Design and Production of a Recombinant Hybrid Toxin to Raise Protective Antibodies against Loxosceles Spider Venom
Abstract
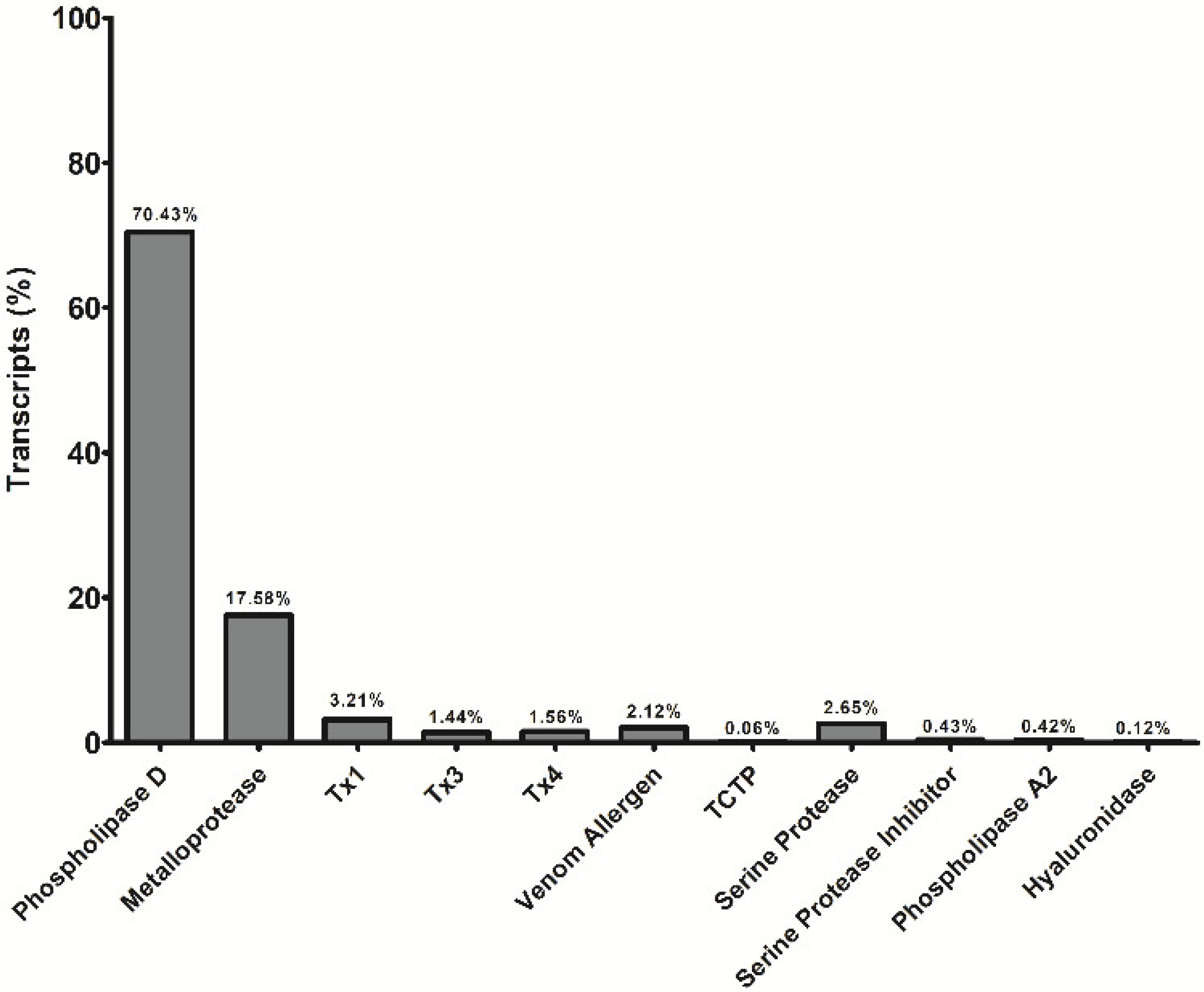
:1. Introduction
2. Results
2.1. Construction of the Hybrid Molecule LgRec1ALP1
2.2. Expression and Purification of the Hybrid Immunogen LgRec1ALP1
2.3. Immunogenicity and Cross-Reactivity of Anti-LgRec1ALP1
2.4. Neutralization Assays
2.4.1. Neutralization of Fibrinogen Degradation Caused by Loxosceles spp. Venoms
2.4.2. Neutralization of Platelets Aggregation Caused by Loxosceles spp. Venoms
2.4.3. Neutralization of Dermonecrosis and Edema Caused by Loxosceles spp. Venoms
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Ethics Committees
5.2. Animals and Venoms
5.3. Sequences and Analysis of Sequenced Transcripts
5.4. Construction of the Hybrid Immunogen
5.5. Recombinant LgRec1ALP1 Expression
5.6. LgRec1ALP1 Purification
5.7. SDS-Polyacrylamide Gel Electrophoresis
5.8. Quantification of Recombinant Proteins and Venoms
5.9. Production of Anti-LgRec1ALP1 in Mice
5.10. Immunoenzymatic Assay (ELISA)
5.11. Western Blot Analysis
5.12. Neutralization of Fibrinogen Degradation
5.13. Neutralization of Platelet Aggregation
5.14. Neutralization of Dermonecrotic and Edema Activities by the Anti-LgRec1ALP1
5.15. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hogan, C.J.; Barbaro, K.C.; Winkel, K. Loxoscelism: Old obstacles, new directions. Ann. Emerg. Med. 2004, 44, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Swanson, D.L.; Vetter, R.S. Loxoscelism. Clin. Dermatol. 2006, 24, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Gremski, L.H.; Trevisan-Silva, D.; Ferrer, V.P.; Matsubara, F.H.; Meissner, G.O.; Wille, A.C.M.; Vuitika, L.; Dias-Lopes, C.; Ullah, A.; de Moraes, F.R.; et al. Recent advances in the understanding of brown spider venoms: From the biology of spiders to the molecular mechanisms of toxins. Toxicon 2014, 83, 91–120. [Google Scholar] [CrossRef] [PubMed]
- Málaque, C.M.; Castro-Valencia, J.E.; Cardoso, J.L.; Francca, F.O.; Barbaro, K.C.; Fan, H.W. Clinical and epidemiological features of definitive and presumed loxoscelism in São Paulo, Brazil. Rev. Inst. Med. Trop. Sao Paulo 2002, 44, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Pauli, I.; Puka, J.; Gubert, I.C.; Minozzo, J.C. The efficacy of antivenom in loxoscelism treatment. Toxicon 2006, 48, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Chaim, O.M.; Trevisan-Silva, D.; Chaves-Moreira, D.; Wille, A.C.M.; Ferrer, V.P.; Matsubara, F.H.; Mangili, O.C.; da Silveira, R.B.; Gremski, L.H.; Gremski, W.; et al. Brown Spider (Loxosceles genus) Venom Toxins: Tools for Biological Purposes. Toxins 2011, 3, 309–344. [Google Scholar] [CrossRef] [PubMed]
