The Presence of Toxic and Non-Toxic Cyanobacteria in the Sediments of the Limpopo River Basin: Implications for Human Health
Abstract
:1. Introduction
2. Results
2.1. The Physical-Chemical Characteristics of the River Sediments
2.2. The Presence of Cyanobacteria in the River Sediments
2.3. PCR Analysis of the 16S rRNA Gene
2.4. Detection of Genes Involved in Toxin Production
2.5. Phylogenetic Relationship
3. Discussion
4. Conclusions
5. Future Research Work
6. Materials and Methods
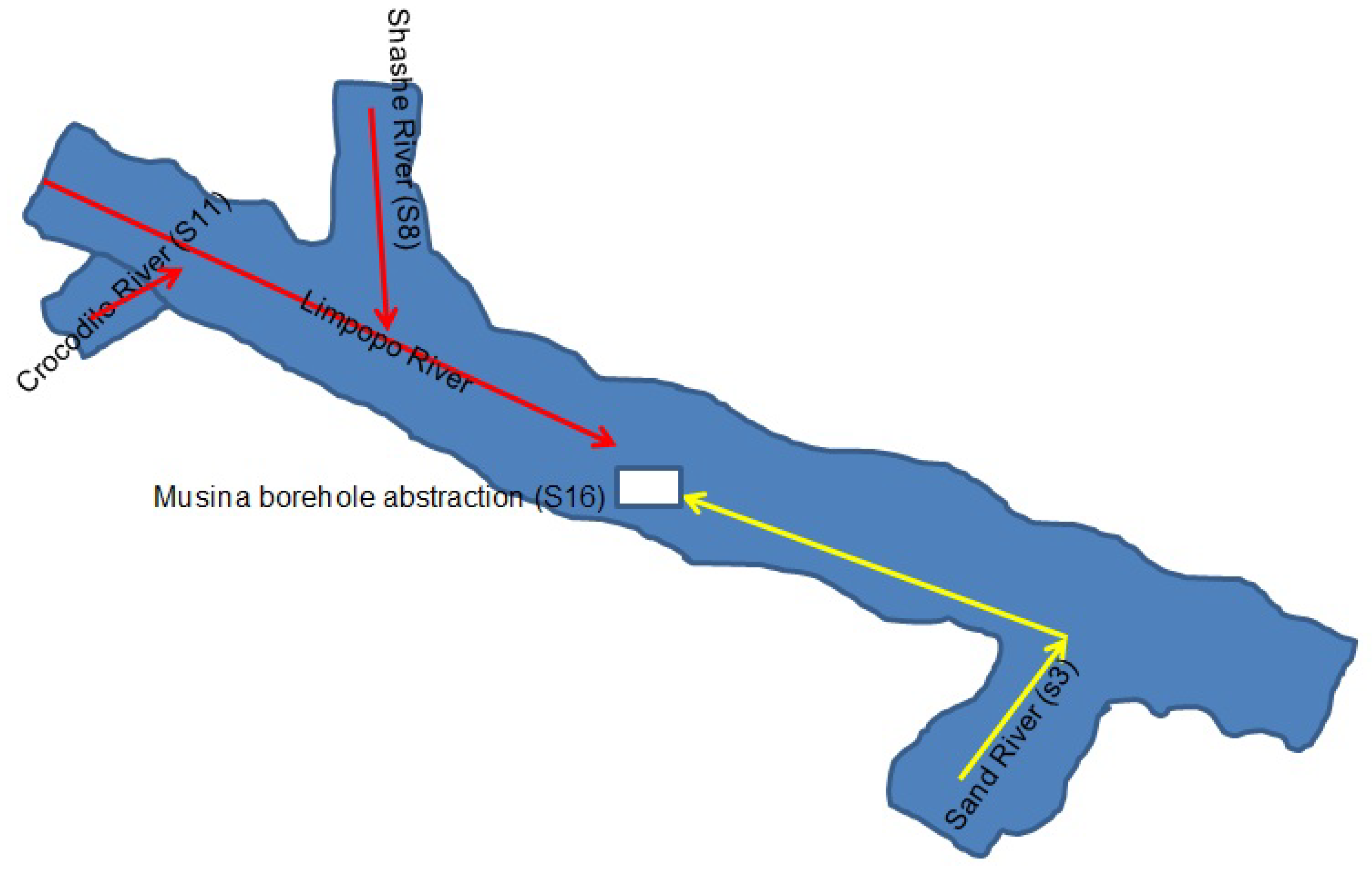
6.1. The Study Area
6.2. Sampling Sites and Sampling Methods
6.3. Physical-Chemical Measurements
6.4. Nutrient Analysis
6.4.1. Total Phosphorus Analysis
6.4.2. Total Nitrogen Analysis
6.5. Data Analysis
6.6. The Culture of Cyanobacteria Species in River Sediments
6.7. The Identification of Cyanobacterial Species Using the FlowCam
6.8. The Identification of Cyanobacterial Species Using Molecular Characterization
6.8.1. DNA Extraction and Purification
6.8.2. Detection and Amplification of 16S rRNA by Polymerase Chain Reaction
6.8.3. Toxin Gene Detection
Electrophoresis
6.8.4. PCR Purification and Sequencing
Primers
6.8.5. Phylogenetic Relationship
6.8.6. Divergence Matrix
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Christiansen, G.; Molitor, C.; Philmus, B.; Kurmayer, R. Nontoxic strains of cyanobacteria are the result of major gene deletion events induced by a transposable element. Mol. Biol. Evol. 2008, 25, 1695–1704. [Google Scholar] [CrossRef] [PubMed]
- Janse, I.; Kardinaal, W.E.A.; Meima, M.; Fastner, J.; Visser, P.M.; Zwart, G. Toxic and nontoxic Microcystis colonies in natural populations can be differentiated on the basis of rRNA gene internal transcribed spacer diversity. Appl. Environ. Microbiol. 2004, 70, 3979–3987. [Google Scholar] [CrossRef] [PubMed]
- Frazao, B.; Martins, R.; Vasconcelos, V. Are Known Cyanotoxins Involved in the Toxicity of Picoplanktonic and Filamentous North Atlantic Marine Cyanobacteria? Mar. Drugs 2010, 8, 1908–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, L.A.; Dittmann, E.; Mazmouz, R.; Ongley, S.E.; D’Agostino, P.M.; Neilan, B.A. The genetics, biosynthesis and regulation of toxic specialized metabolites of cyanobacteria. Harmful Algae 2016, 54, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Pearson, L.A.; Davis, T.W.; Muenchhoff, J.; Pratama, R.; Jex, A.; Neilan, B.A. Comparative genomics of Cylindrospermopsis raciborskii strains with differential toxicities. BMC Genom. 2014, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Gumbo, R.J.; Ross, G.; Cloete, E.T. Biological control of Microcystis dominated harmful algal blooms. Afr. J. Biotechnol. 2008, 7, 4765–4773. [Google Scholar]
- Paerl, H.W.; Fulton, R.S.; Moisander, P.H.; Dyble, J. Harmful freshwater algal blooms with an emphasis on cyanobacteria. Sci. World 2001, 1, 76–113. [Google Scholar] [CrossRef] [PubMed]
- Boyer, G.L. Toxic Cyanobacteria in the Great Lakes: More than just the Western Basin of Lake Erie. GLRC Great Lakes Res. Rev. 2006, 7, 2–7. [Google Scholar]
- O’Neil, J.M.; Davis, T.W.; Burford, M.A.; Gobler, C.J. The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harmful Algae 2012, 14, 313–334. [Google Scholar] [CrossRef]
- Vitousek, P.M.J.; Aber, R.W.; Howarth, G.E.; Likens, P.A.; Matson, D.W.; Schindler, W.H.; Tilman, G.D. Human alteration of the global nitrogen cycle: Causes and consequences. Issues Ecol. 1997, 1, 1–17. [Google Scholar]
