Innovative Immunization Strategies for Antivenom Development
Abstract
:1. Introduction
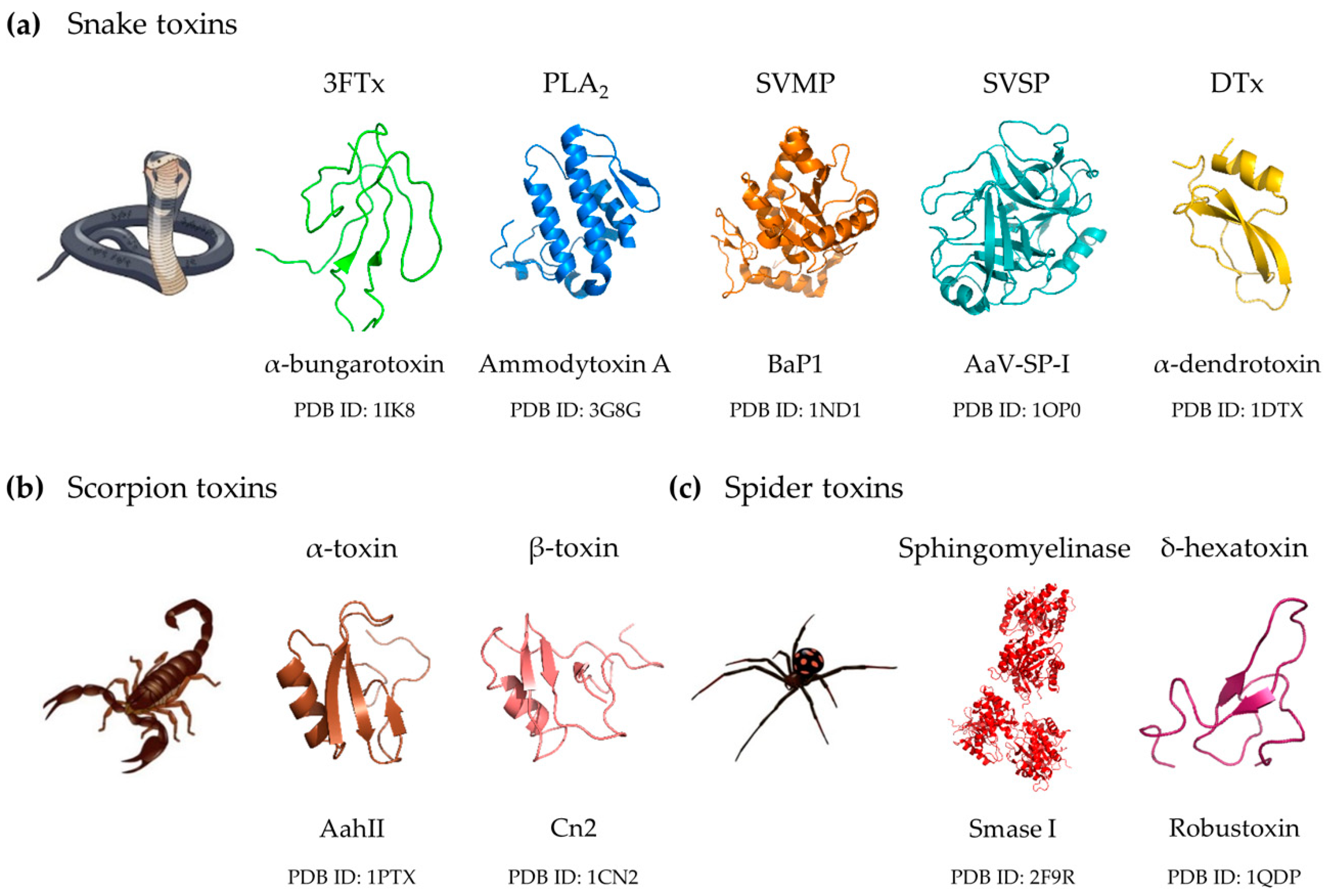
2. Clinically Important Toxin Families
2.1. Snake Venom Toxin Families
2.1.1. Three-Finger Toxins
2.1.2. Phospholipases A2
2.1.3. Snake Venom Metalloproteinases
2.1.4. Snake Venom Serine Proteinases
2.1.5. Dendrotoxins
2.1.6. Minor Snake Venom Toxin Families
2.2. Scorpion Venom Toxins
2.3. Spider Venom Toxins
2.3.1. α-Latrotoxin
2.3.2. Sphingomyelinases D
2.3.3. δ-Hexatoxins
2.3.4. Tx2-6
3. Innovative Venom-Independent Immunization Strategies
3.1. Studies within Snake Antivenom Development
3.2. Studies within Scorpion Antivenom Development
3.3. Studies within Spider Antivenom Development
4. Alternative Venom-Dependent Immunization Approaches
5. Alternative Immunization Approaches from Other Research Fields
6. Biochemical, Bioinformatic, and Omics Tools that Could Aid Antivenom Development
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gutiérrez, J.M.; Theakston, R.D.G.; Warrell, D.A. Confronting the neglected problem of snake bite envenoming: The need for a global partnership. PLoS Med. 2006, 3, e150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Rabies and Envenomings: A Neglected Public Health Issue: Report of A Consultative Meeting; World Health Organization: Geneva, Switzerland, 2007; ISBN 9789241563482. [Google Scholar]
- Diaz, J.H. The global epidemiology, syndromic classification, management, and prevention of spider bites. Am. J. Trop. Med. Hyg. 2004, 71, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Chippaux, J.P. Snake-bites: Appraisal of the global situation. Bull. World Health Organ. 1998, 76, 515–524. [Google Scholar] [PubMed]
- Kasturiratne, A.; Wickremasinghe, A.R.; de Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; de Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, e218. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primer 2017, 3, 17063. [Google Scholar] [CrossRef] [PubMed]
- Chippaux, J.-P.; Goyffon, M. Epidemiology of scorpionism: A global appraisal. Acta Trop. 2008, 107, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Bochner, R. Paths to the discovery of antivenom serotherapy in France. J. Venom. Anim. Toxins Trop. Dis. 2016, 22, 20. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; León, G.; Burnouf, T. Antivenoms for the treatment of snakebite envenomings: The road ahead. Biologicals 2011, 39, 129–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theakston, R.D.; Warrell, D.A. Crisis in snake antivenom supply for Africa. Lancet 2000, 356, 2104. [Google Scholar] [CrossRef]
- Brown, N.; Landon, J. Antivenom: The most cost-effective treatment in the world? Toxicon 2010, 55, 1405–1407. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.J.; Gutiérrez, J.-M.; Calvete, J.J.; Wüster, W.; Ratanabanangkoon, K.; Paiva, O.; Brown, N.I.; Casewell, N.R.; Harrison, R.A.; Rowley, P.D.; et al. Ending the drought: New strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J. Proteomics 2011, 74, 1735–1767. [Google Scholar] [CrossRef] [PubMed]
- Chacón, D.; Rodríguez, S.; Arias, J.; Solano, G.; Bonilla, F.; Gómez, A. Maintaining Coral Snakes (Micrurus nigrocinctus, Serpentes: Elapidae) for venom production on an alternative fish-based diet. Toxicon 2012, 60, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Lalloo, D.G.; Theakston, R.D.G. Snake antivenoms. J. Toxicol. Clin. Toxicol. 2003, 41, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Oukkache, N.; Chgoury, F.; Lalaoui, M.; Cano, A.A.; Ghalim, N. Comparison between two methods of scorpion venom milking in Morocco. J. Venom. Anim. Toxins Trop. Dis. 2013, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- Meadows, P.E.; Russell, F.E. Milking of arthropods. Toxicon 1970, 8, 311–312. [Google Scholar] [CrossRef]
- Rojas, G.; Jiménez, J.M.; Gutiérrez, J.M. Caprylic acid fractionation of hyperimmune horse plasma: Description of a simple procedure for antivenom production. Toxicon 1994, 32, 351–363. [Google Scholar] [CrossRef]
- World Health Organization. WHO Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins; Blood Products and Related Biologicals Quality and Safety: Essential Medicines and Pharmaceutical Policies Health Systems and Services; World Health Organization: Geneva, Switzerland, 2010; ISBN 2105-0678. [Google Scholar]
- León, G.; Vargas, M.; Segura, Á.; Herrera, M.; Villalta, M.; Sánchez, A.; Solano, G.; Gómez, A.; Sánchez, M.; Estrada, R.; et al. Current technology for the industrial manufacture of snake antivenoms. Toxicon 2018, 151, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; León, G.; Lomonte, B.; Angulo, Y. Antivenoms for snakebite envenomings. Inflamm. Allergy Drug Targets 2011, 10, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Segura, A.; Herrera, M.; Villalta, M.; Vargas, M.; Gutiérrez, J.M.; León, G. Assessment of snake antivenom purity by comparing physicochemical and immunochemical methods. Biologicals 2013, 41, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Rawat, S.; Laing, G.; Smith, D.C.; Theakston, D.; Landon, J. A new antivenom to treat eastern coral snake (Micrurus fulvius fulvius) envenoming. Toxicon 1994, 32, 185–190. [Google Scholar] [CrossRef]
- BTG International Inc. CroFab crotalidae polyvalent immune fab (ovine)—Highlights of Prescribing Information 2017. Available online: https://www.fda.gov/downloads/BloodBloodProducts/ucm117573.pdf (accessed on 8 August 2018).
