Structural Basis for the Specific Neutralization of Stx2a with a Camelid Single Domain Antibody Fragment
Abstract
:1. Introduction
2. Results
2.1. Production of Recombinant Stx2a B Domain and Construction of an Immune Nb Phage Display Library
2.2. Selection of rStx2aB-Binding Nanobodies
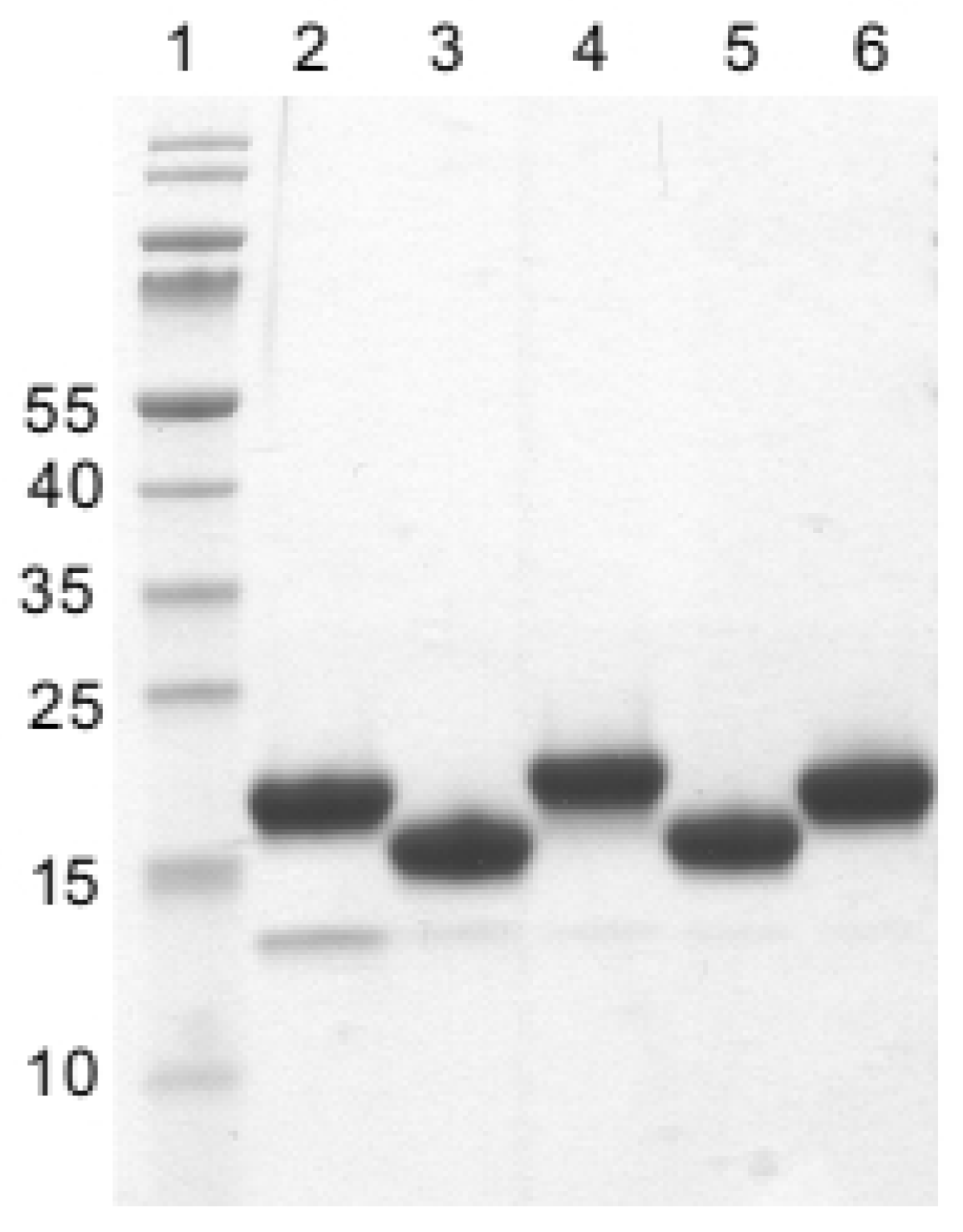
2.3. Specificity of Nanobodies Assessed via Western Blot
2.4. Interaction between Nbs and rStx2aB Measured via Surface Plasmon Resonance
2.4.1. In Vivo Biotinylation of rStx2aB Protein
2.4.2. Affinity Measurement
2.4.3. Epitope Binning
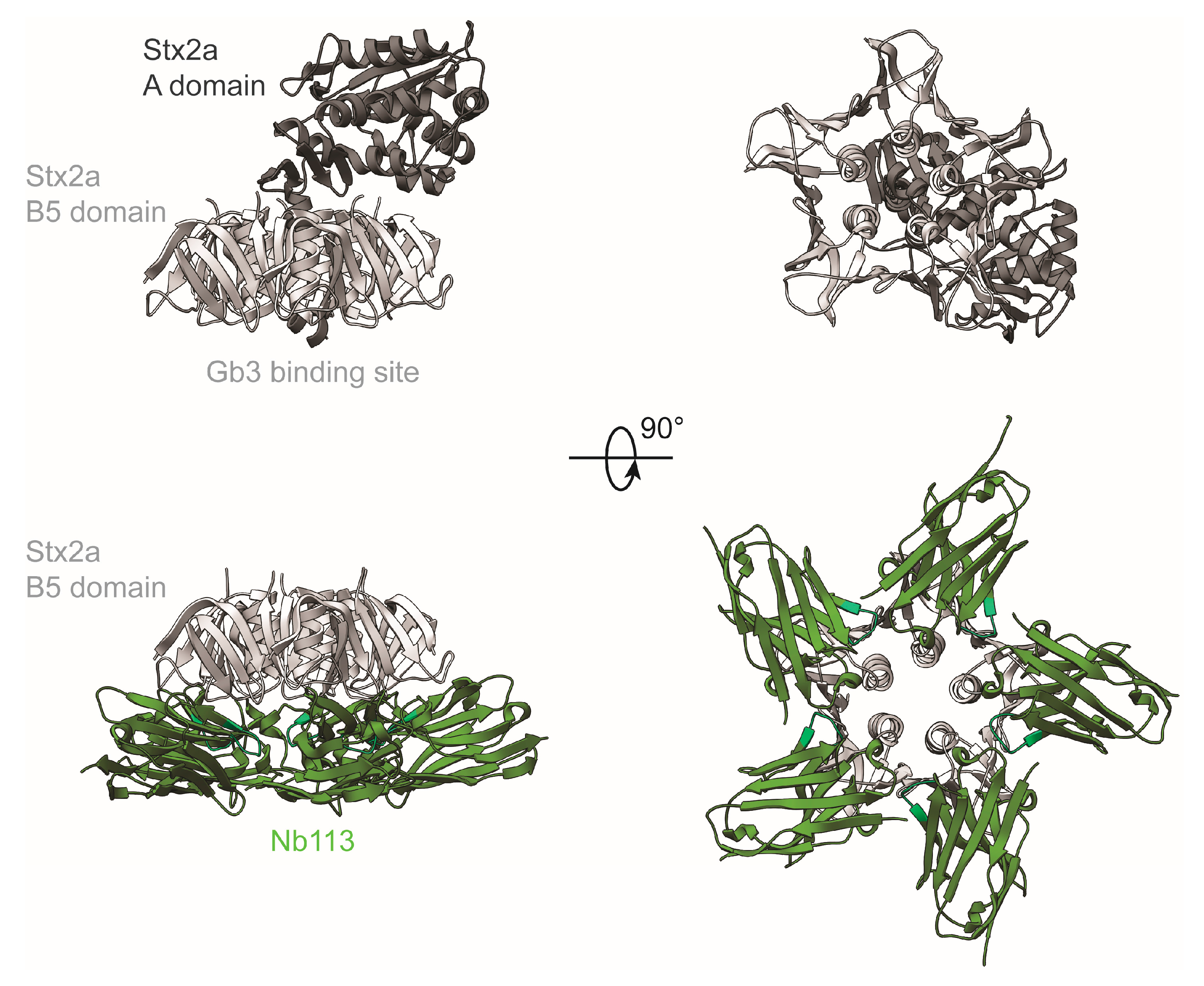
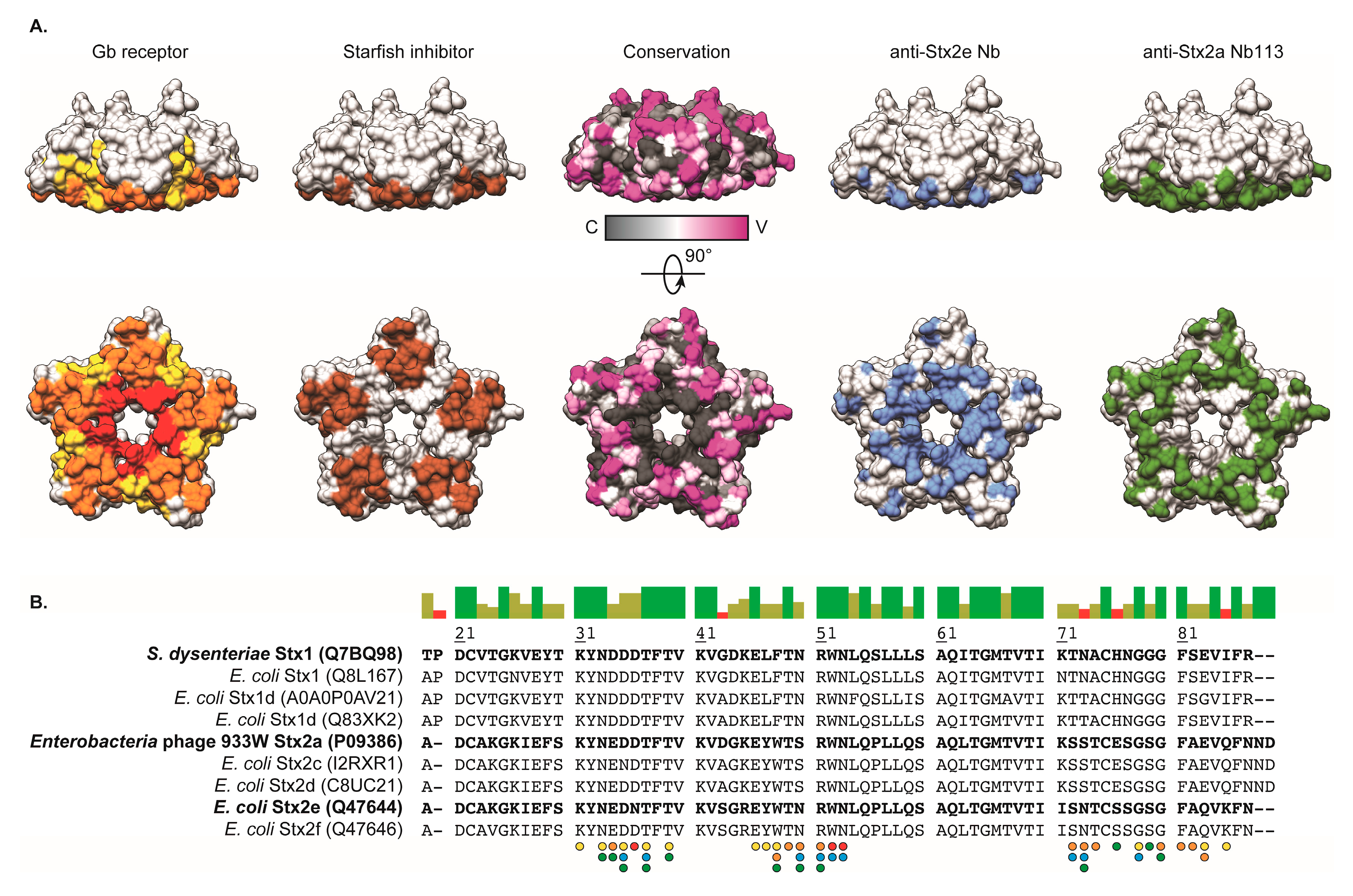
2.5. Crystal Structure of Nb113–rStx2aB Complex
2.6. Multimerization of Nanobodies and Neutralization of Cytotoxicity in Cell Cultures
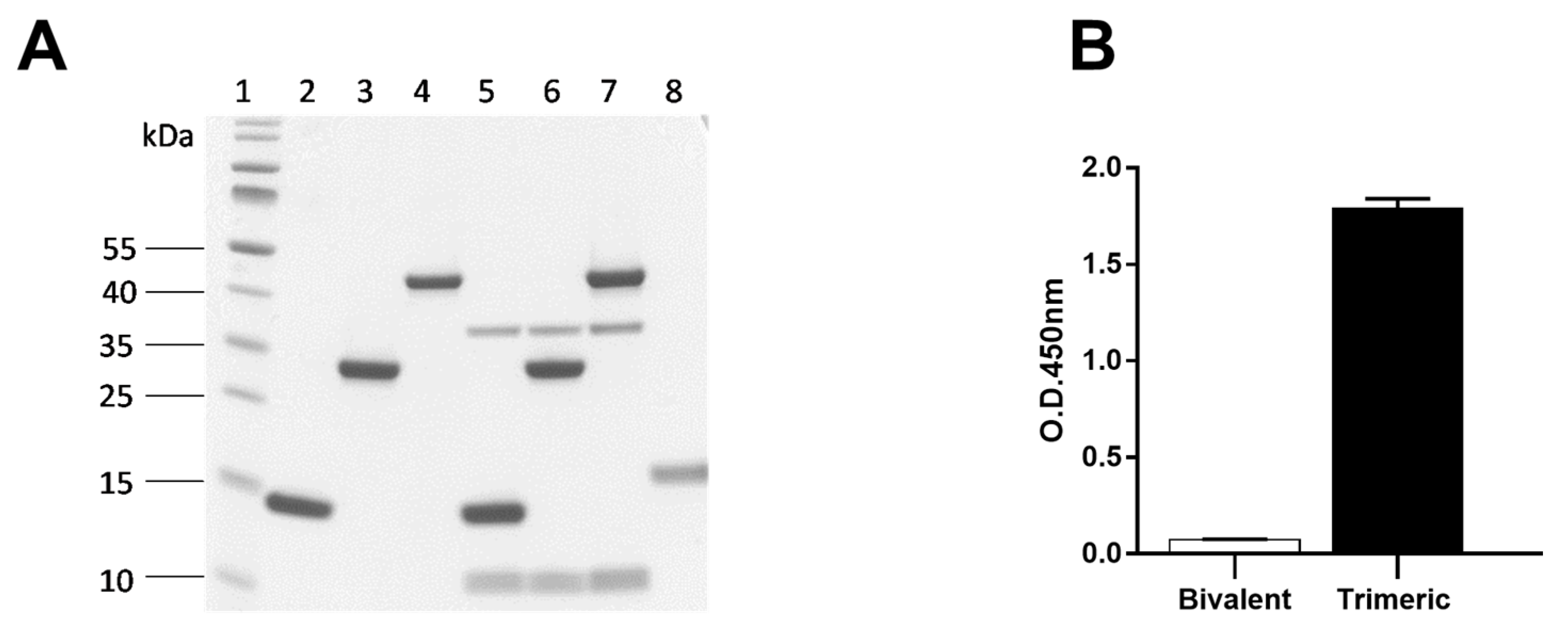
2.6.1. Specificity of Bivalent Nb1132 and Trimeric Nb1132–NbSA1
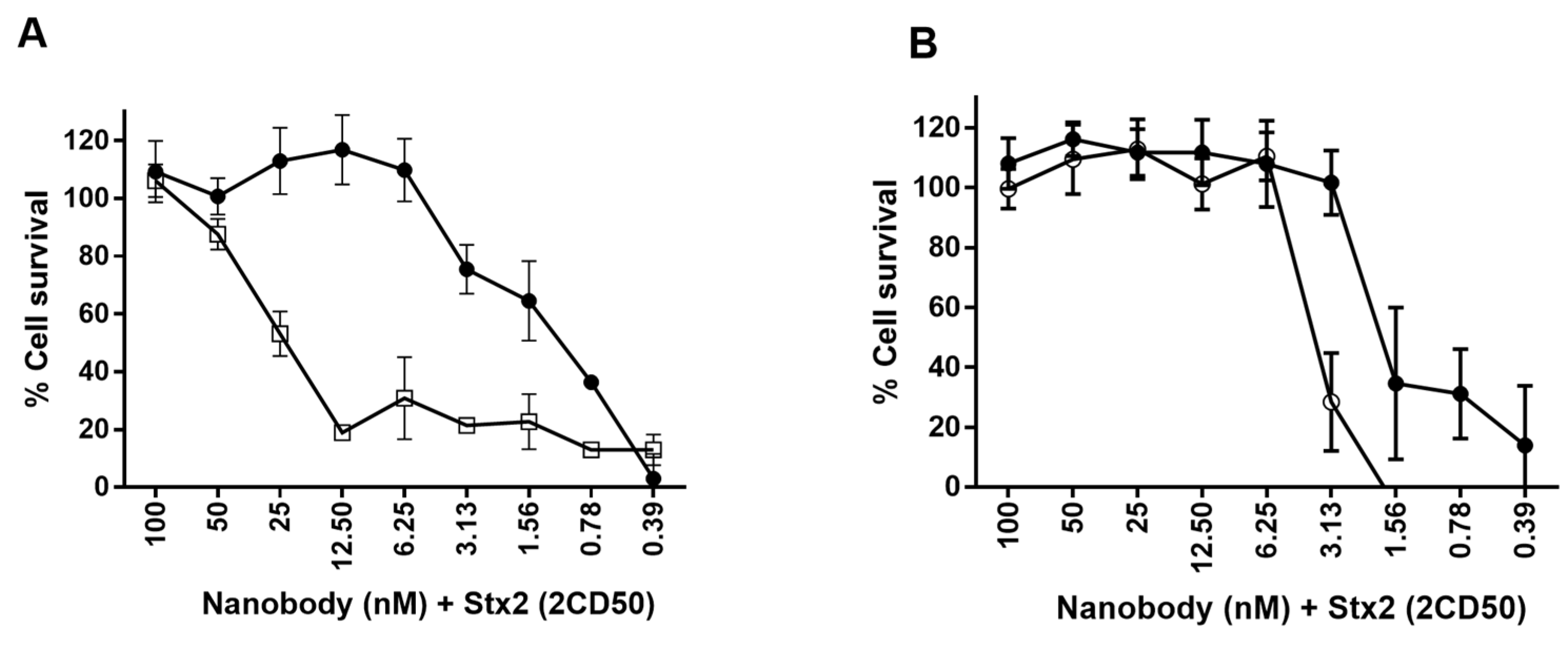
2.6.2. Neutralization of Stx2
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Immunogen Production
4.3. Alpaca Immunization
4.4. Construction of the Immune Phage Display Library
4.5. Selection of rStx2aB-Binding Nanobodies
4.6. Expression and Purification of rStx2aB-Binding Nanobodies
4.7. Western Blot Using Nanobodies as Probe
4.8. Western Blot Using rStx2aB as Probe
4.9. In Vivo Biotinylation of rStx2aB for Surface Plasmon Resonance Assays
4.10. Affinity Measurement and Epitope Binning
4.11. Crystallization, Data Collection, Data Processing, and Structure Determination of Nb113–rStx2aB Complex
4.12. Construction of a Bivalent Nanobody
4.13. Construction of Trimeric Bispecific Nb1132–NbSA1
4.14. Expression and Purification of Multimerised Nanobodies
4.15. Immunocapturing Experiments
4.16. Serum Albumin Binding ELISA
4.17. Neutralization of Stx2 Cytotoxicity in Cell Cultures