- Scheffer, M. The story of some shallow lakes. In Ecology of Shallow Lakes; Springer: Dordrecht, The Netherlands, 2004; pp. 1–19. [Google Scholar]
- Reynolds, C.S. Ecology of Phytoplankton; Cambridge University Press: Cambridge, UK, 2006; 550p. [Google Scholar]
- Smith, V.H. Low nitrogen to phosphorus ratios favor dominance by blue-green algae in lake Phytoplankton. Science 1983, 221, 669–671. [Google Scholar] [CrossRef] [PubMed]
- Hyenstrand, P. Factors Influencing the Success of Pelagic Cyanobacteria; Uppsala University: Uppsala, Sweden, 1999. [Google Scholar]
- Berman, T. The role of DON and the effect of N: P ratios on occurrence of cyanobacterial blooms: Implications from the outgrowth of Aphanizomenon in Lake Kinneret. Limnol. Oceanogr. 2001, 46, 443–447. [Google Scholar] [CrossRef]
- Downing, J.A.; Watson, S.B.; McCauley, E. Predicting cyanobacteria dominance in lakes. Can. J. Fish. Aquat. Sci. 2001, 58, 1905–1908. [Google Scholar] [CrossRef]
- Von Ruckert, G.; Giani, A. Effect of nitrate and ammonium on the growth and protein concentration of Microcystis viridis Lemmermann (Cyanobacteria). Rev. Brasiliera Bot. 2004, 27, 325–331. [Google Scholar]
- McCarthy, M.J.; Gardner, W.S.; Lavrentyev, P.J.; Moats, K.M.; Joehem, F.J.; Klarer, D.M. Effects of hydrological flow regime on sediment-water interface and water column nitrogen dynamics in a great lakes coastal wetland (Old Woman Creek, Lake Erie). J. Great Lakes Res. 2007, 33, 219–231. [Google Scholar] [CrossRef]
- Andersen, K.; Shanmugam, K. Energetics of biological nitrogen fixation: Determination of the ratio of formation of H2 to NH4+ catalyzed by nitrogenase of Klebsiella pneumoniae in vivo. J. Gen. Microbiol. 1977, 103, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Visser, P.M. Growth and Vertical Movement of the Cyanobacterium Microcystis in Stable and Artificially Mixed Water Columns. Ph.D. Thesis, University of Amsterdam, Amsterdam, The Netherland, 1995. [Google Scholar]
- Thiel, T.; Pratte, B. Effect on heterocyst differentiation of nitrogen fixation in vegetative cells of the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 2001, 183, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.; Pace, M.L.; Howarth, R.W.; Marino, R.M. Bloom formation in heterocystic nitrogen-fixing cyanobacteria: The dependence on colony size and zooplankton grazing. Limnol. Oceanogr. 2004, 49, 2171–2178. [Google Scholar] [CrossRef] [Green Version]
- Mbukwa, E.; Msagati, T.A.; Mamba, B.B.; Boussiba, S.; Wepener, V.; Leu, S.; Kaye, Y. Toxic Microcystis novacekii T20-3 from Phakalane Ponds, Botswana: PCR Amplifications of Microcystin Synthetase (mcy) Genes, Extraction and LC-ESI-MS Identification of Microcystins. J. Environ. Anal. Toxicol. 2015. [Google Scholar] [CrossRef]
- Mbukwa, E.A.; Boussiba, S.; Wepener, V.; Leu, S.; Kaye, Y.; Msagati, T.A.; Mamba, B.B. PCR amplification and DNA sequence of mcyA gene: The distribution profile of a toxigenic Microcystis aeruginosa in the Hartbeespoort Dam, South Africa. J. Water Health 2013, 11, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Pedro, O.; Rundberget, T.; Lie, E.; Correia, D.; Skaare, J.U.; Berdal, K.G.; Neves, L.; Sandvik, M. Occurrence of microcystins in freshwater bodies in Southern Mozambique. J. Res. Environ. Sci. Toxicol. 2012, 1, 58–65. [Google Scholar]