- Antúnez, J.; Fernández, J.; Lomonte, B.; Angulo, Y.; Sanz, L.; Pérez, A.; Calvete, J.J.; Gutiérrez, J.M. Antivenomics of Atropoides mexicanus and Atropoides picadoi snake venoms: Relationship to the neutralization of toxic and enzymatic activities. J. Venom Res. 2010, 1, 8–17. [Google Scholar] [PubMed]
- Fernández, J.; Alape-Girón, A.; Angulo, Y.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.J.; Lomonte, B. Venomic and antivenomic analyses of the Central American coral snake, Micrurus nigrocinctus (Elapidae). J. Proteome Res. 2011, 10, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Escolano, J.; Fernández, J.; Sanz, L.; Angulo, Y.; Gutiérrez, J.M.; Calvete, J.J. Snake venomics and antivenomics of the arboreal neotropical pitvipers Bothriechis lateralis and Bothriechis schlegelii. J. Proteome Res. 2008, 7, 2445–2457. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Lomonte, B.; Lohse, B.; Fernández, J.; Gutiérrez, J.M. Unveiling the nature of black mamba (Dendroaspis polylepis) venom through venomics and antivenom immunoprofiling: Identification of key toxin targets for antivenom development. J. Proteomics 2015, 119, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Judge, R.K.; Henry, P.J.; Mirtschin, P.; Jelinek, G.; Wilce, J.A. Toxins not neutralized by brown snake antivenom. Toxicol. Appl. Pharmacol. 2006, 213, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Carmo, A.O.; Chatzaki, M.; Horta, C.C.R.; Magalhães, B.F.; Oliveira-Mendes, B.B.R.; Chávez-Olórtegui, C.; Kalapothakis, E. Evolution of alternative methodologies of scorpion antivenoms production. Toxicon 2015, 97, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H. Guiding recombinant antivenom development by omics technologies. New Biotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, L.M.; Zahid, M.; di Tommaso, A.; Juste, M.O.; Aubrey, N.; Billiald, P.; Muzard, J. Engineering venom’s toxin-neutralizing antibody fragments and its therapeutic potential. Toxins 2014, 6, 2541–2567. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Engmark, M.; Milbo, C.; Johannesen, J.; Lomonte, B.; Gutiérrez, J.M.; Lohse, B. From Fangs to Pharmacology: The Future of Snakebite Envenoming Therapy. Curr. Pharm. Des. 2016, 22, 5270–5293. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Solà, M.; Jappe, E.C.; Oscoz, S.; Lauridsen, L.P.; Engmark, M. Biotechnological Trends in Spider and Scorpion Antivenom Development. Toxins 2016, 8, 226. [Google Scholar] [CrossRef] [PubMed]
- El-Aziz, T.M.A.; Ravelet, C.; Molgo, J.; Fiore, E.; Pale, S.; Amar, M.; Al-Khoury, S.; Dejeu, J.; Fadl, M.; Ronjat, M.; et al. Efficient functional neutralization of lethal peptide toxins in vivo by oligonucleotides. Sci. Rep. 2017, 7, 7202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laustsen, A.H.; Gutiérrez, J.M.; Knudsen, C.; Johansen, K.H.; Bermúdez-Méndez, E.; Cerni, F.A.; Jürgensen, J.A.; Ledsgaard, L.; Martos-Esteban, A.; Øhlenschlæger, M.; et al. Pros and cons of different therapeutic antibody formats for recombinant antivenom development. Toxicon 2018, 146, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, C.; Laustsen, A.H. Recent Advances in Next Generation Snakebite Antivenoms. Trop. Med. Infect. Dis. 2018, 3, 42. [Google Scholar] [CrossRef] [PubMed]
- Bulfone, T.C.; Samuel, S.P.; Bickler, P.E.; Lewin, M.R. Developing Small Molecule Therapeutics for the Initial and Adjunctive Treatment of Snakebite. J. Trop. Med. 2018, 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Sannaningaiah, D.; Subbaiah, G.K.; Kempaiah, K. Pharmacology of spider venom toxins. Toxin Rev. 2014, 33, 206–220. [Google Scholar] [CrossRef]
- Isbister, G.K.; Bawaskar, H.S. Scorpion Envenomation. N. Engl. J. Med. 2014, 371, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Sanz, L.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M. Venoms, venomics, antivenomics. FEBS Lett. 2009, 583, 1736–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Rahman, M.A.; Harrison, P.L.; Strong, P.N. Snapshots of scorpion venomics. J. Arid Environ. 2015, 112, 170–176. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Lohse, B.; Lomonte, B.; Engmark, M.; Gutiérrez, J.M. Selecting key toxins for focused development of elapid snake antivenoms and inhibitors guided by a Toxicity Score. Toxicon 2015, 104, 43–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laustsen, A.H.; Engmark, M.; Clouser, C.; Timberlake, S.; Vigneault, F.; Gutiérrez, J.M.; Lomonte, B. Exploration of immunoglobulin transcriptomes from mice immunized with three-finger toxins and phospholipases A2 from the Central American coral snake, Micrurus nigrocinctus. PeerJ 2017, 5, e2924. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Sanz, L.; Flores-Díaz, M.; Figueroa, L.; Madrigal, M.; Herrera, M.; Villalta, M.; León, G.; Estrada, R.; Borges, A.; et al. Impact of Regional Variation in Bothrops asper Snake Venom on the Design of Antivenoms: Integrating Antivenomics and Neutralization Approaches. J. Proteome Res. 2010, 9, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Santos, A.C.; Mota, I. Effect of heating on the toxic, immunogenic and immunosuppressive activities of Crotalus durissus terrificus venom. Toxicon 2000, 38, 1451–1457. [Google Scholar] [CrossRef]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J. Snake venomics: From the inventory of toxins to biology. Toxicon 2013, 75, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Fernández, J.; Sanz, L.; Angulo, Y.; Sasa, M.; Gutiérrez, J.M.; Calvete, J.J. Venomous snakes of Costa Rica: Biological and medical implications of their venom proteomic profiles analyzed through the strategy of snake venomics. J. Proteomics 2014, 105, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L. Twenty years of dendrotoxins. Toxicon 2001, 39, 15–26. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Utkin, Y.N. Cobra Cytotoxins: Structural Organization and Antibacterial Activity. Acta Naturae 2014, 6, 11–18. [Google Scholar] [PubMed]
- Montecucco, C.; Gutiérrez, J.M.; Lomonte, B. Cellular pathology induced by snake venom phospholipase A2 myotoxins and neurotoxins: Common aspects of their mechanisms of action. Cell Mol. Life Sci. 2008, 65, 2897–2912. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 2003, 42, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C.; Fox, J.W. A Comprehensive View of the Structural and Functional Alterations of Extracellular Matrix by Snake Venom Metalloproteinases (SVMPs): Novel Perspectives on the Pathophysiology of Envenoming. Toxins 2016, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Serrano, S.M.T. The long road of research on snake venom serine proteinases. Toxicon 2013, 62, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Robertson, B. Dendrotoxins: Structure-activity relationships and effects on potassium ion channels. Curr. Med. Chem. 2004, 11, 3065–3072. [Google Scholar] [CrossRef] [PubMed]
- Boldrini-França, J.; Cologna, C.T.; Pucca, M.B.; Bordon, K.D.C.F.; Amorim, F.G.; Anjolette, F.A.P.; Cordeiro, F.A.; Wiezel, G.A.; Cerni, F.A.; Pinheiro-Junior, E.L.; et al. Minor snake venom proteins: Structure, function and potential applications. Biochim. Biophys. Acta BBA-Gen. Subj. 2017, 1861, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernández, V.; Jiménez-Vargas, J.M.; Gurrola, G.B.; Valdivia, H.H.F.; Possani, L.D. Scorpion venom components that affect ion-channels function. Toxicon 2013, 76, 328–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isbister, G.K.; White, J. Clinical consequences of spider bites: Recent advances in our understanding. Toxicon 2004, 43, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Isbister, G.K.; Fan, H.W. Spider bite. Lancet 2011, 378, 2039–2047. [Google Scholar] [CrossRef]
- Nicholson, G.M.; Graudins, A. Spiders of medical importance in the Asia–Pacific: Atracotoxin, latrotoxin and related spider neurotoxins. Clin. Exp. Pharmacol. Physiol. 2002, 29, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Tambourgi, D.V.; Gonçalves-de-Andrade, R.M.; van den Berg, C.W. Loxoscelism: From basic research to the proposal of new therapies. Toxicon 2010, 56, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, G.M.; Little, M.J.; Birinyi-Strachan, L.C. Structure and function of δ-atracotoxins: Lethal neurotoxins targeting the voltage-gated sodium channel. Toxicon 2004, 43, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, M.D.N.; Diniz, C.R.; Valentim, A.D.C.; von Eickstedt, V.R.D.; Gilroy, J.; Richardson, M. The purification and amino acid sequences of four Tx2 neurotoxins from the venom of the Brazilian ‘armed’ spider Phoneutria nigriventer (Keys). FEBS Lett. 1992, 310, 153–156. [Google Scholar] [CrossRef]
- Matavel, A.; Cruz, J.S.; Penaforte, C.L.; Araújo, D.A.M.; Kalapothakis, E.; Prado, V.F.; Diniz, C.R.; Cordeiro, M.N.; Beirão, P.S.L. Electrophysiological characterization and molecular identification of the Phoneutria nigriventer peptide toxin PnTx2-6 1. FEBS Lett. 2002, 523, 219–223. [Google Scholar] [CrossRef]
