh, P.A. Monoclonal antibody-mediated neutralization of infectious human papillomavirus type 11. J. Virol. 1990, 64, 5678–5681. [Google Scholar] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Sinkovits, R.S.; Baker, T.S. AUTO3DEM—An automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007, 157, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mooney, P.; Zheng, S.; Booth, C.R.; Braunfeld, M.B.; Gubbens, S.; Agard, D.A.; Cheng, Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 2013, 10, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Dryden, K.A.; Tang, J.; Baker, T.S. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 2007, 157, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.J.; Luque, D.; Castón, J.R.; Carrascosa, J.L. Sharpening high resolution information in single particle electron cryomicroscopy. J. Struct. Biol. 2008, 164, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, P.B.; Henderson, R. Optimal determination of particle orientation, absolute hand and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003, 333, 721–745. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.W.; Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 2012, 9, 853–854. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, J.; Bienkowska-Haba, M.; Ortega, M.E.; Patel, H.D.; Bodevin, S.; Spillmann, D.; Bishop, B.; Sapp, M.; Chen, X.S. Structural basis of oligosaccharide receptor recognition by human papillomavirus. J. Biol. Chem. 2011, 286, 2617–2624. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Cardone, G.; Moyer, A.L.; Cheng, N.; Thompson, C.D.; Dvoretzky, I.; Lowy, D.R.; Schiller, J.T.; Steven, A.C.; Buck, C.B.; Trus, B.L. Maturation of the human papillomavirus 16 capsid. MBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Wriggers, W.; Milligan, R.A.; McCammon, J.A. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999, 125, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Wriggers, W. Conventions and workflows for using Situs. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [PubMed]
- Hafenstein, S.; Bowman, V.D.; Sun, T.; Nelson, C.D.; Palermo, L.M.; Chipman, P.R.; Battisti, A.J.; Parrish, C.R.; Rossmann, M.G. Structural comparison of different antibodies interacting with parvovirus capsids. J. Virol. 2009, 83, 5556–5566. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Olson, N.H.; Cheng, R.H.; Chase, E.S.; Baker, T.S. Structure of a human rhinovirus-bivalently bound antibody complex: Implications for viral neutralization and antibody flexibility. Proc. Natl. Acad. Sci. USA 1993, 90, 7015–7018. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.A.; Stanfield, R.L. Antibody-antigen interactions: New structures and new conformational changes. Curr. Opin. Struct. Biol. 1994, 4, 857–867. [Google Scholar] [CrossRef]
- Hewat, E.A.; Blaas, D. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 1996, 15, 1515–1523. [Google Scholar] [PubMed]
- Janeway, C.A. Jr.; Travers, P.; Walport, M. The structure of a typical antibody molecule. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Kilár, F.; Simon, I.; Lakatos, S.; Vonderviszt, F.; Medgyesi, G.A.; Závodszky, P. Conformation of human IgG subclasses in solution. Small-angle X-ray scattering and hydrodynamic studies. Eur. J. Biochem. FEBS 1985, 147, 17–25. [Google Scholar] [CrossRef]
- Yang, R.; Day, P.M.; Yutzy, W.H.; Lin, K.-Y.; Hung, C.-F.; Roden, R.B.S. Cell surface-binding motifs of L2 that facilitate papillomavirus infection. J. Virol. 2003, 77, 3531–3541. [Google Scholar] [CrossRef] [PubMed]
- Selinka, H.-C.; Giroglou, T.; Nowak, T.; Christensen, N.D.; Sapp, M. Further evidence that papillomavirus capsids exist in two distinct conformations. J. Virol. 2003, 77, 12961–12967. [Google Scholar] [CrossRef] [PubMed]








 remains the same regardless of antibody concentration, while it is apparent that the amount of U4 detected by the anti-IgG2a secondary mAb titrates
remains the same regardless of antibody concentration, while it is apparent that the amount of U4 detected by the anti-IgG2a secondary mAb titrates  ; (C) capture of the HPV-V5 complex by the U4 mAb
; (C) capture of the HPV-V5 complex by the U4 mAb  was ablated at high concentrations of V5. The mAb interference and the inability to capture HPV is also evident by the failure to directly detect V5 mAb
was ablated at high concentrations of V5. The mAb interference and the inability to capture HPV is also evident by the failure to directly detect V5 mAb  ; (D) schematic of pentavalent and hexavalent L1 pentamers (blue) demonstrating steric hindrance of V5 mAb (black) with the U4 mAb (purple). Preliminary binding of the V5 mab to the vertices of hexavalent capsomers creates an “umbrella” over the U4 epitope and thus prevents the secondary binding of U4 mAb.
; (D) schematic of pentavalent and hexavalent L1 pentamers (blue) demonstrating steric hindrance of V5 mAb (black) with the U4 mAb (purple). Preliminary binding of the V5 mab to the vertices of hexavalent capsomers creates an “umbrella” over the U4 epitope and thus prevents the secondary binding of U4 mAb.
 remains the same regardless of antibody concentration, while it is apparent that the amount of U4 detected by the anti-IgG2a secondary mAb titrates
remains the same regardless of antibody concentration, while it is apparent that the amount of U4 detected by the anti-IgG2a secondary mAb titrates  ; (C) capture of the HPV-V5 complex by the U4 mAb
; (C) capture of the HPV-V5 complex by the U4 mAb  was ablated at high concentrations of V5. The mAb interference and the inability to capture HPV is also evident by the failure to directly detect V5 mAb
was ablated at high concentrations of V5. The mAb interference and the inability to capture HPV is also evident by the failure to directly detect V5 mAb  ; (D) schematic of pentavalent and hexavalent L1 pentamers (blue) demonstrating steric hindrance of V5 mAb (black) with the U4 mAb (purple). Preliminary binding of the V5 mab to the vertices of hexavalent capsomers creates an “umbrella” over the U4 epitope and thus prevents the secondary binding of U4 mAb.
