The Gut Microbiome Correlated to Chemotherapy Efficacy in Diffuse Large B-Cell Lymphoma Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection of Patients and Healthy Controls
2.2. Treatment and Response
2.3. 16S rRNA Sequencing
2.4. Metagenomic Sequencing
3. Results
3.1. Patients Characteristics
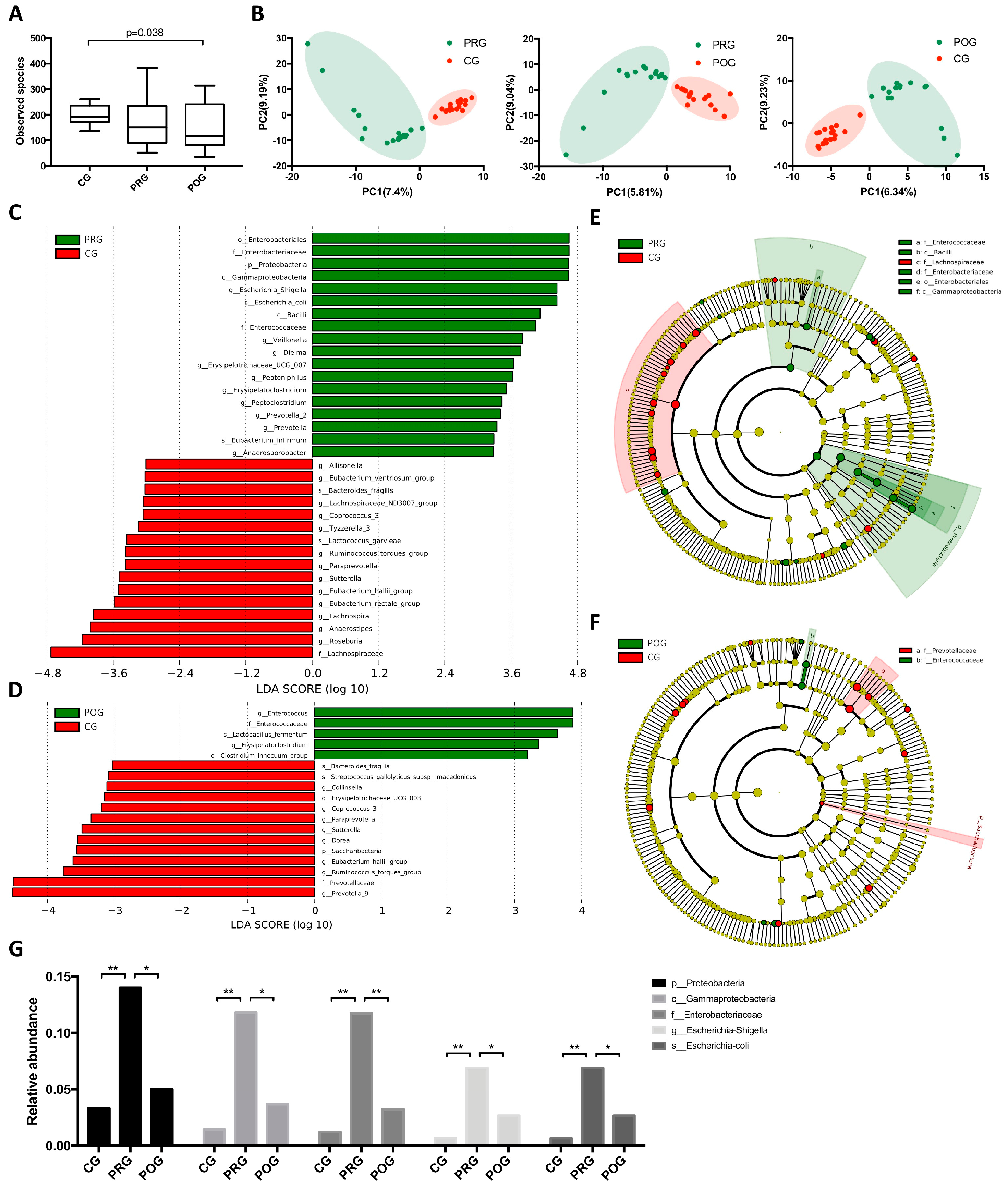
3.2. GMB Composition Was Altered in DLBCL Patients
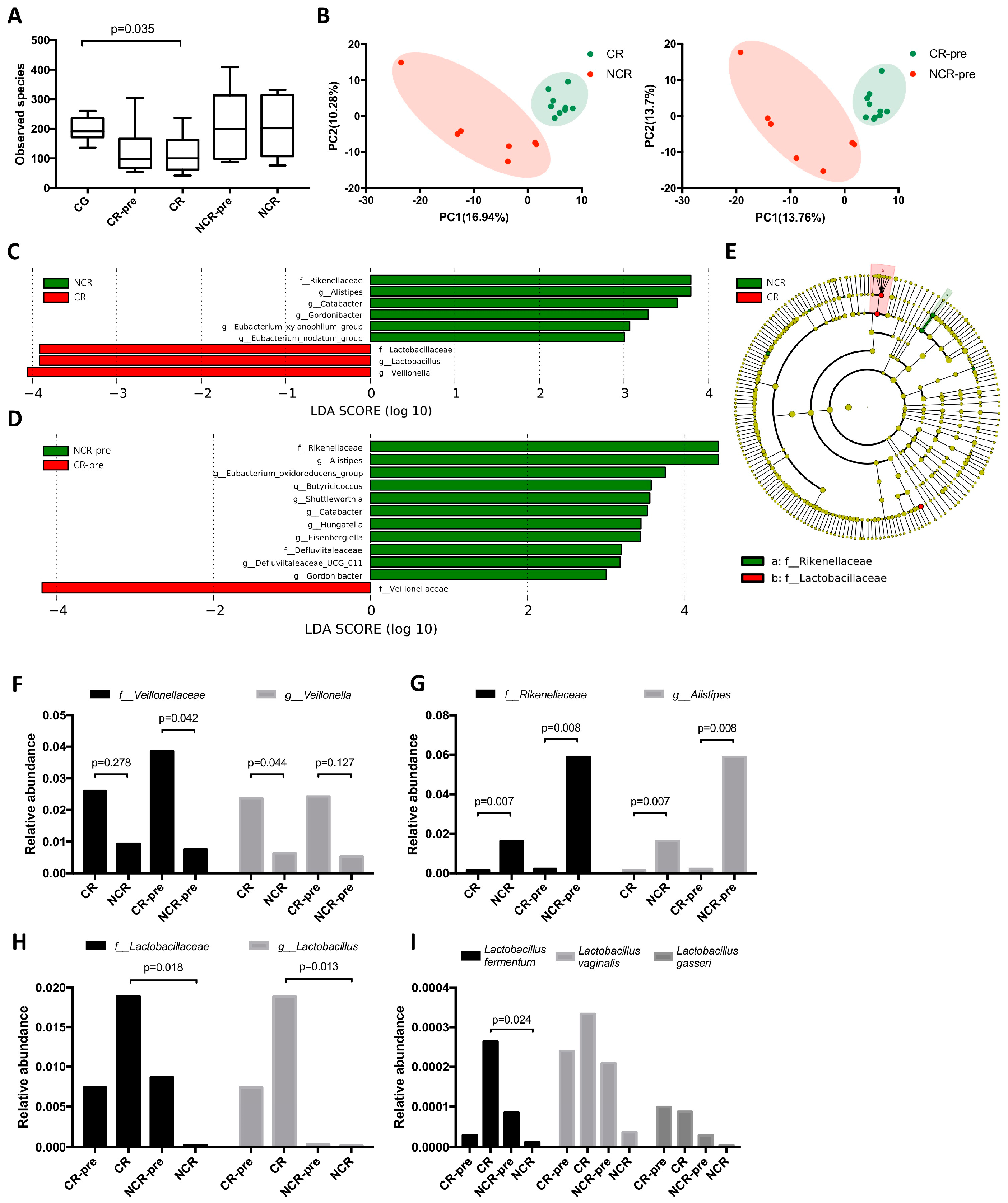
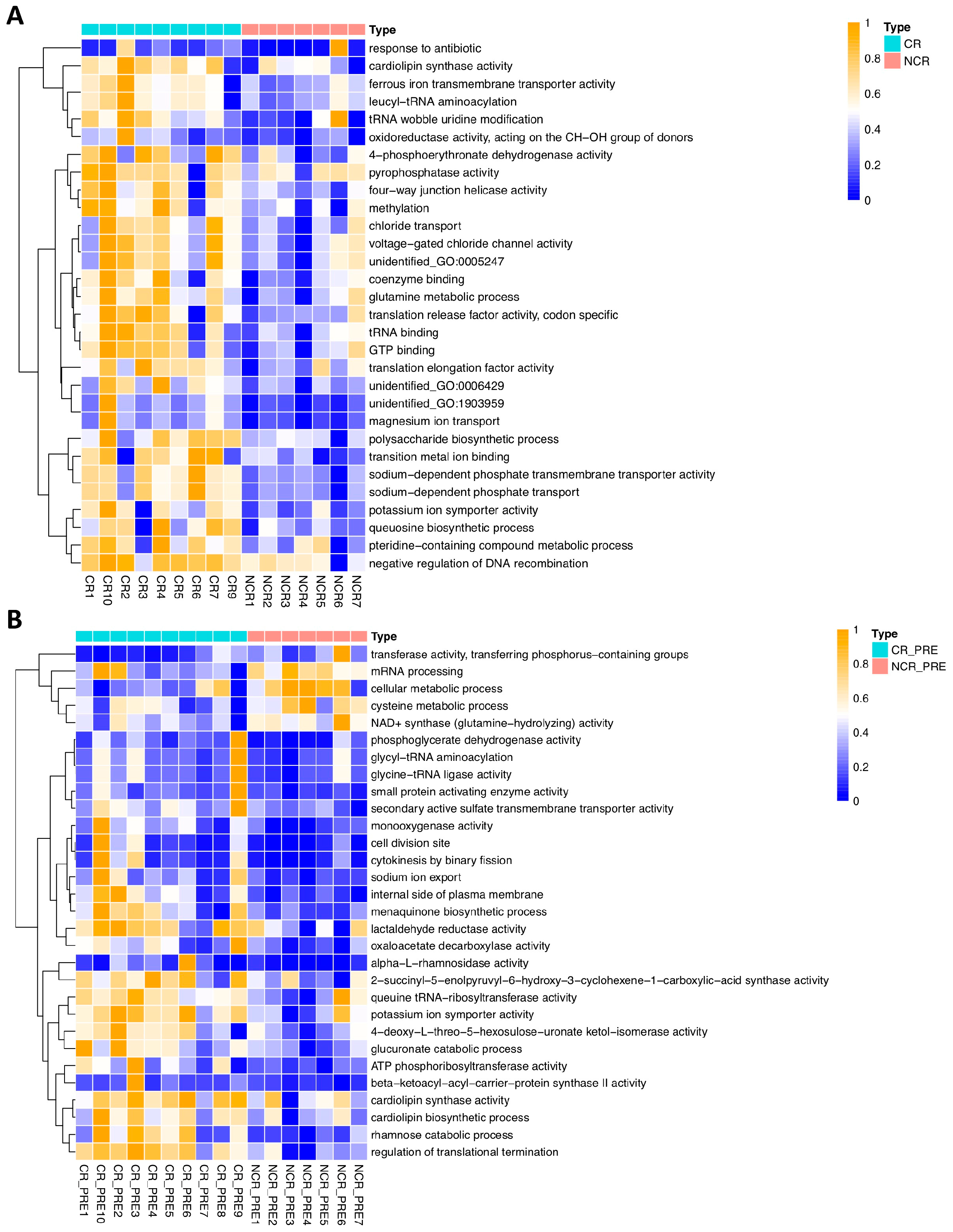
3.3. GMB Composition Was Associated with Chemotherapy Efficacy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Peled, J.U.; Gomes, A.L.; Devlin, S.M.; Littmann, E.R.; Taur, Y.; Sung, A.D.; Weber, D.; Hashimoto, D.; Slingerland, A.E.; Slingerland, J.B.; et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N. Engl. J. Med. 2020, 382, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Panebianco, C.; Andriulli, A.; Pazienza, V. Pharmacomicrobiomics: Exploiting the drug-microbiota interactions in anticancer therapies. Microbiome 2018, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Wotherspoon, A.C.; Ortiz-Hidalgo, C.; Falzon, M.R.; Isaacson, P.G. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 1991, 338, 1175–1176. [Google Scholar] [CrossRef] [PubMed]
- Rajagopala, S.V.; Yooseph, S.; Harkins, D.M.; Moncera, K.J.; Zabokrtsky, K.B.; Torralba, M.G.; Tovchigrechko, A.; Highlander, S.K.; Pieper, R.; Sender, L.; et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis. BMC Genom. 2016, 17, 635. [Google Scholar] [CrossRef]