- Ramos, H.R.; Junqueira-de-Azevedo, I.D.L.M.; Novo, J.B.; Castro, K.; Duarte, C.G.; Machado-de-Ávila, R.A.; Chavez-Olortegui, C.; Ho, P.L. A Heterologous Multiepitope DNA Prime/Recombinant Protein Boost Immunisation Strategy for the Development of an Antiserum against Micrurus corallinus (Coral Snake) Venom. PLoS Negl. Trop. Dis. 2016, 10, e0004484. [Google Scholar] [CrossRef] [PubMed]
- Calderón, L.; Lomonte, B. Inhibition of the myotoxic activity of Bothrops asper myotoxin II in mice by immunization with its synthetic 13-mer peptide 115–129. Toxicon 1999, 37, 683–687. [Google Scholar] [CrossRef]
- Čurin-Šerbec, V.; Délot, E.; Faure, G.; Saliou, B.; Gubenšek, F.; Bon, C.; Choumet, V. Antipeptide antibodies directed to the C-terminal part of ammodytoxin A react with the PLA2 subunit of crotoxin and neutralize its pharmacological activity. Toxicon 1994, 32, 1337–1348. [Google Scholar] [CrossRef]
- Ferreira, R.N.; Machado de Avila, R.A.; Sanchez, E.F.; Maria, W.S.; Molina, F.; Granier, C.; Chávez-Olórtegui, C. Antibodies against synthetic epitopes inhibit the enzymatic activity of mutalysin II, a metalloproteinase from bushmaster snake venom. Toxicon 2006, 48, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Čurin-Šerbec, V.; Novak, D.; Babnik, J.; Turk, D.; Gubenšek, F. Immunological studies of the toxic site in ammodytoxin A. FEBS Lett. 1991, 280, 175–178. [Google Scholar] [CrossRef] [Green Version]
- Dolimbek, B.Z.; Atassi, M.Z. Protection against alpha-bungarotoxin poisoning by immunization with synthetic toxin peptides. Mol. Immunol. 1996, 33, 681–689. [Google Scholar] [CrossRef]
- Cao, Y.-L.; Guo, G.-N.; Zhu, G.-Y.; Tian, Z.; Gou, Y.-J.; Chen, C.; Liu, M.-H. Bioinformatics-based design of novel antigenic B-cell linear epitopes of Deinagkistrodon acutus venom. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 781–787. [Google Scholar] [PubMed]
- Suntrarachun, S.; Tirawatnapong, T.; Khunsap, S.; Puempumpanich, S. cDNA cloning, sequencing, and expression of α- and β-neurotoxins from Thai-Malayan krait. Indian J. Biotechnol. 2010, 9, 31–37. [Google Scholar]
- Guerrero-Garzón, J.F.; Bénard-Valle, M.; Restano-Cassulini, R.; Zamudio, F.; Corzo, G.; Alagón, A.; Olvera-Rodríguez, A. Cloning and sequencing of three-finger toxins from the venom glands of four Micrurus species from Mexico and heterologous expression of an alpha-neurotoxin from Micrurus diastema. Biochimie 2018, 147, 114–121. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, G.; Corrales-García, L.L.; Rodriguez-Ruiz, X.; López-Vera, E.; Corzo, G. Short-chain consensus alpha-neurotoxin: A synthetic 60-mer peptide with generic traits and enhanced immunogenic properties. Amino Acids 2018. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Rey-Suárez, P.; Fernández, J.; Sasa, M.; Pla, D.; Vargas, N.; Bénard-Valle, M.; Sanz, L.; Corrêa-Netto, C.; Núñez, V.; et al. Venoms of Micrurus coral snakes: Evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon 2016, 122, 7–25. [Google Scholar] [CrossRef] [PubMed]
- De Avila, R.M.; Stransky, S.; Velloso, M.; Castanheira, P.; Schneider, F.S.; Kalapothakis, E.; Sanchez, E.F.; Nguyen, C.; Molina, F.; Granier, C.; et al. Mimotopes of mutalysin-II from Lachesis muta snake venom induce hemorrhage inhibitory antibodies upon vaccination of rabbits. Peptides 2011, 32, 1640–1646. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.; Homsi-Brandeburgo, M.I.; Rodrigues, V.M.; Santos, W.B.; Souza, G.L.R.; Prudencio, C.R.; Siquieroli, A.C.S.; Goulart, L.R. Peptide mimicking antigenic and immunogenic epitope of neuwiedase from Bothrops neuwiedi snake venom. Toxicon 2009, 53, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.A.; Moura-Da-Silva, A.M.; Laing, G.D.; Wu, Y.; Richards, A.; Broadhead, A.; Bianco, A.E.; Theakston, R.D.G. Antibody from mice immunized with DNA encoding the carboxyl-disintegrin and cysteine-rich domain (JD9) of the haemorrhagic metalloprotease, Jararhagin, inhibits the main lethal component of viper venom. Clin. Exp. Immunol. 2000, 121, 358–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, R.A.; Richards, A.; Laing, G.D.; Theakston, R.D.G. Simultaneous GeneGun immunisation with plasmids encoding antigen and GM-CSF: Significant enhancement of murine antivenom IgG1 titres. Vaccine 2002, 20, 1702–1706. [Google Scholar] [CrossRef]
- Hasson, S.S.A.A. Generation of antibodies against disintegrin and cysteine-rich domains by DNA immunization: An approach to neutralize snake venom-induced haemorrhage. Asian Pac. J. Trop. Biomed. 2017, 7, 198–207. [Google Scholar] [CrossRef]
- Arce-Estrada, V.; Azofeifa-Cordero, G.; Estrada, R.; Alape-Girón, A.; Flores-Díaz, M. Neutralization of venom-induced hemorrhage by equine antibodies raised by immunization with a plasmid encoding a novel P-II metalloproteinase from the lancehead pitviper Bothrops asper. Vaccine 2009, 27, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, S.C.; Laing, G.D.; Theakston, R.D.G.; Papaspyridis, C.; Harrison, R.A. Bioinformatics and multiepitope DNA immunization to design rational snake antivenom. PLoS Med. 2006, 3, e184. [Google Scholar] [CrossRef] [PubMed]
- Azofeifa-Cordero, G.; Arce-Estrada, V.; Flores-Díaz, M.; Alape-Girón, A. Immunization with cDNA of a novel P-III type metalloproteinase from the rattlesnake Crotalus durissus durissus elicits antibodies which neutralize 69% of the hemorrhage induced by the whole venom. Toxicon 2008, 52, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Pergolizzi, R.G.; Dragos, R.; Ropper, A.; Menez, A.; Crystal, R.G. Development of a Genetic Vaccine Conferring Protective Immunity Against α-Cobratoxin Following a Single Administration of an Adenovirus Vector Encoding a Modified, Non-Toxic Cobratoxin Variant. Mol. Ther. 2004, 9, S143. [Google Scholar] [CrossRef]
- Bolhassani, A.; Yazdi, S.R. DNA Immunization as an Efficient Strategy for Vaccination. Avicenna J. Med. Biotechnol. 2009, 1, 71–88. [Google Scholar] [PubMed]
- Harrison, R.A. Development of venom toxin-specific antibodies by DNA immunisation: Rationale and strategies to improve therapy of viper envenoming. Vaccine 2004, 22, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, S.; Lu, S. DNA immunization as a technology platform for monoclonal antibody induction. Emerg. Microbes Infect. 2016, 5, e33. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wong, Y.C.; Chen, S.M.Y.; Tang, J.; Wang, H.; Cheung, A.K.L.; Chen, Z. DNA prime/MVTT boost regimen with HIV-1 mosaic Gag enhances the potency of antigen-specific immune responses. Vaccine 2018, 36, 4621–4632. [Google Scholar] [CrossRef] [PubMed]
- Perrie, Y.; Frederik, P.M.; Gregoriadis, G. Liposome-mediated DNA vaccination: The effect of vesicle composition. Vaccine 2001, 19, 3301–3310. [Google Scholar] [CrossRef]
- Pérez-Herrero, E.; Fernández-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fotoran, W.L.; Santangelo, R.; Miranda, B.N.M.D.; Irvine, D.J.; Wunderlich, G. DNA-Loaded Cationic Liposomes Efficiently Function as a Vaccine against Malarial Proteins. Mol. Ther. Methods Clin. Dev. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Reyes, M.; Ramírez, C.; Ñancucheo, I.; Villegas, R.; Schaffeld, G.; Kriman, L.; Gonzalez, J.; Oyarzun, P. A novel “in-feed” delivery platform applied for oral DNA vaccination against IPNV enables high protection in Atlantic salmon (Salmon salar). Vaccine 2017, 35, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Liu, H.; Sun, X.; Qin, X.; Xu, Z.; Wang, B. Construction and expression of DNA vaccine against reddish body iridovirus and evaluation of immune efficacy in turbot (Scophthalmus maximus). Aquac. Res. 2017, 48, 4174–4183. [Google Scholar] [CrossRef]
- Leão, L.I.; Ho, P.L.; Junqueira-de-Azevedo, I.D.L.M. Transcriptomic basis for an antiserum against Micrurus corallinus (coral snake) venom. BMC Genomics 2009, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Clement, H.; Flores, V.; De la Rosa, G.; Zamudio, F.; Alagon, A.; Corzo, G. Heterologous expression, protein folding and antibody recognition of a neurotoxin from the Mexican coral snake Micrurus laticorallis. J. Venom. Anim. Toxins Trop. Dis. 2016, 22. [Google Scholar] [CrossRef] [PubMed]
- Bahraoui, E.M.; Granier, C.; Rietschoten, J.V.; Rochat, H.; El Ayeb, M. Specificity and neutralizing capacity of antibodies elicited by a synthetic peptide of scorpion toxin. J. Immunol. 1986, 136, 3371–3377. [Google Scholar] [PubMed]
- Calderón-Aranda, E.S.; Olamendi-Portugal, T.; Possani, L.D. The use of synthetic peptides can be a misleading approach to generate vaccines against scorpion toxins. Vaccine 1995, 13, 1198–1206. [Google Scholar] [CrossRef]