- Vézie, C.; Rapala, J.; Vaitomaa, J.; Seitsonen, J.; Sivonen, K. Effect of nitrogen and phosphorus on growth of toxic and nontoxic Microcystis strains and on intracellular microcystin concentrations. Microb. Ecol. 2002, 43, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Beversdorf, L.J.; Miller, T.R.; McMahon, K.D. The role of nitrogen fixation in cyanobacterial bloom toxicity in a temperate, eutrophic lake. PLoS ONE 2013, 8, e56103. [Google Scholar] [CrossRef] [PubMed]
- Celeste, C.M.M.; Lorena, R.; Oswaldo, A.J.; Sandro, G.; Daniela, S.; Dario, A.; Leda, G. Mathematical modeling of Microcystis aeruginosa growth and [D-Leu1] microcystin-LR production in culture media at different temperatures. Harmful Algae 2017, 67, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Van der Westhuizen, A.J.; Eloff, J.N. Effect of temperature and light on the toxicity and growth of the blue-green alga Microcystis aeruginosa (UV-006). Planta 1985, 163, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Geada, P.; Pereira, R.N.; Vasconcelos, V.; Vicente, A.A.; Fernandes, B.D. Assessment of synergistic interactions between environmental factors on Microcystis aeruginosa growth and microcystin production. Algal Res. 2017, 27, 235–243. [Google Scholar] [CrossRef]
- Ndlela, L.L.; Oberholster, P.J.; Van Wyk, J.H.; Cheng, P.H. An overview of cyanobacterial bloom occurrences and research in Africa over the last decade. Harmful Algae 2016, 60, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Staden, R.; Judge, D.P.; Bonfield, J.K. Analysing sequences using the Staden package and EMBOSS. In Introduction to Bioinformatics. A Theoretical and Practical Approach; Human Press Inc.: Totawa, NJ, USA, 2003. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1990, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Oberholster, P.J.; Botha, A.M.; Myburgh, J.G. Linking climate change and progressive eutrophication to incidents of clustered animal mortalities in different geographical regions of South Africa. Afr. J. Biotechnol. 2009, 8, 5825–5832. [Google Scholar]
- Peperzak, L. Climate change and harmful algal bllom in the North Sea. Acta Oecol. 2003, 24, 139–144. [Google Scholar] [CrossRef]
- Rantala, A.; Rajaniemi-Wacklin, P.; Lyra, C.; Lepisto, L.; Rintala, J.; Mankiewiez-Boczek, J.; Sivonen, K. Detection of microcystin-producing cyanobacteria in Finnish Lakes with genus-specific microcystin synthetase Gene E (mcyE) PCR and associations with environmental factors. Appl. Environ. Microbiol. 2006, 72, 6101–6110. [Google Scholar] [CrossRef] [PubMed]
- Jahnichen, S.; Petzoldt, T.; Benndorf, J. Evidence for control of microcystin dynamics in BautzenReservoir (Germany) by cyanobacterial population growth rates and dissolved inorganic carbon. Arch. Hydrobiol. 2001, 150, 177–196. [Google Scholar] [CrossRef]
- Bartram, J.; Chorus, I.; Carmichael, W.W.; Jones, G.; Skulberg, O.M. Toxic Cyanobacteria in Water. A Guide to Their Public Health Consequences, Monitoring and Management; Chorus, I., Bartram, J., Eds.; World Health Organization: Geneva, Switzerland, 1999; pp. 1–14. [Google Scholar]
- Xu, H.; Paerl, H.W.; Qin, B.; Zhu, G.; Gaoa, G. Nitrogen and phosphorus inputs control phytoplankton growth in eutrophic Lake Taihu, China. Limnol. Oceanogr. 2010, 55, 420–432. [Google Scholar] [CrossRef]