- Diefenbach, C.S.; Peters, B.A.; Li, H.; Raphael, B.; Moskovits, T.; Hymes, K.; Schluter, J.; Chen, J.; Bennani, N.N.; Witzig, T.E.; et al. Microbial dysbiosis is associated with aggressive histology and adverse clinical outcome in B-cell non-Hodgkin lymphoma. Blood Adv. 2021, 5, 1194–1198. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, W.; Zhang, W.; Zhang, Y.; Wei, C.; Li, J.; Zhou, D. Gut Microbiota in Untreated Diffuse Large B Cell Lymphoma Patients. Front. Microbiol. 2021, 12, 646361. [Google Scholar] [CrossRef]
- Lin, Z.; Mao, D.; Jin, C.; Wang, J.; Lai, Y.; Zhang, Y.; Zhou, M.; Ge, Q.; Zhang, P.; Sun, Y.; et al. The gut microbiota correlate with the disease characteristics and immune status of patients with untreated diffuse large B-cell lymphoma. Front. Immunol. 2023, 14, 1105293. [Google Scholar] [CrossRef]
- Zelenetz, A.D.; Gordon, L.I.; Chang, J.E.; Christian, B.; Abramson, J.S.; Advani, R.H.; Bartlett, N.L.; Budde, L.E.; Caimi, P.F.; De Vos, S.; et al. NCCN Guidelines(R) Insights: B-Cell Lymphomas, Version 5.2021. J. Natl. Compr. Canc. Netw. 2021, 19, 1218–1230. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; LoCascio, P.F.; Hauser, L.J.; Uberbacher, E.C. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 2012, 28, 2223–2230. [Google Scholar] [CrossRef]
- Li, W.; Jaroszewski, L.; Godzik, A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 2001, 17, 282–283. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Buc, E.; Dubois, D.; Sauvanet, P.; Raisch, J.; Delmas, J.; Darfeuille-Michaud, A.; Pezet, D.; Bonnet, R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 2013, 8, e56964. [Google Scholar] [CrossRef]
- Johnson, J.R.; Johnston, B.; Kuskowski, M.A.; Nougayrede, J.-P.; Oswald, E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 2008, 46, 3906–3911. [Google Scholar] [CrossRef]