- Calderón-Aranda, E.S.; Selisko, B.; York, E.J.; Gurrola, G.B.; Stewart, J.M.; Possani, L.D. Mapping of an epitope recognized by a neutralizing monoclonal antibody specific to toxin Cn2 from the scorpion Centruroides noxius, using discontinuous synthetic peptides. Eur. J. Biochem. 1999, 264, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, L.M.; Diniz, C.R.; Granier, C.; Chávez-Olórtegui, C. Induction of neutralizing antibodies against Tityus serrulatus scorpion toxins by immunization with a mixture of defined synthetic epitopes. Toxicon 2002, 40, 89–95. [Google Scholar] [CrossRef]
- Inceoglu, B.; Lango, J.; Rabinovich, A.; Whetstone, P.; Hammock, B.D. The neutralizing effect of a polyclonal antibody raised against the N-terminal eighteen-aminoacid residues of birtoxin towards the whole venom of Parabuthus transvaalicus. Toxicon 2006, 47, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Duarte, C.G.; Alvarenga, L.M.; Dias-Lopes, C.; Machado-de-Ávila, R.A.; Nguyen, C.; Molina, F.; Granier, C.; Chávez-Olórtegui, C. In vivo protection against Tityus serrulatus scorpion venom by antibodies raised against a discontinuous synthetic epitope. Vaccine 2010, 28, 1168–1176. [Google Scholar] [CrossRef] [PubMed]
- Devaux, C.; Juin, M.; Mansuelle, P.; Granier, C. Fine molecular analysis of the antigenicity of the Androctonus australis hector scorpion neurotoxin II: A new antigenic epitope disclosed by the pepscan method. Mol. Immunol. 1993, 30, 1061–1068. [Google Scholar] [CrossRef]
- Chavez-Olortegui, C.; Molina, F.; Granier, C. Molecular basis for the cross-reactivity of antibodies elicited by a natural anatoxin with α- and β-toxins from the venom of Tityus serrulatus scorpion. Mol. Immunol. 2001, 38, 867–876. [Google Scholar] [CrossRef]
- Jarząb, A.; Witkowska, D.; Ziomek, E.; Setner, B.; Czajkowska, A.; Dorot, M.; Szewczuk, Z.; Gamian, A. Cyclic OmpC peptidic epitope conjugated to tetanus toxoid as a potential vaccine candidate against shigellosis. Vaccine 2018, 36, 4641–4649. [Google Scholar] [CrossRef] [PubMed]
- Zenouaki, I.; Kharrat, R.; Sabatier, J.-M.; Devaux, C.; Karoui, H.; Van Rietschoten, J.; El Ayeb, M.; Rochat, H. In vivo protection against Androctonus australis hector scorpion toxin and venom by immunization with a synthetic analog of toxin II. Vaccine 1997, 15, 187–194. [Google Scholar] [CrossRef]
- Fischer, P.; Comis, A.; Tyler, M.; Howden, M. Oral and parenteral immunization with synthetic retro-inverso peptides induce antibodies that cross-react with native peptides and parent antigens. Indian J. Biochem. Biophys. 2007, 44, 5. [Google Scholar]
- Devaux, C.; Clot-Faybesse, O.; Juin, M.; Mabrouk, K.; Sabatier, J.-M.; Rochat, H. Monoclonal antibodies neutralizing the toxin II from Androctonus australis hector scorpion venom: Usefulness of a synthetic, non-toxic analog. FEBS Lett. 1997, 412, 456–460. [Google Scholar] [CrossRef]
- Guatimosim, S.C.F.; Kalapothakis, E.; Diniz, C.R.; Chávez-Olórtegui, C. Induction of neutralizing antibodies against Tityus serrulatus toxins by immunization with a recombinant nontoxic protein. Toxicon 2000, 38, 113–121. [Google Scholar] [CrossRef]
- Legros, C.; Kaabi, H.; El Ayeb, M.; Céard, B.; Vacher, H.; Bougis, P.E.; Martin-Eauclaire, M.-F. Use of fusion protein constructs to generate potent immunotherapy and protection against scorpion toxins. Vaccine 2001, 20, 934–942. [Google Scholar] [CrossRef]
- Benkhadir, K.; Mejri, T.; Bel Haj Rhouma, R.; El Ayeb, M.; Karoui, H. Induction de protection in vivo et in vitro contre l’activite letale du veni de Buthus occitanus tunetanus avec une proteine recombinante. Arch. Inst. Pasteur Tunis 2002, 79, 19–26. [Google Scholar] [PubMed]
- Garcia, C.; Calderón-Aranda, E.S.; Anguiano, G.A.V.; Becerril, B.; Possani, L.D. Analysis of the immune response induced by a scorpion venom sub-fraction, a pure peptide and a recombinant peptide, against toxin Cn2 of Centruroides noxius Hoffmann. Toxicon 2003, 41, 417–427. [Google Scholar] [CrossRef]
- Corona Villegas, M.; Garcia Rodríguez, M.C.; Gurrola Briones, G.; Valdez Cruz, N.A.; Becerril Luján, B.; Possani Postay, L.D. Recombinant immunogens for the generation of antivenoms to the venom of scorpions of the genus Centruroides. U.S. Patent 7,335,759 B2, 26 February 2008. [Google Scholar]
- Mendes, T.M.; Dias, F.; Horta, C.C.R.; Pena, I.F.; Arantes, E.C.; Kalapothakis, E. Effective Tityus serrulatus anti-venom produced using the Ts1 component. Toxicon 2008, 52, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Salgado, K.; Estrada, G.; Olvera, A.; Coronas, F.I.; Possani, L.D.; Corzo, G. Heterologous expressed toxic and non-toxic peptide variants of toxin CssII are capable to produce neutralizing antibodies against the venom of the scorpion Centruroides suffusus suffusus. Immunol. Lett. 2009, 125, 93–99. [Google Scholar] [CrossRef] [PubMed]
- García-Gómez, B.I.; Olamendi-Portugal, T.C.; Paniagua, J.; van der Walt, J.; Dyason, K.; Possani, L.D. Heterologous expression of a gene that codes for Pg8, a scorpion toxin of Parabuthus granulatus, capable of generating protecting antibodies in mice. Toxicon 2009, 53, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, G.; Jolodar, A.; Seyfiabad Shapouri, M.R.; Bahmainmehr, A.; Navidpour, S. Production of Recombinant Alpha Neurotoxin of Scorpion Venom Mesobuthus eupeus and Analysis of its Immunogenicity. Iran. Red Crescent Med. J. 2014, 16. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Vargas, J.M.; Quintero-Hernández, V.; González-Morales, L.; Ortiz, E.; Possani, L.D. Design and expression of recombinant toxins from Mexican scorpions of the genus Centruroides for production of antivenoms. Toxicon 2017, 128, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Safari Foroushani, N.; Modarressi, M.H.; Behdani, M.; Torabi, E.; Pooshang Bagheri, K.; Shahbazzadeh, D. Developing recombinant phospholipase D1 (rPLD1) toxoid from Iranian Hemiscorpius lepturus scorpion and its protective effects in BALB/c mice. Toxicon 2018, 152, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Uawonggul, N.; Sukprasert, S.; Incamnoi, P.; Patramanon, R.; Thammasirirak, S.; Preecharram, S.; Bunyatratchata, W.; Kuaprasert, B.; Daduang, J.; Daduang, S. Bacterial Overexpression of Recombinant Heteroscorpine-1 (rHS-1), a Toxin from Heterometrus laoticus Scorpion Venom: Trends for Antibacterial Application and Antivenom Production. Biochem. Genet. 2014, 52, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Bouhaouala-Zahar, B.; Ducancel, F.; Zenouaki, I.; Khalifa, R.B.; Borchani, L.; Pelhate, M.; Boulain, J.-C.; Ayeb, M.E.; Ménez, A.; Karoui, H. A Recombinant Insect-Specific α-Toxin of Buthus occitanus tunetanus Scorpion Confers Protection Against Homologous Mammal Toxins. Eur. J. Biochem. 1996, 238, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Gazarian, T.; Selisko, B.; Hérion, P.; Gazarian, K. Isolation and structure–functional characterization of phage display library-derived mimotopes of noxiustoxin, a neurotoxin of the scorpion Centruroides noxius Hoffmann. Mol. Immunol. 2000, 37, 755–766. [Google Scholar] [CrossRef]
- Tambourgi, D.V.; Pedrosa, M.d.F.F.; van den Berg, C.W.; Gonçalves-de-Andrade, R.M.; Ferracini, M.; Paixão-Cavalcante, D.; Morgan, B.P.; Rushmere, N.K. Molecular cloning, expression, function and immunoreactivities of members of a gene family of sphingomyelinases from Loxosceles venom glands. Mol. Immunol. 2004, 41, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, G.S.; Caporrino, M.C.; Della-Casa, M.S.; Kimura, L.F.; Prezotto-Neto, J.P.; Fukuda, D.A.; Portes-Junior, J.A.; Neves-Ferreira, A.G.C.; Santoro, M.L.; Barbaro, K.C. Cloning, expression and characterization of a phospholipase D from Loxosceles gaucho venom gland. Biochimie 2013, 95, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.D.A.; Guerra-Duarte, C.; Costal-Oliveira, F.; Mendes, T.M.; Figueiredo, L.F.M.; Oliveira, D.; Machado de Avila, R.A.; Ferrer, V.P.; Trevisan-Silva, D.; Veiga, S.S.; et al. Recombinant Protein Containing B-Cell Epitopes of Different Loxosceles Spider Toxins Generates Neutralizing Antibodies in Immunized Rabbits. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Comis, A.; Tyler, M.; Mylecharane, E.; Spence, I.; Howden, M. Immunization with a synthetic robustoxin derivative lacking disulphide bridges protects against a potentially lethal challenge with funnel-web spider (Atrax robustus) venom. J. Biosci. 2009, 34, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Araujo, S.C.; Castanheira, P.; Alvarenga, L.M.; Mangili, O.C.; Kalapothakis, E.; Chávez-Olórtegui, C. Protection against dermonecrotic and lethal activities of Loxosceles intermedia spider venom by immunization with a fused recombinant protein. Toxicon 2003, 41, 261–267. [Google Scholar] [CrossRef]