- Malaque, C.M.S.; Santoro, M.L.; Cardoso, J.L.C.; Conde, M.R.; Novaes, C.T.G.; Risk, J.Y.; Franca, F.O.S.; de Medeiros, C.R.; Fan, H.W. Clinical picture and laboratorial evaluation in human loxoscelism. Toxicon 2011, 58, 664–671. [Google Scholar] [CrossRef]
- Da Silva, P.H.; da Silveira, R.B.; Appel, M.H.; Mangili, O.C.; Gremski, W.; Veiga, S.S. Brown spiders and loxoscelism. Toxicon 2004, 44, 693–709. [Google Scholar] [CrossRef]
- Da Silveira, R.B.; Dos Santos Filho, J.F.; Mangili, O.C.; Veiga, S.S.; Gremski, W.; Nader, H.B.; Von Dietrich, C.P. Identification of proteases in the extract of venom glands from brown spiders. Toxicon 2002, 40, 815–822. [Google Scholar] [CrossRef]
- Appel, M.H.; da Silveira, R.B.; Gremski, W.; Veiga, S.S. Insights into brown spider and loxoscelism. Invertebr. Surviv. J. 2005, 2, 152–158. [Google Scholar]
- Barbaro, K.C.; Knysak, I.; Martins, R.; Hogan, C.; Winkel, K. Enzymatic characterization, antigenic cross-reactivity and neutralization of dermonecrotic activity of five Loxosceles spider venoms of medical importance in the Americas. Toxicon 2005, 45, 489–499. [Google Scholar] [CrossRef]
- Barbaro, K.C.; Sousa, M.V.; Morhy, L.; Eickstedt, V.R.D.; Mota, I. Compared chemical properties of dermonecrotic and lethal toxins from spiders of the genus Loxosceles (araneae). J. Protein Chem. 1996, 15, 337–343. [Google Scholar] [CrossRef]
- Tambourgi, D.V.; Magnoli, F.C.; van den Berg, C.W.; Morgan, B.P.; de Araujo, P.S.; Alves, E.W.; Da Silva, W.D. Sphingomyelinases in the venom of the spider Loxosceles intermedia are responsible for both dermonecrosis and complement-dependent hemolysis. Biochem. Biophys. Res. Commun. 1998, 251, 366–373. [Google Scholar] [CrossRef]
- Chaim, O.M.; Sade, Y.B.; da Silveira, R.B.; Toma, L.; Kalapothakis, E.; Chavez-Olortegui, C.; Mangili, O.C.; Gremski, W.; von Dietrich, C.P.; Nader, H.B.; et al. Brown spider dermonecrotic toxin directly induces nephrotoxicity. Toxicol. Appl. Pharmacol. 2006, 211, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Gremski, L.H.; da Silveira, R.B.; Chaim, O.M.; Probst, C.M.; Ferrer, V.P.; Nowatzki, J.; Weinschutz, H.C.; Madeira, H.M.; Gremski, W.; Nader, H.B.; et al. A novel expression profile of the Loxosceles intermedia spider venomous gland revealed by transcriptome analysis. Mol. Biosyst. 2010, 6, 2403–2416. [Google Scholar] [CrossRef]