- Edokpayi, J.N.; Odiyo, J.O.; Popoola, E.O.; Msagati, T.A. Evaluation of Microbiological and Physicochemical Parameters of Alternative Source of Drinking Water: A Case Study of Nzhelele River, South Africa. Open Microbiol. J. 2018, 12, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Burne, C. Macro-Nutrient and Hydrological Trends in Some Streams of the Waterberg, Limpopo: Investigating the Effects of Land-Use Change on Catchment Water Quality. Master’s Dissertation, University of Witwatersrand, Johannesburg, South Africa, 2016. [Google Scholar]
- Correll, D.L. Phosphorus: A rate limiting nutrient in surface waters. Poultry Sci. 1999, 78, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Oberholster, P.J.; Dabrowski, J.; Botha, A.M. Using modified multiple phosphorus sensitivity indices for mitigation and management of phosphorus loads on a catchment level. Fundam. Appl. Limnol. Arch. Hydrobiol. 2013, 182, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mosimanegape, K. Integration of Physicochemical Assessment of Water Quality with Remote Sensing Techniques for the Dikgathong Damin Botswana. Master’s Dissertation, University of Zimbabwe, Harare, Zimbabwe, 2016. [Google Scholar]
- Matthews, M.W. Eutrophication and cyanobacterial blooms in South African inland waters: 10 years of MERIS observations. Remote Sens. Environ. 2014, 155, 161–177. [Google Scholar] [CrossRef]
- Swanepoel, A.; Du Preez, H.H.; Cloete, N. The occurrence and removal of algae (including cyanobacteria) and their related organic compounds from source water in Vaalkop Dam with conventional and advanced drinking water treatment processes. Water SA 2017, 43, 67–80. [Google Scholar] [CrossRef]
- Seanego, K.G.; Moyo, N.A.G. The effect of sewage effluent on the physico-chemical and biological characteristics of the Sand River, Limpopo, South Africa. Phys. Chem. Earth Parts A/B/C 2013, 66, 75–82. [Google Scholar] [CrossRef]
- Musa, R.; Greenfield, R. Nutrient loads on an important watercourse. Pre-and Post-Acid spill. In Proceedings of the 7th International Toxicology Symposium, Johannesburg, South Africa, 31 August 2015; p. 89. [Google Scholar]
- Basima, L.B. An Assessment of Plankton Diversity as a Water Quality Indicator in Small Man-m Ade Reservoirs in the Mzingwane Catchment, Limpopo Basin, Zimbabwe. Master’s Dissertation, University of Zimbabwe, Harare, Zimbabwe, 2005. [Google Scholar]
- Mupfiga, E.T.; Munkwakwata, R.; Mudereri, B.; Nyatondo, U.N. Assessment of sedimentation in Tuli Makwe Dam using remotely sensed data. J. Soil Sci. Environ. Manag. 2016, 7, 230–238. [Google Scholar]
- Mavhunga, M. The Presence of Cyanobacteria & Diatoms in Limpopo River Sediment Profile: Implications for Human Health. Unpublished Hons mini Thesis, University of Venda, Johannesburg, South Africa, 2015. [Google Scholar]
- Oberholster, P.J.; Jappie, S.; Cheng, P.H.; Botha, A.M.; Matthews, M.W. First report of an Anabaena Bory strain containing microcystin-LR in a freshwater body in Africa. Afr. J. Aquat. Sci. 2015, 40, 21–36. [Google Scholar] [CrossRef]
- Kim, Y.J.; Baek, J.S.; Youn, S.J.; Kim, H.N.; Lee, B.C.; Kim, G.; Park, S.; You, K.A.; Lee, J.K. Cyanobacteria Community and Growth Potential Test in Sediment of Lake Paldang. J. Korean Soc. Water Environ. 2016, 32, 261–270. [Google Scholar] [CrossRef]
- Dzebu, W. Statement in Relation to Water Supply Challenges in Musina, 14 March 2017. Available online: www.musina.gov.za/index.php/public-notices?download=1028...in...to...musina (accessed on 26 March 2014).