- Olvera, A.; Ramos-Cerrillo, B.; Estévez, J.; Clement, H.; de Roodt, A.; Paniagua-Solís, J.; Vázquez, H.; Zavaleta, A.; Salas Arruz, M.; Stock, R.P.; et al. North and South American Loxosceles spiders: Development of a polyvalent antivenom with recombinant sphingomyelinases D as antigens. Toxicon 2006, 48, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Felicori, L.; Araujo, S.C.; Machado de Ávila, R.A.; Sanchez, E.F.; Granier, C.; Kalapothakis, E.; Chávez-Olórtegui, C. Functional characterization and epitope analysis of a recombinant dermonecrotic protein from Loxosceles intermedia spider. Toxicon 2006, 48, 509–519. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, D.M.; Fernandes-Pedrosa, M.D.F.; de Andrade, R.M.G.; Marcelino, J.R.; Gondo-Higashi, H.; de Azevedo, I.D.L.M.J.; Ho, P.L.; van den Berg, C.; Tambourgi, D.V. A new anti-loxoscelic serum produced against recombinant sphingomyelinase D: Results of preclinical trials. Am. J. Trop. Med. Hyg. 2008, 79, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Mendes, T.M.; Oliveira, D.; Figueiredo, L.F.M.; Machado-de-Avila, R.A.; Duarte, C.G.; Dias-Lopes, C.; Guimarães, G.; Felicori, L.; Minozzo, J.C.; Chávez-Olortegui, C. Generation and characterization of a recombinant chimeric protein (rCpLi) consisting of B-cell epitopes of a dermonecrotic protein from Loxosceles intermedia spider venom. Vaccine 2013, 31, 2749–2755. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.F.M.; Dias-Lopes, C.; Alvarenga, L.M.; Mendes, T.M.; Machado-de-Ávila, R.A.; McCormack, J.; Minozzo, J.C.; Kalapothakis, E.; Chávez-Olórtegui, C. Innovative immunization protocols using chimeric recombinant protein for the production of polyspecific loxoscelic antivenom in horses. Toxicon 2014, 86, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Dias-Lopes, C.; Felicori, L.; Rubrecht, L.; Cobo, S.; Molina, L.; Nguyen, C.; Galéa, P.; Granier, C.; Molina, F.; Chávez-Olortegui, C. Generation and molecular characterization of a monoclonal antibody reactive with conserved epitope in sphingomyelinases D from Loxosceles spider venoms. Vaccine 2014, 32, 2086–2092. [Google Scholar] [CrossRef] [PubMed]
- Duarte, C.G.; Bonilla, C.; Guimarães, G.; Machado de Avila, R.A.; Mendes, T.M.; Silva, W.; Tintaya, B.; Yarleque, A.; Chávez-Olórtegui, C. Anti-loxoscelic horse serum produced against a recombinant dermonecrotic protein of Brazilian Loxosceles intermedia spider neutralize lethal effects of Loxosceles laeta venom from Peru. Toxicon 2015, 93, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.F.B.; Vilela, A.; Coura, L.A.M.; Rodrigues, F.T.G.; Nagem, R.A.P.; Chávez-Olortegui, C.; Maioli, T.U.; Felicori, L.F. Protective antibodies against a sphingomyelinase D from Loxosceles intermedia spider venom elicited in mice with different genetic background. Vaccine 2016, 34, 3828–3834. [Google Scholar] [CrossRef] [PubMed]
- Felicori, L.; Fernandes, P.B.; Giusta, M.S.; Duarte, C.G.; Kalapothakis, E.; Nguyen, C.; Molina, F.; Granier, C.; Chávez-Olórtegui, C. An in vivo protective response against toxic effects of the dermonecrotic protein from Loxosceles intermedia spider venom elicited by synthetic epitopes. Vaccine 2009, 27, 4201–4208. [Google Scholar] [CrossRef] [PubMed]
- Dias-Lopes, C.; Guimarães, G.; Felicori, L.; Fernandes, P.; Emery, L.; Kalapothakis, E.; Nguyen, C.; Molina, F.; Granier, C.; Chávez-Olórtegui, C. A protective immune response against lethal, dermonecrotic and hemorrhagic effects of Loxosceles intermedia venom elicited by a 27-residue peptide. Toxicon 2010, 55, 481–487. [Google Scholar] [CrossRef] [PubMed]
- De Moura, J.; Felicori, L.; Moreau, V.; Guimarães, G.; Dias-Lopes, C.; Molina, L.; Alvarenga, L.M.; Fernandes, P.; Frézard, F.; Ribeiro, R.R.; et al. Protection against the toxic effects of Loxosceles intermedia spider venom elicited by mimotope peptides. Vaccine 2011, 29, 7992–8001. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Pedrosa, M.F.; Junqueira de Azevedo, I.L.; Gonçalves-de-Andrade, R.M.; van den Berg, C.; Ramos, C.R.; Ho, P.L.; Tambourgi, D.V. Molecular cloning and expression of a functional dermonecrotic and haemolytic factor from Loxosceles laeta venom. Biochem. Biophys. Res. Commun. 2002, 298, 638–645. [Google Scholar] [CrossRef]
- Chen, S.-W.W.; Van Regenmortel, M.H.V.; Pellequer, J.-L. Structure-activity relationships in peptide-antibody complexes: Implications for epitope prediction and development of synthetic peptide vaccines. Curr. Med. Chem. 2009, 16, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zaro, J.L.; Shen, W.-C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaim, O.M.; da Silveira, R.B.; Trevisan-Silva, D.; Ferrer, V.P.; Sade, Y.B.; Bóia-Ferreira, M.; Gremski, L.H.; Gremski, W.; Senff-Ribeiro, A.; Takahashi, H.K.; et al. Phospholipase-D activity and inflammatory response induced by brown spider dermonecrotic toxin: Endothelial cell membrane phospholipids as targets for toxicity. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2011, 1811, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- New, R.R.C.; Theakston, R.D.G.; Zumbuehl, O.; Iddon, D.; Friend, J. Liposomal immunisation against snake venoms. Toxicon 1985, 23, 215–219. [Google Scholar] [CrossRef]
- Laing, G.; Theakston, R.D.G.; New, R.R.C. Use of liposomes incorporating immunostimulants for immunisation against snake venom. Toxicon 1988, 26, 29. [Google Scholar] [CrossRef]
- Freitas, T.V.; Tavares, A.P.; Theakston, R.D.G.; Laing, G.; New, R.R.C. Use of liposomes for protective immunisation against Crotalus durissus (tropical rattlesnake) venom. Toxicon 1989, 27, 341–347. [Google Scholar] [CrossRef]
- Chavez-Olortegui, C.; Amara, D.A.; Rochat, H.; Diniz, C.; Granier, C. In vivo protection against scorpion toxins by liposomal immunization. Vaccine 1991, 9, 907–910. [Google Scholar] [CrossRef]
- Laing, G.D.; Theakston, R.D. Immunization against Echis ocellatus (carpet viper) venom using liposomes incorporating immunostimulants: Role of lipopolysaccharide in conferring protection in a mouse model. Toxicon 1993, 31, 615–626. [Google Scholar] [CrossRef]
- Fonseca, S.G.; Ferreira, A.M.M.; Diniz, C.R.; Chávez-Olórtegui, C. Induction of neutralizing antibodies in mice immunized with scorpion toxins detoxified by liposomal entrapment. Braz. J. Med. Biol. Res. 1997, 30, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, V.T.; Gomes, R.T.; Viotti, A.P.; Freitas, T.V. Immunization with liposome-encapsulated Bothrops jararaca venom. Toxicon 2000, 38, 881–886. [Google Scholar] [CrossRef]
- Rocha Soares, K.S.; Cardozo Fonseca, J.L.; Oliveira Bitencourt, M.A.; Santos, K.S.C.R.; Silva-Júnior, A.A.; Fernandes-Pedrosa, M.F. Serum production against Tityus serrulatus scorpion venom using cross-linked chitosan nanoparticles as immunoadjuvant. Toxicon 2012, 60, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Nait Mohamed, F.A.; Laraba-Djebari, F. Development and characterization of a new carrier for vaccine delivery based on calcium-alginate nanoparticles: Safe immunoprotective approach against scorpion envenoming. Vaccine 2016, 34, 2692–2699. [Google Scholar] [CrossRef] [PubMed]
- Ayari-Riabi, S.; Trimaille, T.; Mabrouk, K.; Bertin, D.; Gigmes, D.; Benlasfar, Z.; Zaghmi, A.; Bouhaouala-Zahar, B.; Elayeb, M. Venom conjugated polylactide applied as biocompatible material for passive and active immunotherapy against scorpion envenomation. Vaccine 2016, 34, 1810–1815. [Google Scholar] [CrossRef] [PubMed]
- Soares, K.S.R.; Gláucia-Silva, F.; Daniele-Silva, A.; Torres-Rêgo, M.; Araújo, N.K.D.; Menezes, Y.A.S.D.; Damasceno, I.Z.; Tambourgi, D.V.; da Silva-Júnior, A.A.; Fernandes-Pedrosa, M.D.F. Antivenom Production against Bothrops jararaca and Bothrops erythromelas Snake Venoms Using Cross-Linked Chitosan Nanoparticles as an Immunoadjuvant. Toxins 2018, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Valverde, J.M.; Rodríguez, K.; Herrera, M.; Segura, Á.; Vargas, M.; Villalta, M.; Montero, M.; Gutiérrez, J.M.; León, G. Comparison of the adjuvant activity of emulsions with different physicochemical properties on the antibody response towards the venom of West African carpet viper (Echis ocellatus). Toxicon 2017, 127, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, C.; Solano, S.; Herrera, M.; Segura, Á.; Estrada, R.; Vargas, M.; Villalta, M.; Gutiérrez, J.M.; León, G. Lachesis stenophrys venom reduces the equine antibody response towards Bothrops asper venom used as co-immunogen in the production of polyspecific snake antivenom. Toxicon 2015, 103, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Hati, R.N.; Mandal, M.; Hati, A.K. Active immunization of rabbit with gamma irradiated Russell’s viper venom toxoid. Toxicon 1990, 28, 895–902. [Google Scholar] [CrossRef]
- Clissa, P.B.; do Nascimento, N.; Rogero, J.R. Toxicity and immunogenicity of Crotalus durissus terrificus venom treated with different doses of gamma rays. Toxicon 1999, 37, 1131–1141. [Google Scholar] [CrossRef]