; (D) schematic of pentavalent and hexavalent L1 pentamers (blue) demonstrating steric hindrance of V5 mAb (black) with the U4 mAb (purple). Preliminary binding of the V5 mab to the vertices of hexavalent capsomers creates an “umbrella” over the U4 epitope and thus prevents the secondary binding of U4 mAb.

| HPV-V5 | HPV-U4 | |
|---|---|---|
| Number of Micrographs | 1899 | 8899 |
| Defocus level range (µm) | 0.65–3.56 | 0.26–5.18 |
| Number of Particles Selected from Micrograph | 27,095 | 58,571 |
| Number of Particles Selected for reconstruction | 17,612 | 35,151 |
| Final Resolution | 4.7 | 5.8 |
| Asymmetric Unit | Six Subunits (L1 Protein) | Cryo-Electron Microscopy (Cryo-EM) Model 3732 Residues at 4.7 Å Resolution after Phenix Global Refinement |
|---|---|---|
| Density agreement | Correlation Coefficient | 0.85 |
| All-atom contacts | Clash score * (all atoms) | 31.04 (14th percentile) |
| Protein geometry | Poor rotamers | 0.13% |
| Favored rotamers | 99.25% | |
| Ramachandran outliers | 1.05% | |
| Ramachandran favored | 90.52% | |
| Molprobity Score | 2.51% (47nd percentile) | |
| Cβ deviations | 0.03% | |
| Bad backbone | 0.06% | |
| Bad backbone angles | 0.19% |
| Asymmetric Unit | Six Subunits (L1 Protein) | Cryo-EM Model 3004 Residues at 5.8 Å Resolution after Phenix Global Refinement |
|---|---|---|
| Density agreement | Correlation Coefficient | 0.82 |
| All-atom contacts | Clash score * (all atoms) | 49.58 (4th percentile) |
| Protein geometry | Poor rotamers | 0.25% |
| Favored rotamers | 98.75% | |
| Ramachandran outliers | 1.33% | |
| Ramachandran favored | 91.09% | |
| Molprobity Score | 2.69% (47nd percentile) | |
| Cβ deviations | 0.13% | |
| Bad backbone | 0.18% | |
| Bad backbone angles | 0.34% |
| HPV-V5 | Pentavalent Capsomer L1 | Hexavalent Capsomer L1 | ||||||||
| #1 | #2 | #3 | #4 | #5 | #a | #b | #c | #d | #e | |
| Angle (◦) | 0.21 | 0.22 | 0.22 | 0.21 | 0.55 | 0.20 | 0.54 | 0.42 | 0.48 | 0.27 |
| Distance (Å) | 2.99 | 2.28 | 0.99 | 1.61 | 2.51 | 1.57 | 1.50 | 2.45 | 2.57 | 2.25 |
| HPV-U4 | Pentavalent Capsomer L1 | Hexavalent Capsomer L1 | ||||||||
| #1 | #2 | #3 | #4 | #5 | #a | #b | #c | #d | #e | |
| Angle (◦) | 0.96 | 0.96 | 0.96 | 0.96 | 0.97 | 1.39 | 1.00 | 0.64 | 0.93 | 1.37 |
| Distance (Å) | 1.98 | 1.98 | 1.99 | 2.00 | 3.45 | 1.25 | 1.68 | 2.36 | 2.86 | 1.82 |
| L1 Protein Surface Loop and C-Terminal Arm (C-Ter) | V5 Fab Heavy Chain Outwards | U4 Fab Heavy Chain Count Clockwise | ||
|---|---|---|---|---|
| BC | 55 | PRO | X | |
| DE | 139 | ALA | X′ | |
| 142 | ASP | X | ||
| 143 | ASN | X | ||
| EF | 176 | X | ||
| 179 | X | |||
| 180 | X | |||
| 181 | ASN | X′ | ||
| 182 | PRO | X′ | ||
| FG | 267 | VAL | X′ | |
| 278 | LYS | X′ | ||
| 280 | SER | X′ | ||
| 282 | SER | X′ | ||
| 285 | ASN | X′ | ||
| HI | 348 | ILE | X | |
| 357 | ASN | X | ||
| 358 | THR | X | ||
| C-TER | 431 | HIS | X′ | |
| 437 | LYS | X′ | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, J.; Bywaters, S.M.; Brendle, S.A.; Ashley, R.E.; Makhov, A.M.; Conway, J.F.; Christensen, N.D.; Hafenstein, S. High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16. Viruses 2017, 9, 374. https://doi.org/10.3390/v9120374
Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16. Viruses. 2017; 9(12):374. https://doi.org/10.3390/v9120374
Chicago/Turabian StyleGuan, Jian, Stephanie M. Bywaters, Sarah A. Brendle, Robert E. Ashley, Alexander M. Makhov, James F. Conway, Neil D. Christensen, and Susan Hafenstein. 2017. "High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16" Viruses 9, no. 12: 374. https://doi.org/10.3390/v9120374
APA StyleGuan, J., Bywaters, S. M., Brendle, S. A., Ashley, R. E., Makhov, A. M., Conway, J. F., Christensen, N. D., & Hafenstein, S. (2017). High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16. Viruses, 9(12), 374. https://doi.org/10.3390/v9120374