- Matsubara, F.H.; Gremski, L.H.; Meissner, G.O.; Constantino Lopes, E.S.; Gremski, W.; Senff-Ribeiro, A.; Chaim, O.M.; Veiga, S.S. A novel ICK peptide from the Loxosceles intermedia (brown spider) venom gland: Cloning, heterologous expression and immunological cross- reactivity approaches. Toxicon 2013, 71, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Manzoni-de-Almeida, D.; Squaiella-BaptistãO, C.C.; Lopes, P.H.; van Den Berg, C.W.; Tambourgi, D.V. Loxosceles venom Sphingomyelinase D activates human blood leukocytes: Role of the complement system. Mol. Immunol. 2018, 94, 45–53. [Google Scholar] [CrossRef]
- Barbaro, K.C.; Cardoso, J.L.C.; Eickstedt, V.R.D.; Mota, I. Dermonecrotic and lethal components of loxosceles-gaucho spider venom. Toxicon 1992, 30, 331–338. [Google Scholar] [CrossRef]
- Futrell, J.M. Loxoscelism. Am. J. Med. Sci. 1992, 304, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, G.S.; Caporrino, M.C.; Della-Casa, M.S.; Kimura, L.F.; Prezotto-Neto, J.P.; Fukuda, D.A.; Portes, J.A.; Neves-Ferreira, A.G.C.; Santoro, M.L.; Barbaro, K.C. Cloning, expression and characterization of a phospholipase D from Loxosceles gaucho venom gland. Biochimie 2013, 95, 1773–1783. [Google Scholar] [CrossRef]
- Shimokawa-Falcao, L.; Caporrino, M.C.; Barbaro, K.C.; Della-Casa, M.S.; Magalhaes, G.S. Toxin Fused with SUMO Tag: A New Expression Vector Strategy to Obtain Recombinant Venom Toxins with Easy Tag Removal inside the Bacteria. Toxins 2017, 9, 82. [Google Scholar] [CrossRef]
- Kurpiewski, G.; Forrester, L.J.; Barrett, J.T.; Campbell, B.J. platelet-aggregation and sphingomyelinase d activity of a purified toxin from the venom of loxosceles-reclusa. Biochim. Biophys. Acta 1981, 678, 467–476. [Google Scholar] [CrossRef]
- Fukuda, D.A.; Caporrino, M.C.; Barbaro, K.C.; Della-Casa, M.S.; Faquim-Mauro, E.L.; Magalhaes, G.S. Recombinant phospholipase D from Loxosceles gaucho binds to platelets and promotes phosphatidylserine exposure. Toxins 2017, 9, 191. [Google Scholar] [CrossRef]
- Forrester, L.J.; Barrett, J.T.; Campbell, B.J. Red blood cell lysis induced by the venom of the brown recluse spider: The role of sphingomyelinase D. Arch. Biochem. Biophys. 1978, 187, 355–365. [Google Scholar] [CrossRef]