- Abib, L.; Laraba-Djebari, F. Effect of gamma irradiation on toxicity and immunogenicity of Androctonus australis hector venom. Can. J. Physiol. Pharmacol. 2003, 81, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Abdou, F.; Denshary, E.E.; Shaaban, E.; Mohamed, M. Assessment of the neutralizing potency of antisera raised against native and γ-irradiated Naja nigricollis (black-necked spitting cobra) venom in rabbits, concerning its cardiotoxic effect. Hum. Exp. Toxicol. 2017, 36, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Possani, L.D.; de Castro, J.F.; Juliá, J.Z. Detoxification with glutaraldehyde of purified scorpion (Centruroides noxius Hoffmann) venom. Toxicon 1981, 19, 323–329. [Google Scholar] [CrossRef]
- Kuo, K.W.; Chang, C.C. High affinity antibody to cobrotoxin prepared from the derivatives of glutaraldehyde-detoxified cobrotoxin. J. Biochem. 1991, 110, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.G.; Duarte, C.G.; Oliveira, M.S.; Castro, K.L.P.; Teixeira, M.S.; Reis, L.P.G.; Zambrano, J.A.; Kalapothakis, E.; Michel, A.F.R.M.; Soto-Blanco, B.; et al. Toxicity of crude and detoxified Tityus serrulatus venom in anti-venom-producing sheep. J. Vet. Sci. 2016, 17, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Kaabi, H.; Kharrat, R.; el Ayeb, M. Study of the protective capacity of scorpion venom Buthus occitanus tunetanus polymerised to glutaraldehyde in mice strains with different haplotypes. Arch. Inst. Pasteur Tunis 2001, 78, 17–23. [Google Scholar] [PubMed]
- Heneine, L.G.D.; Cardoso, V.N.; Daniel, J.; Heneine, I.F. Detoxification of the T2 fraction from a scorpion (Tityus serrulatus, Lutz and Mello) venom by iodination and some immunogenic properties of the derivatives. Toxicon 1986, 24, 501–505. [Google Scholar] [CrossRef]
- Heneine, I.F.; Heneine, L.G. Stepwise iodination. A general procedure for detoxification of proteins suitable for vaccine development and antiserum production. Biologicals 1998, 26, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.M.; Sestito, W.P.; Marcussi, S.; Stábeli, R.G.; Andrião-Escarso, S.H.; Cunha, O.A.B.; Vieira, C.A.; Giglio, J.R. Alkylation of myotoxic phospholipases A2 in Bothrops moojeni venom: A promising approach to an enhanced antivenom production. Int. J. Biochem. Cell Biol. 2004, 36, 258–270. [Google Scholar] [CrossRef]
- Chulpanova, D.S.; Kitaeva, K.V.; James, V.; Rizvanov, A.A.; Solovyeva, V.V. Therapeutic Prospects of Extracellular Vesicles in Cancer Treatment. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskienė, N.; Pašukonienė, V.; Darinskas, A.; Kraśko, J.A.; Žilionytė, K.; Mlynska, A.; Gudlevičienė, Ž.; Mišeikytė-Kaubrienė, E.; Schijns, V.; Lubitz, W.; et al. Tumor lysate-loaded Bacterial Ghosts as a tool for optimized production of therapeutic dendritic cell-based cancer vaccines. Vaccine 2018, 36, 4171–4180. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Yang, H.; Zhao, D.; Chen, J.; Zhang, Q.; Liang, J.; Yin, Y.; Kong, G.; Li, G. Design and immune characterization of a novel Neisseria gonorrhoeae DNA vaccine using bacterial ghosts as vector and adjuvant. Vaccine 2018, 36, 4532–4539. [Google Scholar] [CrossRef] [PubMed]
- Pulido, M.R.; García-Quintanilla, M.; Pachón, J.; McConnell, M.J. Immunization with lipopolysaccharide-free outer membrane complexes protects against Acinetobacter baumannii infection. Vaccine 2018, 36, 4153–4156. [Google Scholar] [CrossRef] [PubMed]
- Estanqueiro, M.; Amaral, M.H.; Conceição, J.; Sousa Lobo, J.M. Nanotechnological carriers for cancer chemotherapy: The state of the art. Colloids Surf. B Biointerfaces 2015, 126, 631–648. [Google Scholar] [CrossRef] [PubMed]
- Biswaro, L.S.; Sousa, G.M.D.C.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial Peptides and Nanotechnology, Recent Advances and Challenges. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kou, Y.; Zhang, X.; Cheng, H.; Chen, X.; Mao, S. Strategies and industrial perspectives to improve oral absorption of biological macromolecules. Expert Opin. Drug Deliv. 2018, 15, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Schetters, S.T.; Kruijssen, L.J.; Crommentuijn, M.H.; Kalay, H.; Ochando, J.; Den Haan, J.M.; Garcia-Vallejo, J.J.; Van Kooyk, Y. Mouse DC-SIGN/CD209a as Target for Antigen Delivery and Adaptive Immunity. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Howarth, M. New Routes and Opportunities for Modular Construction of Particulate Vaccines: Stick, Click, and Glue. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Palladini, A.; Thrane, S.; Janitzek, C.M.; Pihl, J.; Clemmensen, S.B.; Jongh, W.A.D.; Clausen, T.M.; Nicoletti, G.; Landuzzi, L.; Penichet, M.L.; Balboni, T.; et al. Virus-like particle display of HER2 induces potent anti-cancer responses. OncoImmunology 2018, 7, e1408749. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Thrane, S.; Janitzek, C.M.; Nielsen, M.A.; Theander, T.G.; Theisen, M.; Salanti, A.; Sander, A.F. Improving the malaria transmission-blocking activity of a Plasmodium falciparum 48/45 based vaccine antigen by SpyTag/SpyCatcher mediated virus-like display. Vaccine 2017, 35, 3726–3732. [Google Scholar] [CrossRef] [PubMed]
- Thrane, S.; Janitzek, C.M.; Matondo, S.; Resende, M.; Gustavsson, T.; de Jongh, W.A.; Clemmensen, S.; Roeffen, W.; van de Vegte-Bolmer, M.; van Gemert, G.J.; et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines. J. Nanobiotechnology 2016, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, H.; Wang, W.; Tan, W.; Fu, Y.-X.; Zhu, M. A novel method for synthetic vaccine construction based on protein assembly. Sci. Rep. 2014, 4, 7266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chenal, A.; Ladant, D. Bioengineering of Bordetella pertussis Adenylate Cyclase Toxin for Antigen-Delivery and Immunotherapy. Toxins 2018, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.-R.; Hildebrand, W.H.; Mardis, E.R.; et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H. Toxin-centric development approach for next-generation antivenoms. Toxicon 2018, 150, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Lomonte, B.; León, G.; Alape-Girón, A.; Flores-Díaz, M.; Sanz, L.; Angulo, Y.; Calvete, J.J. Snake venomics and antivenomics: Proteomic tools in the design and control of antivenoms for the treatment of snakebite envenoming. J. Proteomics 2009, 72, 165–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvete, J.J. Proteomic tools against the neglected pathology of snake bite envenoming. Expert Rev. Proteomics 2011, 8, 739–758. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Petras, D.; Calderón-Celis, F.; Lomonte, B.; Encinar, J.R.; Sanz-Medel, A. Protein-species quantitative venomics: Looking through a crystal ball. J. Venom. Anim. Toxins Trop. Dis. 2017, 23. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Calvete, J.J. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Trop. Dis. 2017, 23. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Lomonte, B. A bright future for integrative venomics. Toxicon 2015, 107, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Pal, R. Integrated Analysis of Transcriptomic and Proteomic Data. Curr. Genomics 2013, 14, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Gutiérrez, J.M.; Rasmussen, A.R.; Engmark, M.; Gravlund, P.; Sanders, K.L.; Lohse, B.; Lomonte, B. Danger in the reef: Proteome, toxicity, and neutralization of the venom of the olive sea snake, Aipysurus laevis. Toxicon 2015, 107, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauridsen, L.P.; Laustsen, A.H.; Lomonte, B.; Gutiérrez, J.M. Toxicovenomics and antivenom profiling of the Eastern green mamba snake (Dendroaspis angusticeps). J. Proteomics 2016, 136, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Lauridsen, L.P.; Laustsen, A.H.; Lomonte, B.; Gutiérrez, J.M. Exploring the venom of the forest cobra snake: Toxicovenomics and antivenom profiling of Naja melanoleuca. J. Proteomics 2017, 150, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Flower, D.R. Towards in silico prediction of immunogenic epitopes. Trends Immunol. 2003, 24, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, X. An introduction to epitope prediction methods and software. Rev. Med. Virol. 2008, 19, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Ju, H.; Liu, Z.; Ning, Q.; Zhang, J.; Zhao, X.; Huang, Y.; Ma, Z.; Li, Y. Bioinformatics Resources and Tools for Conformational B-Cell Epitope Prediction. Comput. Math. Methods Med. 2013, 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-W.; Pai, T.-W. Machine Learning-Based Methods for Prediction of Linear B-Cell Epitopes. In Immunoinformatics; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2014; pp. 217–236. ISBN 978-1-4939-1114-1. [Google Scholar]
- Backert, L.; Kohlbacher, O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Soria-Guerra, R.E.; Nieto-Gomez, R.; Govea-Alonso, D.O.; Rosales-Mendoza, S. An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J. Biomed. Inform. 2015, 53, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Potocnakova, L.; Bhide, M.; Pulzova, L.B. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J. Immunol. Res. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- EL-Manzalawy, Y.; Dobbs, D.; Honavar, V.G. In Silico Prediction of Linear B-Cell Epitopes on Proteins. In Prediction of Protein Secondary Structure; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; pp. 255–264. ISBN 978-1-4939-6404-8. [Google Scholar]