- Nougayrède, J.-P.; Homburg, S.; Taieb, F.; Boury, M.; Brzuszkiewicz, E.; Gottschalk, G.; Buchrieser, C.; Hacker, J.; Dobrindt, U.; Oswald, E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Kasper, D.L. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 2006, 6, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides produced by lactic acid bacteria: From health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Croxen, M.A.; Finlay, B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.; Hogan, A.M.; Winter, D.C. Microbial and viral pathogens in colorectal cancer. Lancet Oncol. 2011, 12, 504–512. [Google Scholar] [CrossRef]
- Reddy, A.; Zhang, J.; Davis, N.S.; Moffitt, A.; Love, C.L.; Waldrop, A.; Leppä, S.; Pasanen, A.; Meriranta, L.; Karjalainen-Lindsberg, M.-L.; et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017, 171, 481–494.e15. [Google Scholar] [CrossRef] [PubMed]
- van den Bogert, B.; Meijerink, M.; Zoetendal, E.G.; Wells, J.M.; Kleerebezem, M. Immunomodulatory properties of Streptococcus and Veillonella isolates from the human small intestine microbiota. PLoS ONE 2014, 9, e114277. [Google Scholar] [CrossRef]
- Zhan, Z.; Liu, W.; Pan, L.; Bao, Y.; Yan, Z.; Hong, L. Overabundance of Veillonella parvula promotes intestinal inflammation by activating macrophages via LPS-TLR4 pathway. Cell Death Discov. 2022, 8, 251. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar]