- Botha, A.M.; Oberholster, P.J. PCR-Based Markers for Detection and Identification of Toxic Cyanobacteria; WRC Report No. K5/1502/01/07; Water Research Commission: Pretoria, South Africa, 2007; p. 70. [Google Scholar]
- Su, Y.; You, X.; Lin, H.; Zhuang, H.; Weng, Y.; Zhang, D. Recruitment of cyanobacteria from the sediments in the eutrophic Shanzi Reservoir. Environ. Technol. 2016, 37, 641–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldener, I.; Summers, M.L.; Sukenik, A. Cellular differentiation in filamentous cyanobacteria. In The Cell Biology of Cyanobacteria; Flores, E., Herrero, A., Eds.; Caister Academic Press: Norwich, UK, 2014; pp. 263–291. [Google Scholar]
- Perez, R.; Forchhammer, K.; Salerno, G.; Maldener, I. Clear differences in metabolic and morphological adaptations of akinetes of two Nostocales living in different habitats. Microbiology 2016, 162, 214–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, D.G.; Duggan, P.S. Heterocyst and akinete differentiation in cyanobacteria. New Phytol. 1999, 144, 3–33. [Google Scholar] [CrossRef]
- Scherer, P.I.; Raeder, U.; Geist, J.; Zwirglmaier, K. Influence of temperature, mixing, and addition of microcystin-LR on microcystin gene expression in Microcystis aeruginosa. Microbiol. Open 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Mikalsen, B.; Boison, G.; Skulberg, O.M.; Fastner, J.; Davies, W.; Gabrielsen, T.M.; Rudi, K.; Jakobsen, K.S. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 2003, 185, 2774–2785. [Google Scholar] [CrossRef] [PubMed]
- Davis, T.W.; Berry, D.L.; Boyer, G.L.; Gobler, C.J. The effects of temperature and nutrients on the growth and dynamics of toxic and non-toxic strains of Microcystis during cyanobacteria blooms. Harmful Algae 2009, 8, 715–725. [Google Scholar] [CrossRef]
- Yamamoto, Y. Effect of temperature on recruitment of cyanobacteria from the sediment and bloom formation in a shallow pond. Plankton Benthos Res. 2009, 4, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Oberholster, P.J.; Botha, A.-M.; Cloete, T.E. Use of molecular markers as indicators for winter zooplankton grazing on toxic benthic cyanobacteria colonies in an urban Colorado lake. Harmful Algae 2006, 5, 705–716. [Google Scholar] [CrossRef] [Green Version]
- Mosase, E.; Ahiablame, L. Rainfall and Temperature in the Limpopo River Basin, Southern Africa: Means, Variations, and Trends from 1979 to 2013. Water 2018, 10, 364. [Google Scholar] [CrossRef]
- Hoeger, S.J.; Dietrick, D.R.; Hitzfeld, B.C. Effect of ozonation on the removal of cyanobacteria toxins during drinking water treatment. Environ. Health Perspect. 2002, 110, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Brittain, S.M.; Wang, J.; Babcock-Jackson, L.; Carmichael, W.W.; Rinehart, K.L.; Culver, D.A. Isolation and characterization of microcystins, cyclic heptapeptide hepatotoxins from Lake Erie strain of Microcystis aeruginosa. J. Great Lakes Res. 2000, 26, 241–249. [Google Scholar] [CrossRef]
- Andersson, J.A.; de Garine-Wichatitsky, M.; Cumming, D.H.; Dzingirai, V.; Giller, K.E. People at wildlife frontiers in Southern Africa. In Transfrontier Conservation Area: People Living on the Edge; Routledge: Abingdon-on-Thames, UK, 2013; pp. 1–11. [Google Scholar]
- Oberholster, P.J.; Myburgh, J.G.; Govender, D.; Bengis, R.; Botha, A.M. Identification of toxigenic Microcystis strains after incidents of wild animal mortalities in the Kruger National Park, South Africa. Ecotoxicol. Environ. Saf. 2009, 72, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- McCollough, B. Toxic Algae and Other Marine Biota: Detection, Mitigation, Prevention and Effects on the Food Industry. Master’s Dissertation, Kansas State University, Manhattan, KS, USA, 2016. [Google Scholar]