- Larsen, J.E.P.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Raghava, G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, H.; Yang, J.; Chou, K.-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef] [PubMed]
- EL-Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit. 2008, 21, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, N.D.; Mayrose, I.; Martz, E.; Pupko, T. Epitopia: A web-server for predicting B-cell epitopes. BMC Bioinform. 2009, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Ansari, H.R.; Raghava, G.P.S. Improved Method for Linear B-Cell Epitope Prediction Using Antigen’s Primary Sequence. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Cao, Y.; Cha, L.; Zhang, X.; Ying, X.; Zhang, W.; Ge, K.; Li, W.; Zhong, L. Predicting linear B-cell epitopes using amino acid anchoring pair composition. BioData Min. 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Haste Andersen, P.; Nielsen, M.; Lund, O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. Publ. Protein Soc. 2006, 15, 2558–2567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponomarenko, J.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Kringelum, J.V.; Lundegaard, C.; Lund, O.; Nielsen, M. Reliable B Cell Epitope Predictions: Impacts of Method Development and Improved Benchmarking. PLoS Comput. Biol. 2012, 8, e1002829. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, K.; Liu, X.; Baker, T.; Shi, J.; Deane, C.M. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics 2014, 30, 2288–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledsgaard, L.; Jenkins, T.P.; Davidsen, K.; Krause, K.E.; Martos-Esteban, A.; Engmark, M.; Rørdam Andersen, M.; Lund, O.; Laustsen, A.H. Antibody Cross-Reactivity in Antivenom Research. Toxins 2018, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; Gutiérrez, J.M.; Calvete, J.J. Second generation snake antivenomics: Comparing immunoaffinity and immunodepletion protocols. Toxicon 2012, 60, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Lomonte, B.; Sanz, L.; Calvete, J.J.; Pla, D. Immunological profile of antivenoms: Preclinical analysis of the efficacy of a polyspecific antivenom through antivenomics and neutralization assays. J. Proteomics 2014, 105, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Sanz, L.; Pla, D.; Lomonte, B.; Gutiérrez, J.M. Omics Meets Biology: Application to the Design and Preclinical Assessment of Antivenoms. Toxins 2014, 6, 3388–3405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pla, D.; Rodríguez, Y.; Calvete, J.J. Third Generation Antivenomics: Pushing the Limits of the In Vitro Preclinical Assessment of Antivenoms. Toxins 2017, 9, 158. [Google Scholar] [CrossRef]
- Engmark, M.; Andersen, M.R.; Laustsen, A.H.; Patel, J.; Sullivan, E.; Masi, F.D.; Hansen, C.S.; Kringelum, J.V.; Lomonte, B.; Gutiérrez, J.M.; et al. High-throughput immuno-profiling of mamba (Dendroaspis) venom toxin epitopes using high-density peptide microarrays. Sci. Rep. 2016, 6, 36629. [Google Scholar] [CrossRef] [PubMed]
- Engmark, M.; Lomonte, B.; Gutiérrez, J.M.; Laustsen, A.H.; Masi, F.D.; Andersen, M.R.; Lund, O. Cross-recognition of a pit viper (Crotalinae) polyspecific antivenom explored through high-density peptide microarray epitope mapping. PLoS Negl. Trop. Dis. 2017, 11, e0005768. [Google Scholar] [CrossRef] [PubMed]
- Engmark, M.; Jespersen, M.C.; Lomonte, B.; Lund, O.; Laustsen, A.H. High-density peptide microarray exploration of the antibody response in a rabbit immunized with a neurotoxic venom fraction. Toxicon 2017, 138, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.A.; Hargreaves, A.; Wagstaff, S.C.; Faragher, B.; Lalloo, D.G. Snake Envenoming: A Disease of Poverty. PLoS Negl. Trop. Dis. 2009, 3, e569. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.A.; Gutiérrez, J.M. Priority Actions and Progress to Substantially and Sustainably Reduce the Mortality, Morbidity and Socioeconomic Burden of Tropical Snakebite. Toxins 2016, 8, 351. [Google Scholar] [CrossRef] [PubMed]
- Bawaskar, H.S.; Bawaskar, P.H.; Bawaskar, P.H. Snake bite in India: A neglected disease of poverty. Lancet 2017, 390, 1947–1948. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Dorrestijn, N. Integrating Engineering, Manufacturing, and Regulatory Considerations in the Development of Novel Antivenoms. Toxins 2018, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H. Snakebites: Costing recombinant antivenoms. Nature 2016, 538, 41. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Johansen, K.H.; Engmark, M.; Andersen, M.R. Recombinant snakebite antivenoms: A cost-competitive solution to a neglected tropical disease? PLoS Negl. Trop. Dis. 2017, 11, e0005361. [Google Scholar] [CrossRef] [PubMed]
- Longbottom, J.; Shearer, F.M.; Devine, M.; Alcoba, G.; Chappuis, F.; Weiss, D.J.; Ray, S.E.; Ray, N.; Warrell, D.A.; de Castañeda, R.R.; et al. Vulnerability to snakebite envenoming: A global mapping of hotspots. Lancet 2018. [Google Scholar] [CrossRef]
- Brüggemann, M.; Osborn, M.J.; Ma, B.; Hayre, J.; Avis, S.; Lundstrom, B.; Buelow, R. Human Antibody Production in Transgenic Animals. Arch. Immunol. Ther. Exp. 2015, 63, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Cervenak, J.; Kurrle, R.; Kacskovics, I. Accelerating antibody discovery using transgenic animals overexpressing the neonatal Fc receptor as a result of augmented humoral immunity. Immunol. Rev. 2015, 268, 269–287. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.L.; Gilliland, G.L. Engineering antibody therapeutics. Curr. Opin. Struct. Biol. 2016, 38, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Murawsky, C.M. Strategies for Generating Diverse Antibody Repertoires Using Transgenic Animals Expressing Human Antibodies. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]


| Authors & Year | Immunization Strategy | Species | Target Toxin(s) | Antivenom | Challenge Toxin(s) | Effect(s) Neutralized | Ref. |
|---|---|---|---|---|---|---|---|
| Čurin-Šerbec et al., 1991 | Synthetic epitope | Vipera ammodytes | Ammodytoxin A | Rabbit antiserum | Ammodytoxin A | Lethality | [70] |
| Čurin-Šerbec et al., 1994 | Synthetic epitope | Crotalus durissus terrificus, Vipera ammodytes | Crotoxin and ammodytoxin A | Murine IgG, IgM | Crotoxin | Prolonged survival time | [68] |
| Dolimbek and Atassi, 1996 | Synthetic epitope | Bungarus multicinctus | α-bungarotoxin | Murine antiserum | α-bungarotoxin | Lethality | [71] |
| Calderón et al., 1999 | Synthetic epitope | Bothrops asper | Myotoxin II | Murine antiserum | Myotoxin II | Myotoxicity | [67] |
| Harrison et al., 2000 | DNA | Bothrops jararaca | Jararhagin | Murine antiserum | B. jararaca venom | Myotoxicity | [79] |
| Harrison et al., 2002 | DNA | Bothrops jararaca | Jararhagin | Murine antiserum | N/A | Not Evaluated | [80] |
| Pergolizzi et al., 2004 | DNA | Naja kaouthia | α-cobratoxin | Murine antiserum | α-cobratoxin | Lethality | [85] |
| Wagstaff et al., 2006 | DNA | Echis ocellatus | SVMPs | Murine IgG | E. ocellatus and Cerastes cerastes venom | Hemotoxicity | [83] |
| Ferreira et al., 2006 | Synthetic epitope | Lachesis muta muta | Mutalysin II | Rabbit IgG | Mutalysin II | Hemotoxicity | [69] |
| Azofeifa-Cordero et al., 2008 | DNA | Crotalus durissus durissus | P-III SVMP | Murine antiserum | C. d. durissus venom | Hemotoxicity | [84] |
| Leão et al., 2009 | DNA | Micrurus corallinus | 3FTx and PLA2s | Murine antiserum | N/A | Not evaluated | [95] |
| Cardoso et al., 2009 | Recombinant mimotope | Bothrops neuwiedi | Neuwiedase | Murine antiserum | N/A | Not evaluated | [78] |
| Arce-Estrada et al., 2009 | DNA | Bothrops asper | P-II SVMP | Equine antiserum | B. asper and C. d. durissus venom | Hemotoxicity | [82] |
| Suntrarachun et al., 2010 | Recombinant toxin | Bungarus candidus | α and β-neurotoxins | Murine antiserum | B. candidus venom | Lethality | [73] |
| Machado de Avila et al., 2011 | Synthetic mimotope | Lachesis muta | Mutalysin II | Rabbit antiserum | L. muta venom | Hemotoxicity | [77] |
| Ramos et al., 2016 | DNA + Recombinant protein | Micrurus corallinus | 3FTxs and PLA2 | Murine antiserum | M. corallinus venom | Lethality | [66] |
| Cao et al., 2016 | Recombinant protein | Deinagkistrodon acutus | SVSPs, SVMPs and PLA2s | Murine antiserum | D. acutus venom | Hemotoxicity | [72] |
| Clement et al., 2016 | Recombinant toxin | Micrurus laticorallis | Cysteine-rich neurotoxins (Mlat1) | Rabbit antiserum | M. laticorallis venom | Not neutralizing | [96] |
| Hasson, 2017 | DNA | Echis ocellatus | Disintegrin | Murine antiserum | Crotalus atrox, E. ocellatus and Bitis arietans venom | Hemotoxicity | [81] |