- Iebba, V.; Guerrieri, F.; Di Gregorio, V.; Levrero, M.; Gagliardi, A.; Santangelo, F.; Sobolev, A.P.; Circi, S.; Giannelli, V.; Mannina, L.; et al. Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Sci. Rep. 2018, 8, 8210. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Park, S.Y.; Kashyap, D.R.; Dowd, S.E.; Gupta, D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS ONE 2016, 11, e0146162. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sung, C.Y.J.; Lee, N.; Ni, Y.; Pihlajamäki, J.; Panagiotou, G.; El-Nezami, H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E1306–E1315. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef] [PubMed]
- Moschen, A.R.; Gerner, R.R.; Wang, J.; Klepsch, V.; Adolph, T.E.; Reider, S.J.; Hackl, H.; Pfister, A.; Schilling, J.; Moser, P.L.; et al. Lipocalin 2 Protects from Inflammation and Tumorigenesis Associated with Gut Microbiota Alterations. Cell Host Microbe 2016, 19, 455–469. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, J.; O’Toole, P.W. Lactobacillus: Host-microbe relationships. Curr. Top. Microbiol. Immunol. 2013, 358, 119–154. [Google Scholar] [PubMed]
- Ghorbani, E.; Avan, A.; Ryzhikov, M.; Ferns, G.; Khazaei, M.; Soleimanpour, S. Role of lactobacillus strains in the management of colorectal cancer: An overview of recent advances. Nutrition 2022, 103–104, 111828. [Google Scholar] [CrossRef]
- Aindelis, G.; Tiptiri-Kourpeti, A.; Lampri, E.; Spyridopoulou, K.; Lamprianidou, E.; Kotsianidis, I.; Ypsilantis, P.; Pappa, A.; Chlichlia, K. Immune Responses Raised in an Experimental Colon Carcinoma Model Following Oral Administration of Lactobacillus casei. Cancers 2020, 12, 368. [Google Scholar] [CrossRef] [PubMed]