- Zhu, T.; Ringler, C. Climate change impacts on water availability and use in the Limpopo River Basin. Water 2012, 4, 63–84. [Google Scholar] [CrossRef]
- Department of Water Affairs. Joint Water Quality Baseline Report for Limpopo Basin between Botswana and South Africa; Department of Water Affairs: Pretoria, South Africa, 2011. [Google Scholar]
- Islam, M.S.; Ahmed, M.K.; Raknuzzaman, M.; Habibullah-Al-Mamun, M.; Islam, M.K. Heavy metal pollution in surface water and sediment: A preliminary assessment of an urban river in a developing country. Ecol. Indic. 2015, 48, 282–291. [Google Scholar] [CrossRef]
- APHA; AWWA; WPCF. Standard Methods for Examination of Water and Wastewater, 20th ed.; American Publishing Health Association: Washington, DC, USA, 1998. [Google Scholar]
- Gumbo, J.R.; Ross, G.; Cloete, T.E. The Isolation and identification of Predatory Bacteria from a Microcystis algal Bloom. Afr. J. Biotechnol. 2010, 9, 663–671. [Google Scholar]
- Van Vuuren, S.J.; Taylor, J.; Gerber, A. A Guide for the Identification of Microscopic Algae in South Africa Freshwaters; Department of Water Affairs and Forestry, North-West University: Potchefstroom, South Africa, 2006. [Google Scholar]
- Moffitt, M.C.; Neilan, B.A. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl. Environ. Microbiol. 2004, 70, 6353–6362. [Google Scholar] [CrossRef] [PubMed]
- Pearson, L.A.; Neilan, B.A. The molecular genetics of cyanobacterial toxicity as a basis for monitoring water quality and public health risk. Curr. Opin. Biotechnol. 2008, 19, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, A.D.; Neilan, B.A. Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria. Arch. Microbiol. 2006, 185, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Fergusson, K.M.; Saint, C.P. Multiplex PCR assay for Cylindrospermopsis raciborskii and cylindrospermopsin-producing cyanobacteria. Environ. Toxicol. 2003, 18, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Hisbergues, M.; Christiansen, G.; Rouhiainen, L.; Sivonen, K.; Borner, T. PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch. Microbiol. 2003, 180, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Schembri, M.A.; Neilan, B.A.; Saint, C.P. Identification of genes implicated in toxin production in the cyanobacterium Cylindrospermopsis raciborskii. Environ. Toxicol. 2001, 16, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Neilan, B.A.; Jacobs, D.; Del Dot, T.; Blackall, L.L.; Hawkins, P.R.; Cox, P.T.; Goodman, A.E. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Bacteriol. 1997, 47, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, A.D.; Hawes, I.; Mountfort, D.; Hitzfeld, B.; Dietrich, D.R.; Burns, B.P.; Neilan, B.A. Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol. 2005, 7, 519–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. “Gapped Blast and PSI-BLAST” A generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]










| Cyanobacteria Species/Sample Sites | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S15 | S16 | S17 | S18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aphanizomenon sp. | + * | |||||||||||||||||
| Raphidiopsis curvata | + * | |||||||||||||||||
| Microcystis aeruginosa | + | + | + | + | ++ * | + | + | + | ++ * | + | + | + | ||||||
| Microcystis panniformis | + * | |||||||||||||||||
| Synechocystis PCC 6803 | + * | + | + | |||||||||||||||
| Cylindrospermopsis sp. | + * | + * | ||||||||||||||||
| Lyngbya sp. | + | |||||||||||||||||
| Leptolyngbya sp. | + | + | ||||||||||||||||
| Leptolyngbya boryana | + | + | ||||||||||||||||