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Majowicz, S.E.; Scallan, E.; Jones-bitton, A.; Jan, M.; Stapleton, J.; Angulo, F.J.; Yeung, D.H.; Kirk, M.D. Global Incidence of Human Shiga Toxin–Producing Escherichia coli Infections and Deaths: A Systematic Review and Knowledge Synthesis. Food Microbiol. 2014, 11, 447–455. [Google Scholar] [CrossRef]
- Pennington, H. Escherichia coli O157. Lancet 2017, 376, 1428–1435. [Google Scholar] [CrossRef]
- Frenzen, P.D.; Drake, A.; Angulo, F.J. Economic cost of illness due to Escherichia coli O157 infections in the United States. J. Food Prot. 2005, 68, 2623–2630. [Google Scholar] [CrossRef] [PubMed]
- Buzby, J.C.; Roberts, T. The economics of enteric infections: Human foodborne disease costs. Gastroenterology 2009, 136, 1851–1862. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Byrne, L.; Jenkins, C.; Launders, N.; Elson, R.; Adak, G.K. The epidemiology, microbiology and clinical impact of Shiga toxin-producing Escherichia coli in England, 2009–2012. Epidemiol. Infect. 2015, 143, 3475–3487. [Google Scholar] [CrossRef] [PubMed]
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fuller, C.A.; Pellino, C.A.; Flagler, M.J.; Strasser, J.E.; Weiss, A.A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011, 79, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.; Werber, D.; Cramer, J.P.; Askar, M.; Faber, M.; an der Heiden, M.; Bernard, H.; Fruth, A.; Prager, R.; Spode, A.; et al. Epidemic Profile of Shiga-Toxin–Producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 2011, 365, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- Karch, H.; Denamur, E.; Dobrindt, U.; Finlay, B.B.; Hengge, R.; Johannes, L.; Ron, E.Z.; Tønjum, T.; Sansonetti, P.J.; Vicente, M. The enemy within us: Lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol. Med. 2012, 4, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, C.; Amaral, M.M.; Palermo, M.S. Advances in pathogenesis