| de la Rosa et al., 2018 | Recombinant toxin | Acanthophis spp., Oxyuranus spp., Walterinnesia spp., Naja spp., Dendroaspis spp. and Micrurus spp. | Type I α-neurotoxins | Rabbit antiserum | N/A | Not evaluated | [75] |
| Guerrero-Garzón et al., 2018 | Recombinant toxin | Micrurus diastema | Type I α-neurotoxin D.H. | Rabbit antiserum | rD.H, MlatA1, and fraction F5 from M. diastema venom | Lethality | [74] |
| Authors & Year | Immunization Strategy | Species | Target Toxin(s) | Antivenom | Challenge Toxin(s) | Effect(s) Neutralized | Ref. |
|---|---|---|---|---|---|---|---|
| Bahraoui et al., 1986 | Synthetic epitope | Androctonus australis hector | Toxin II (AahII) | Murine antiserum | AahII | Lethality | [97] |
| Devaux et al., 1993 | Synthetic epitope | Androctonus australis hector | Toxin II (AahII) | Rabbit antiserum | N/A | N/A | [103] |
| Calderón-Aranda et al., 1995 | Synthetic epitope | Centruroides noxius | Cn2 | Rabbit and murine antisera | Cn2 | Lethality | [98] |
| Bouhaouala-Zahar et al., 1996 | Recombinant toxin | Buthus occitanus tunetanus | α-toxin | Murine antiserum | Bot and AaHG | Lethality | [121] |
| Devaux et al., 1997 | Synthetic peptide | Androctonus australis hector | Toxin II (AahII) | Murine IgG | AahII | Lethality | [108] |
| Zenouaki et al., 1997 | Synthetic peptide | Androctonus australis hector | Toxin II (AahII) | Rabbit antiserum | AahII | Lethality | [106] |
| Calderón-Aranda et al., 1999 | Synthetic epitope | Centruroides noxius | Cn2 | Rabbit and murine antisera | Cn2 | Lethality | [99] |
| Guatimosim et al., 2000 | Recombinant toxoid | Tityus serrulatus | TsNTxP | Rabbit antiserum | T. serrulatus venom | Lethality | [109] |
| Gazarian et al., 2000 | Mimotopes | Centruroides noxius Hoffmann | Noxiustoxin | Murine antiserum | N/A | N/A | [122] |
| Chávez-Olórtegui et al., 2001 | Synthetic epitope | Tityus serrulatus | TsNTxP | Rabbit antiserum | TstG50 | Lethality | [104] |
| Legros et al., 2001 | Recombinant toxin | Androctonus australis hector | AahI, AahII and AahIII (α-toxins) | Rabbit and murine antisera | AaH-G50 | Lethality | [110] |
| Benkhadir et al., 2002 | Recombinant toxin | Buthus occitanus tunetanus | Bot III (α-toxin) | Murine antiserum | B. occitanus tunetanus venom | Lethality | [111] |
| Alvarenga et al., 2002 | Synthetic epitope | Tityus serrulatus | TsNTxP and TsIV | Rabbit antiserum | TstG50 | Lethality | [100] |
| Garcia et al., 2003 | Recombinant toxin | Centruroides noxius Hoffmann | Cn5 and sub-fraction | Rabbit antiserum | Cn2 | Lethality | [112] |
| Inceoglu et al., 2006 | Synthetic epitope | Parabuthus transvaalicus | Birtoxin | Rabbit polyclonal IgG | P. transvaalicus venom | Lethality | [101] |
| Corona Villegas et al., 2008 | Recombinant toxin | Centruroides spp. | Cex1-13, Cll3-8, Cn4b, Cn10b, Ce3, Ce5-7, Ce13(b), Cg1-3, CsEv1-3, CsEV8-9, CsE1x, CsEIa, CexErg1-4, Cll Erg1-4, Cn Erg3-5, CeErg1-3, CgErg1-3, CsErg1-5 | Rabbit antiserum | Cn2 | Lethality | [113] |
| Mendes et al., 2008 | Recombinant toxin | Tityus serrulatus | Ts1 | Rabbit antiserum | Tst1 and T. serrulatus venom | Lethality | [114] |
| Hernández-Salgado et al., 2009 | Recombinant toxin and toxoid | Centruroides suffusus suffusus | CssII | Rabbit antiserum | CssII, Cn2, and C. suffusus suffusus venom | Lethality | [115] |
| García-Gómez et al., 2009 | Recombinant toxin | Parabuthus granulatus | Pg8 | Murine antiserum | Pg8 and P. granulatus venom | Lethality | [116] |
| Duarte et al., 2010 | Synthetic epitope | Tityus serrulatus | TsNTxP | Murine antiserum | T. serrulatus venom | Lethality | [102] |
| Eskandari et al., 2014 | Recombinant toxin | Mesobuthus eupeus | BMK neurotoxin | Murine antiserum | N/A | Not evaluated | [117] |
| Uawonggul et al., 2014 | Recombinant toxin | Heterometrus laoticus | Heteroscorpine-1 (HS-1) | Murine antiserum | H. laoticus venom | Paralysis | [120] |
| Jiménez-Vargas et al., 2017 | Recombinant toxin | Centruroides spp. | Cn2, Css2, Cll1, and Cll2 | Murine and rabbit antisera | C. noxius, C. suffusus, C. limpidus, C. elegans, C. tecomanus, and C. sculpturatus venom | Lethality | [118] |
| Safari Foroushani et al., 2018 | Recombinant toxoid | Hemiscorpius lepturus | rPLD1 | Murine antiserum | rPLD1 and H. lepturus venom | Lethality | [119] |
| Authors & Year | Immunization Strategy | Species | Target Toxin(s) | Antivenom | Challenge Toxin(s) | Effect(s) Neutralized | Ref. |
|---|---|---|---|---|---|---|---|
| Fernandes Pedrosa et al., 2002 | Recombinant toxin | Loxosceles laeta | Smase I | Rabbit antiserum | rSmase I and L. laeta venom | Dermonecrosis | [139] |
| Araujo et al., 2003 | Recombinant toxin | Loxosceles intermedia | Dermonecrotic toxin | Murine antiserum | L. intermedia venom | Dermonecrosis and lethality | [127] |
| Tambourgi et al., 2004 | Recombinant toxin | Loxosceles intermedia | Sphingomyelinases | Rabbit antiserum | N/A | N/A | [123] |
| Olvera et al., 2006 | Recombinant toxin | Loxosceles reclusa, Loxosceles boneti, Loxosceles laeta | Sphingomyelinase D | Rabbit antiserum and equine F(ab’)2 | rSMD, L. reclusa, L. boneti and L. laeta venom | Lethality | [128] |
| Felicori et al., 2006 | Recombinant toxin | Loxosceles intermedia | Dermonecrotic toxin LiD1 | Murine antiserum | L. intermedia venom | Lethality | [129] |
| Fischer et al., 2007 | Synthetic epitope | Atrax robustus | Robustoxin | Murine antiserum | A. robustus venom | Lethality | [107] |
| de Almeida et al., 2008 | Recombinant toxin | Loxosceles intermedia, Loxosceles laeta and Loxosceles gaucho | Sphingomyelinase D | Equine antiserum | L. intermedia, L. laeta, and L. gaucho venom | Dermonecrosis | [130] |
| Felicori et al., 2009 | Synthetic epitope | Loxosceles intermedia | Dermonecrotic toxin LiD1 | Rabbit IgGs | LiD1 | Dermonecrosis, hemotoxicity, and edema | [136] |
| Comis et al., 2009 | Synthetic toxin | Atrax robustus | Robustoxin | Monkey antiserum | A. robustus venom | Lethality | [126] |
| Dias-Lopes et al., 2010 | Synthetic epitope | Loxosceles intermedia | Dermonecrotic toxin LiD1 | Rabbit and murine antisera | rLiD1 and L. intermedia venom | Dermonecrosis, hemotoxicity, and lethality | [137] |
| Chaim et al., 2011 | Recombinant toxoid | Loxosceles intermedia | Dermonecrotic toxin LiD1 | Rabbit antiserum | N/A | N/A | [142] |
| de Moura et al., 2011 | Synthetic mimotope | Loxosceles intermedia | Dermonecrotic toxin LiD1 | Rabbit antiserum | L. intermedia venom | Dermonecrosis, hemotoxicity | [138] |
| Mendes et al., 2013 | Recombinant toxin | Loxosceles intermedia | Dermonecrotic toxin LiD1 | Rabbit antiserum and IgG | rLiD1 | Dermonecrosis, hemotoxicity | [131] |
| Magalhães et al., 2013 | Recombinant toxin | Loxosceles gaucho | Phospholipase D | Rabbit antiserum | LgRec1 and L. gaucho venom | Dermonecrosis, local reaction | [124] |
| Figueiredo et al., 2014 | Recombinant toxin | Loxosceles intermedia, Loxosceles laeta, and Loxosceles gaucho | Sphingomyelinase D | Equine antiserum | L. intermedia, L. gaucho and L. laeta venom | Dermonecrosis | [132] |
| Dias-Lopes et al., 2014 | Recombinant toxin | Loxosceles intermedia, Loxosceles laeta, and Loxosceles gaucho | Sphingomyelinase D | Murine IgG | rLiD1 | Dermonecrosis, hemotoxicity, and edema | [133] |
| Duarte et al., 2015 | Recombinant toxin | Loxosceles intermedia and Loxosceles laeta | Dermonecrotic toxin LiD1 | Equine antiserum | L. intermedia and L. laeta venom | Dermonecrosis, hemotoxicity | [134] |
| Oliveira et al., 2016 | Recombinant toxin | Loxosceles intermedia | Sphingomyelinase D (SMD) | Murine antiserum | rSMDs, L. reclusa, L. boneti, and L. laeta venom | Lethality | [135] |
| Lima et al., 2018 | Recombinant toxin | Loxosceles intermedia and Loxosceles laeta | Loxosceles astacin-like protease 1, hyaluronidases, SMase-I | Rabbit antiserum | L. intermedia venom | Lethality | [125] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bermúdez-Méndez, E.; Fuglsang-Madsen, A.; Føns, S.; Lomonte, B.; Gutiérrez, J.M.; Laustsen, A.H. Innovative Immunization Strategies for Antivenom Development. Toxins 2018, 10, 452. https://doi.org/10.3390/toxins10110452
Bermúdez-Méndez E, Fuglsang-Madsen A, Føns S, Lomonte B, Gutiérrez JM, Laustsen AH. Innovative Immunization Strategies for Antivenom Development. Toxins. 2018; 10(11):452. https://doi.org/10.3390/toxins10110452
Chicago/Turabian StyleBermúdez-Méndez, Erick, Albert Fuglsang-Madsen, Sofie Føns, Bruno Lomonte, José María Gutiérrez, and Andreas Hougaard Laustsen. 2018. "Innovative Immunization Strategies for Antivenom Development" Toxins 10, no. 11: 452. https://doi.org/10.3390/toxins10110452
APA StyleBermúdez-Méndez, E., Fuglsang-Madsen, A., Føns, S., Lomonte, B., Gutiérrez, J. M., & Laustsen, A. H. (2018). Innovative Immunization Strategies for Antivenom Development. Toxins, 10(11), 452. https://doi.org/10.3390/toxins10110452