| Calothrix sp. | + | + | ++ * | + | + | |||||||||||||
| Oscillatoria sp. | + | + | ++ * | |||||||||||||||
| Phormidium sp. | + | + | + | + | ||||||||||||||
| Phormidium uncinatum | + | |||||||||||||||||
| Nostoc sp. | + * | + | + * | |||||||||||||||
| Anabaena circinalis | + | |||||||||||||||||
| Anabaena oscillarioides | + | |||||||||||||||||
| Chroococcus | + | |||||||||||||||||
| Anabaechopsis circularis | + | |||||||||||||||||
| Spirulina laxissima SAG 256.80 | + | |||||||||||||||||
| Planktothrix rubescens | + * | + * | + | |||||||||||||||
| Alkalinema pantanalense | + | |||||||||||||||||
| Gloeocapsa sp. | + * | + | ||||||||||||||||
| Arthrospira sp. str PCC8005 | + * |
| Samples | Similarity % | Species Similar to | Family | Accession No |
|---|---|---|---|---|
| S2 | 93 | Uncultured Leptolyngbya sp. Clone | Leptolyngbyaceae | KM108695.1 |
| S3 | 94 | Synechocystis PCC 6803 | Oscillatoriophycideae | CP012832.1 |
| S5 | 97 | Anabaena oscillarioides | Nostocaceae | AJ630428.1 |
| S7 | 99 | Synechocystis sp. PCC 6803 | Oscillatoriophycideae | CP012832.1 |
| S8 | 99 | Leptolyngbya boryana | Leptolyngbyaceae | AP014642.1 |
| S9 | 97 | Synechocystis PCC 6803 | Oscillatoriophycideae | CP012832.1 |
| S9 | 100 | Cylindrospermopsis raciborskii CHAB3438 | Oscillatoriophycideae | KJ139743.1 |
| S9 | 100 | Aphanizomenon sp. | Nostocaceae | GQ385961.1 |
| S9 | 100 | Raphidiopsis curvata | Nostocaceae | KJ139745.1 |
| S10 | 96 | Spirulina laxissima SAG 256.80 | Spirulinaceae | DQ393278.1 |
| S11 | 87 | Uncultured Cyanobacterium clone | - | AM159315.1 |
| S12 | 83 | Uncultured Cyanobacterium clone | - | HQ189039.1 |
| S13 | 90 | Uncultured Cyanobacterium clone | - | JX041703.1 |
| S14 | 98 | Leptolyngbya boryana | Leptolyngbyaceae | AP014642.1 |
| S16 | 83 | Leptolyngbya | Leptolyngbyaceae | KJ654311.1 |
| S18 | 96 | Alkalinema pantanalense | Pseudanabaenaceae | KF246497.2 |
| Primers | Sample No | Similarity % | Species Similar to | Accession No |
|---|---|---|---|---|
| PKS | S3 | 100 | Aphanizomenon sp. 10E6 | GQ385961.1 |
| S3 | 100 | Raphidiopsis curvata | KJ139745.1 | |
| S3 | 100 | Cylindrospermopsis raciborskii | AF160254.1 | |
| S3 | 100 | Arthrospira sp. str. PCC 8005 | FO818640.1 | |
| S3 | 100 | Nostoc sp. NIES-4103 | AP018288.1 | |
| S9 | 93 | Calothrix sp. 336/3 | CP011382.1 | |
| S9 | 89 | Oscillatoria nigro-viridis PCC 7112 | CP003614.1 | |
| S9 | 100 | Gloeocapsa sp. PCC 7428, | CP003646.1 | |
| S9 | 100 | Cylindrospermum sp. NIES-4074 | AP018269.1 | |
| HEP | S8 | 100 | Uncultured Microcystis sp. clone msp microcystin synthetase E (mcyE) gene, partial cds | KF687998 |
| S8 | 100 | Microcystis panniformis FACHB-1757 | CP011339.1 | |
| S8 | 100 | Microcystis aeruginosa PCC 7806 | AF183408.1 | |
| S8 | 100 | Nostoc sp. 152 | KC699835.1 | |
| S8 | 100 | Planktothrix rubescens NIVA-CYA 98 | AM990462.1 | |
| S13 | 100 | Nostoc sp. 152 | KC699835.1 | |
| S13 | 100 | Planktothrix rubescens NIVA-CYA 98 | AM990462.1 | |
| S13 | 100 | Uncultured Microcystis sp. from Uganda | FJ429839.2 | |
| S13 | 100 | Microcystis aeruginosa PCC 7806SL | CP020771.1 | |
| S13 | 100 | Uncultured Microcystis sp. clone mw microcystin synthetase E (mcyE) gene, partial cds | KF687997.1 |
| S2 | S3 | S5 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S16 | S18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Notwane River (S2) | ‒ | ||||||||||||
| Sand River (S3) | 0.216 | ‒ | |||||||||||
| Nzhelele River (S5) | 0.191 | 0.187 | ‒ | ||||||||||
| Mokolo River (S7) | 0.167 | 0.064 | 0.130 | ‒ | |||||||||
| Crocodile River (S8) | 0.166 | 0.160 | 0.149 | 0.119 | ‒ | ||||||||