- Chondrou, P.; Karapetsas, A.; Kiousi, D.E.; Tsela, D.; Tiptiri-Kourpeti, A.; Anestopoulos, I.; Kotsianidis, I.; Bezirtzoglou, E.; Pappa, A.; Galanis, A. Lactobacillus paracasei K5 displays adhesion, anti-proliferative activity and apoptotic effects in human colon cancer cells. Benef. Microbes 2018, 9, 975–983. [Google Scholar] [CrossRef]
- Yue, Y.-C.; Yang, B.-Y.; Lu, J.; Zhang, S.-W.; Liu, L.; Nassar, K.; Xu, X.-X.; Pang, X.-Y. Metabolite secretions of Lactobacillus plantarum YYC-3 may inhibit colon cancer cell metastasis by suppressing the VEGF-MMP2/9 signaling pathway. Microb. Cell Fact. 2020, 19, 213. [Google Scholar] [CrossRef]
- Casas-Solís, J.; del Rosario Huizar-López, M.; Irecta-Nájera, C.A.; Pita-López, M.L.; Santerre, A. Immunomodulatory Effect of Lactobacillus casei in a Murine Model of Colon Carcinogenesis. Probiot. Antimicrob. Proteins 2020, 12, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Ye, K.; Lu, J.; Wang, X.; Zhang, S.; Liu, L.; Yang, B.; Nassar, K.; Xu, X.; Pang, X.; et al. Probiotic strain Lactobacillus plantarum YYC-3 prevents colon cancer in mice by regulating the tumour microenvironment. Biomed. Pharmacother. 2020, 127, 110159. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.L.; Maier, I.; Dang, A.T.; Berry, D.; Liu, J.; Ruegger, P.M.; Yang, J.-I.; Soto, P.A.; Presley, L.L.; Reliene, R.; et al. Intestinal bacteria modify lymphoma incidence and latency by affecting systemic inflammatory state, oxidative stress, and leukocyte genotoxicity. Cancer Res. 2013, 73, 4222–4232. [Google Scholar] [CrossRef] [PubMed]

| Patients (n = 17) | Control (n = 18) | |
|---|---|---|
| Median age, yr | 55 | 53 |
| Gender, n (%) | ||
| Male | 8 (47.1) | 10 (55.6) |
| Female | 9 (52.1) | 8 (44.4) |
| Race/region, n | ||