and therapy of hemolytic uremic syndrome caused by Shiga toxin-2. IUBMB Life 2013, 65, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.J.; Dodd, J.E.; Hautbergue, G.M. Ribosome-inactivating proteins potent poisons and molecular tools. Virulence 2013, 4, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.E.; Boodhoo, A.; Tyrrell, G.J.; Brunton, J.L.; Read, R.J. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature 1992, 355, 748–750. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.E.; Chernaia, M.M.; Kozlov, Y.V.; James, M.N. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 A resolution. Nat. Struct. Biol. 1994, 1, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Johannes, L.; Romer, W. Shiga toxins—From cell biology to biomedical applications. Nat. Rev. Microbiol. 2010, 8, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Bergan, J.; Dyve Lingelem, A.B.; Simm, R.; Skotland, T.; Sandvig, K. Shiga toxins. Toxicon 2012, 60, 1085–1107. [Google Scholar] [CrossRef] [PubMed]
- Garred, O.; Van Deurs, B.; Sandvig, K. Furin-induced cleavage and activation of shiga toxin. J. Biol. Chem. 1995, 270, 10817–10821. [Google Scholar] [CrossRef] [PubMed]
- Rosales, A.; Hofer, J.; Zimmerhackl, L.-B.; Jungraithmayr, T.C.; Riedl, M.; Giner, T.; Strasak, A.; Orth-Holler, D.; Wurzner, R.; Karch, H. Need for long-term follow-up in enterohemorrhagic Escherichia coli-associated hemolytic uremic syndrome due to late-emerging sequelae. Clin. Infect. Dis. 2012, 54, 1413–1421. [Google Scholar] [CrossRef] [PubMed]
- Palermo, M.S.; Exeni, R.A.; Fernandez, G.C. Hemolytic uremic syndrome: Pathogenesis and update of interventions. Expert Rev. Anti-Infect. Ther. 2009, 7, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Spinale, J.M.; Ruebner, R.L.; Copelovitch, L.; Kaplan, B.S. Long-term outcomes of Shiga toxin hemolytic uremic syndrome. Pediatr. Nephrol. 