| Nzhelele River (S9) | 0.184 | 0.095 | 0.152 | 0.028 | 0.140 | ‒ | |||||||
| Sand River (S10) | 0.155 | 0.216 | 0.153 | 0.156 | 0.169 | 0.169 | ‒ | ||||||
| Crocodile River (S11) | 0.257 | 0.295 | 0.280 | 0.244 | 0.278 | 0.254 | 0.236 | ‒ | |||||
| Nzhelele River (S12) | 0.391 | 0.394 | 0.365 | 0.351 | 0.350 | 0.361 | 0.364 | 0.492 | ‒ | ||||
| Mzingwane River (S13) | 0.190 | 0.180 | 0.184 | 0.130 | 0.134 | 0.139 | 0.168 | 0.267 | 0.377 | ‒ | |||
| Shashe River (S14) | 0.173 | 0.163 | 0.156 | 0.119 | 0.006 | 0.140 | 0.173 | 0.278 | 0.355 | 0.134 | ‒ | ||
| Musina borehole (S16) | 0.376 | 0.359 | 0.312 | 0.314 | 0.342 | 0.321 | 0.343 | 0.414 | 0.555 | 0.371 | 0.347 | ‒ | |
| Musina borehole (S18) | 0.183 | 0.184 | 0.179 | 0.136 | 0.128 | 0.150 | 0.186 | 0.285 | 0.366 | 0.173 | 0.131 | 0.348 | ‒ |
| River Names | Samples Numbers |
|---|---|
| Limpopo River (Groblers’ bridge) | S1 |
| Notwane River (Odi Bridge-Matabeleng) | S2 |
| Sand River upstream | S3 |
| Mogalakwena River next to Tolwe | S4 |
| Mawoni River downstream Makhado oxidation ponds | S5 |
| Lephalale river | S6 |
| Mokolo River | S7 |
| Crocodile River downstream Hartbeespoort dam | S8 |
| Nzhelele River downstream near Tshipise | S9 |
| Sand River downstream (at bridge on N1 road towards Musina) | S10 |
| Crocodile River downstream (near bridge on road D1235) near Thabazimbi | S11 |
| Nzhelele River upstream near Mphephu resort (downstream of Siloam oxidation ponds) | S12 |
| Mzingwane River (Zimbabwe) | S13 |
| Shashe River (near Irrigation scheme, Zimbabwe) | S14 |
| Limpopo River next to Thuli coal mine | S15 |
| Limpopo River abstraction point @ 0.0 m | S16 |
| Limpopo River abstraction point @ 1.0 m | S17 |
| Limpopo River abstraction point @ 1.68 m | S18 |
| Primers | Target Genes | Sequence (5′-3′) | A | B | Size (bp) | Amplified Gene | Ref. |
|---|---|---|---|---|---|---|---|
| 27F 809R | - | AGAGTTTGATCCTGGCTCAG GCTTCGGCACGGCTCGGGTCGATA | 52 64 | 60 | 780 | 16S rRNA | [85,86] |
| mcyA-Cd F mcyA-Cd R | mcyA | AAAATTAAAAGCCGTATCAAA AAAAGTGTTTTATTAGCGGCTCAT | 51 43 | 59 | 297 | Microcystin synthetase | [83] |
| HEPF HEPR | mcyE/ndaF | TTTGGGGTTAACTTTTTTGGGCATAGTC AATTCTTGAGGCTGTAAATCGGGTTT | 57 55 | 52 | 472 | Microcystin/nodularin synthetase | [81] |
| PKS M4 PKS M5 | cyr | GAAGCTCTGGAATCCGGTAA AATCCTTACGGGATCCGGTGC | 52 56 | 55 | 650 | Cylindrospermopsin polypeptide synthase | [84] |
| M13 M14 | ps | GGCAAATTGTGATAGCCACGAGC GATGGAACATCGCTCACTGGTG | 57 57 | 55 | 597 | Cylindrospermopsin peptide synthetase | [84] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magonono, M.; Oberholster, P.J.; Shonhai, A.; Makumire, S.; Gumbo, J.R. The Presence of Toxic and Non-Toxic Cyanobacteria in the Sediments of the Limpopo River Basin: Implications for Human Health. Toxins 2018, 10, 269. https://doi.org/10.3390/toxins10070269
Magonono M, Oberholster PJ, Shonhai A, Makumire S, Gumbo JR. The Presence of Toxic and Non-Toxic Cyanobacteria in the Sediments of the Limpopo River Basin: Implications for Human Health. Toxins. 2018; 10(7):269. https://doi.org/10.3390/toxins10070269
Chicago/Turabian StyleMagonono, Murendeni, Paul Johan Oberholster, Addmore Shonhai, Stanley Makumire, and Jabulani Ray Gumbo. 2018. "The Presence of Toxic and Non-Toxic Cyanobacteria in the Sediments of the Limpopo River Basin: Implications for Human Health" Toxins 10, no. 7: 269. https://doi.org/10.3390/toxins10070269
APA StyleMagonono, M., Oberholster, P. J., Shonhai, A., Makumire, S., & Gumbo, J. R. (2018). The Presence of Toxic and Non-Toxic Cyanobacteria in the Sediments of the Limpopo River Basin: Implications for Human Health. Toxins, 10(7), 269. https://doi.org/10.3390/toxins10070269