| East Asian/China | 17 | 18 |
| Pathological subtype, n (%) | ||
| GCB | 9 (52.1) | |
| ABC | 8 (47.1) | |
| Organ involved, n (%) | ||
| GI | 6 (35.3) | |
| NGI | 11 (64.7) | |
| IPI * score, n (%) | ||
| 0 | 2 (11.8) | |
| 1 | 5 (29.4) | |
| 2 | 4 (23.5) | |
| 3 | 4 (23.5) | |
| 4 | 1 (5.9) | |
| 5 | 1 (5.9) | |
| Treatment (%) | ||
| R-CHOP + | 17 (100) | |
| Response (%) | ||
| CR | 10 (58.5) | |
| NCR | 7 (41.2) | |
| Antibiotics use (%) | ||
| Received | 13 (76.4) | |
| Not received | 4 (23.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.-F.; Yuan, L.; Zhang, Y.; Zhang, W.; Wei, C.; Wang, W.; Zhao, D.; Zhou, D.; Li, J. The Gut Microbiome Correlated to Chemotherapy Efficacy in Diffuse Large B-Cell Lymphoma Patients. Hematol. Rep. 2024, 16, 63-75. https://doi.org/10.3390/hematolrep16010007
Xu Z-F, Yuan L, Zhang Y, Zhang W, Wei C, Wang W, Zhao D, Zhou D, Li J. The Gut Microbiome Correlated to Chemotherapy Efficacy in Diffuse Large B-Cell Lymphoma Patients. Hematology Reports. 2024; 16(1):63-75. https://doi.org/10.3390/hematolrep16010007
Chicago/Turabian StyleXu, Zhuo-Fan, Li Yuan, Yan Zhang, Wei Zhang, Chong Wei, Wei Wang, Danqing Zhao, Daobin Zhou, and Jingnan Li. 2024. "The Gut Microbiome Correlated to Chemotherapy Efficacy in Diffuse Large B-Cell Lymphoma Patients" Hematology Reports 16, no. 1: 63-75. https://doi.org/10.3390/hematolrep16010007
APA StyleXu, Z.-F., Yuan, L., Zhang, Y., Zhang, W., Wei, C., Wang, W., Zhao, D., Zhou, D., & Li, J. (2024). The Gut Microbiome Correlated to Chemotherapy Efficacy in Diffuse Large B-Cell Lymphoma Patients. Hematology Reports, 16(1), 63-75. https://doi.org/10.3390/hematolrep16010007