2013, 28, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Melton-Celsa, A.R.; O’Brien, A.D. New Therapeutic Developments against Shiga Toxin-Producing Escherichia coli. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Kavaliauskiene, S.; Dyvelingelem, A.B.; Skotland, T.; Sandvig, K. Protection against shiga toxins. Toxins (Basel) 2017, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Kitov, P.I.; Sadowska, J.M.; Mulvey, G.; Armstrong, G.D.; Ling, H.; Pannu, N.S.; Read, R.J.; Bundle, D.R. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature 2000, 403, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Head, S.C.; Karmali, M.A.; Lingwood, C.A. Preparation of VT1 and VT2 hybrid toxins from their purified dissociated subunits: Evidence for B subunit modulation of a subunit function. J. Biol. Chem. 1991, 266, 3617–3621. [Google Scholar] [PubMed]
- Lingwood, C.A. Role of verotoxin receptors in pathogenesis. Trends Microbiol. 1996, 4, 147–153. [Google Scholar] [CrossRef]
- Marcato, P.; Vander Helm, K.; Mulvey, G.L.; Armstrong, G.D. Serum Amyloid P Component Binding to Shiga Toxin 2 Requires Both A Subunit and B Pentamer. Society 2003, 71, 6075–6078. [Google Scholar] [CrossRef]
- Donohue-Rolfe, A.; Jacewicz, M.; Keusch, G.T. Isolation and characterization of functional Shiga toxin subunits and renatured holotoxin. Mol. Microbiol. 1989, 3, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.M.; Mukherjee, J.; Leysath, C.E.; Debatis, M.; Ofori, K.; Baldwin, K.; Boucher, C.; Peters, R.; Beamer, G.; Sheoran, A.; et al. A single VHH-based toxin-neutralizing agent and an effector antibody protect mice against challenge with Shiga toxins 1 and 2. Infect. Immun. 2013, 81, 4592–4603. [Google Scholar] [CrossRef] [PubMed]
- Mejías, M.P.; Hiriart, Y.; Lauché, C.; Fernández-Brando, R.J.; Pardo, R.; Bruballa, A.; Ramos, M.V.; Goldbaum, F.A.; Palermo, M.S.; Zylberman, V. Development of camelid single chain antibodies against Shiga toxin type 2 (Stx2) with therapeutic potential against Hemolytic Uremic Syndrome (HUS). Sci. Rep. 2016, 6, 24913. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.W.H.; Moonens, K.; De Kerpel, M.; Brys, L.; Pardon, E.; Remaut, H.; De Greve, H. The Molecular Mechanism of Shiga Toxin Stx2e Neutralization by a Single-domain Antibody Targeting the Cell Receptor-binding Domain. J. Biol. Chem. 2014, 289, 25374–25381. [Google Scholar] [CrossRef] [PubMed]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally-occurring antibodies devoid of light-chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Vincke, C.; Gutiérrez, C.; Wernery, U.; Devoogdt, N.; Hassanzadeh-Ghassabeh, G.; Muyldermans, S. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol. Biol. 2012, 907, 145–176. [Google Scholar] [PubMed]
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S.; Atarhouch, T.; Saldanha, J.; Barbosa, J.A.R.G.; Hamers, R. Sequence and structure of V H domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. Des. Sel. 1994, 7, 1129–1135. [Google Scholar] [CrossRef]
- Conrath, K.E.; Lauwereys, M.; Wyns, L.; Muyldermans, S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J. Biol. Chem. 2001, 276, 7346–7350. [Google Scholar] [CrossRef] [PubMed]
- Hauswaldt, S.; Nitschke, M.; Sayk, F.; Solbach, W.; Knobloch, J.K.M. Lessons learned from outbreaks of shiga toxin producing Escherichia coli. Curr. Infect. Dis. Rep. 2013, 15, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Steeland, S.; Vandenbroucke, R.E.; Libert, C. Nanobodies as therapeutics: Big opportunities for small antibodies. Drug Discov. Today 2016, 21, 1076–1113. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh-Ghassabeh, G.; Devoogdt, N.; De Pauw, P.; Vincke, C.; Muyldermans, S. Nanobodies and their potential applications. Nanomedicine 2013, 8, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Hmila, I.; Saerens, D.; Ben Abderrazek, R.; Vincke, C.; Abidi, N.; Benlasfar, Z.; Govaert, J.; El Ayeb, M.; Bouhaouala-Zahar, B.; Muyldermans, S. A bispecific nanobody to provide full protection against lethal scorpion envenoming. FASEB J. 2010, 24, 3479–3489. [Google Scholar] [CrossRef] [PubMed]
- Yardehnavi, N.; Behdani, M.; Pooshang Bagheri, K.; Mahmoodzadeh, A.; Khanahmad, H.; Shahbazzadeh, D.; Habibi-Anbouhi, M.; Ghassabeh, G.H.; Muyldermans, S. A camelid antibody candidate for development of a therapeutic agent against Hemiscorpius lepturus envenomation. FASEB J. 2014, 28, 4004–4014. [Google Scholar] [CrossRef] [PubMed]
- Rossotti, M.A.; González-Techera, A.; Guarnaschelli, J.; Yim, L.; Camacho, X.; Fernández, M.; Cabral, P.; Leizagoyen, C.; Chabalgoity, J.A.; González-Sapienza, G. Increasing the potency of neutralizing single- domain antibodies by functionalization with a CD11b/CD18 binding domain. MAbs 2015, 7, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Mejias, M.P.; Ghersi, G.; Craig, P.O.; Panek, C.A.; Bentancor, L.V.; Baschkier, A.; Goldbaum, F.A.; Zylberman, V.; Palermo, M.S. Immunization with a Chimera Consisting of the B Subunit of Shiga Toxin Type 2 and Brucella Lumazine Synthase Confers Total Protection against Shiga Toxins in Mice. J. Immunol. 2013, 191, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G. Nanobodies as novel agents for disease diagnosis and therapy. Int. J. Nanomed. 2013, 8, 4215–4227. [Google Scholar] [CrossRef] [PubMed]
- Hoefman, S.; Ottevaere, I.; Baumeister, J.; Sargentini-Maier, M. Pre-Clinical Intravenous Serum Pharmacokinetics of Albumin Binding and Non-Half-Life Extended Nanobodies®. Antibodies 2015, 4, 141–156. [Google Scholar] [CrossRef]
- Conrath, K.E.; Lauwereys, M.; Galleni, M.; Matagne, A.; Frère, J.-M.; Kinne, J.; Wyns, L.; Muyldermans, S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob. Agents Chemother. 2001, 45, 2807–2812. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Saerens, D.; Frederix, F.; Reekmans, G.; Conrath, K.; Jans, K.; Brys, L.; Huang, L.; Bosmans, E.; Maes, G.; Borghs, G.; et al. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal. Chem. 2005, 77, 7547–7555. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, S.M.; Couty, S.; Terskikh, A.; Marguerat, L.; Crivelli, V.; Pugnieres, M.; Mani, J.C.; Leisinger, H.J.; Mach, J.P.; Deperthes, D. Streptabody, a high avidity molecule made by tetramerization of in vivo biotinylated, phage display-selected scFv fragments on streptavidin. Mol. Immunol. 2000, 37, 1067–1077. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Dahms, S.O.; Creemers, J.W.M.; Schaub, Y.; Bourenkov, G.P.; Zögg, T.; Brandstetter, H.; Than, M.E. The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers. Sci. Rep. 2016, 6, 34303. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Joosten, R.P.; Long, F.; Murshudov, G.N.; Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 2014, 1, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Smart, O.S.; Womack, T.O.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Vonrhein, C.; Bricogne, G. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Painter, J.; Merritt, E.A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 2006, 39, 109–111. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Duc, T.; Peeters, E.; Muyldermans, S.; Charlier, D.; Hassanzadeh-Ghassabeh, G. Nanobody(R)-based chromatin immunoprecipitation/micro-array analysis for genome-wide identification of transcription factor DNA binding sites. Nucleic Acids Res. 2013, 41, e59. [Google Scholar] [CrossRef] [PubMed]
- Oanh, T.K.N.; Nguyen, V.K.; De Greve, H.; Goddeeris, B.M. Protection of piglets against edema disease by maternal immunization with Stx2e toxoid. Infect. Immun. 2012, 80, 469–473. [Google Scholar] [CrossRef] [PubMed]

| kon (105 M−1s−1) | koff (s−1) | KD (nM) | Χ2 | Rmax Expected (RU) | Rmax Observed (RU) | n 3 | |
|---|---|---|---|---|---|---|---|
| Nb29 | 8.7 | 0.2998 | 344.4 | 3.33 | 173 | 357 | 2.06 |
| Nb31 | 11.38 | 0.0812 | 71.33 | 3.58 | 163 | 310 | 1.90 |
| Nb41 | 1.26 | 0.0049 | 39.01 | 51.5 | 182 | 805 | 4.42 |
| Nb140 | 1.32 | 0.0071 | 53.65 | 38.6 | 181 | 852 | 4.70 |
| Nb113 | 1.95 | 0.0019 | 9.6 | 27.0 | 166 | 748 | 4.52 |
| Nb1132 1 | 48.27 | 0.0008 | 0.17 | 18.4 | 302 | 813 | 2.69 |
| Nb1132-SA1 2 | 34.15 | 0.0009 | 0.25 | 50.6 | 466 | 1114 | 2.39 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernedo-Navarro, R.A.; Romão, E.; Yano, T.; Pinto, J.; De Greve, H.; Sterckx, Y.G.-J.; Muyldermans, S. Structural Basis for the Specific Neutralization of Stx2a with a Camelid Single Domain Antibody Fragment. Toxins 2018, 10, 108. https://doi.org/10.3390/toxins10030108
Bernedo-Navarro RA, Romão E, Yano T, Pinto J, De Greve H, Sterckx YG-J, Muyldermans S. Structural Basis for the Specific Neutralization of Stx2a with a Camelid Single Domain Antibody Fragment. Toxins. 2018; 10(3):108. https://doi.org/10.3390/toxins10030108
Chicago/Turabian StyleBernedo-Navarro, Robert Alvin, Ema Romão, Tomomasa Yano, Joar Pinto, Henri De Greve, Yann G.-J. Sterckx, and Serge Muyldermans. 2018. "Structural Basis for the Specific Neutralization of Stx2a with a Camelid Single Domain Antibody Fragment" Toxins 10, no. 3: 108. https://doi.org/10.3390/toxins10030108
APA StyleBernedo-Navarro, R. A., Romão, E., Yano, T., Pinto, J., De Greve, H., Sterckx, Y. G.-J., & Muyldermans, S. (2018). Structural Basis for the Specific Neutralization of Stx2a with a Camelid Single Domain Antibody Fragment. Toxins, 10(3), 108. https://doi.org/10.3390/toxins10030108