- Luciano, M.N.; Da Silva, P.H.; Chaim, O.M.; Dos Santos, V.L.P.; Franco, C.R.C.; Soares, M.F.S.; Zanata, S.M.; Mangili, O.C.; Gremski, W.; Veiga, S.S. Experimental Evidence for a Direct Cytotoxicity of Loxosceles intermedia (Brown Spider) Venom in Renal Tissue. J. Histochem. Cytochem. 2004, 52, 455–467. [Google Scholar] [CrossRef]
- Kusma, J.; Chaim, O.M.; Wille, A.C.M.; Ferrer, V.P.; Sade, Y.B.; Donatti, L.; Gremski, W.; Mangili, O.C.; Veiga, S.S. Nephrotoxicity caused by brown spider venom phospholipase-D (dermonecrotic toxin) depends on catalytic activity. Biochimie 2008, 90, 1722–1736. [Google Scholar] [CrossRef]
- Saude, M.D. Manual de Diagnóstico e Tratamento de Acidentes por Animais Peçonhentos. Available online: http://bvsms.saude.gov.br/bvs/publicacoes/funasa/manu_peconhentos.pdf (accessed on 26 November 2018).
- Guilherme, P.C.; Fernandes, I.; Barbaro, K.C. Neutralization of dermonecrotic and lethal activities and differences among 32–35 kDa toxins of medically important Loxosceles spider venoms in Brazil revealed by monoclonal antibodies. Toxicon 2001, 39, 1333–1342. [Google Scholar] [CrossRef]
- Pauli, I.; Minozzo, J.C.; Henrique Da Silva, P.; Chaim, O.M.; Veiga, S.S. Analysis of therapeutic benefits of antivenin at different time intervals after experimental envenomation in rabbits by venom of the brown spider (Loxosceles intermedia). Toxicon 2009, 53, 660–671. [Google Scholar] [CrossRef]
- Dias-Lopes, C.; Guimarães, G.; Felicori, L.; Fernandes, P.; Emery, L.; Kalapothakis, E.; Nguyen, C.; Molina, F.; Granier, C.; Chávez-Olórtegui, C. A protective immune response against lethal, dermonecrotic and hemorrhagic effects of Loxosceles intermedia venom elicited by a 27-residue peptide. Toxicon 2010, 55, 481–487. [Google Scholar] [CrossRef]
- Figueiredo, L.F.M.; Dias-Lopes, C.; Alvarenga, L.M.; Mendes, T.M.; Machado-de-Ávila, R.A.; McCormack, J.; Minozzo, J.C.; Kalapothakis, E.; Chávez-Olórtegui, C. Innovative immunization protocols using chimeric recombinant protein for the production of polyspecific loxoscelic antivenom in horses. Toxicon 2014, 86, 59–67. [Google Scholar] [CrossRef]
- Karim-Silva, S.; Moura, J.D.; Noiray, M.; Minozzo, J.C.; Aubrey, N.; Alvarenga, L.M.; Billiald, P. Generation of recombinant antibody fragments with toxin-neutralizing potential in loxoscelism. Immunol. Lett. 2016, 176, 90–96. [Google Scholar] [CrossRef]
- Tambourgi, D.V.; Pedrosa, M.D.F.; van den Berg, C.W.; Goncalves-de-Andrade, R.M.; Ferracini, M.; Paixao-Cavalcante, D.; Morgan, B.P.; Rushmere, N.K. Molecular cloning, expression, function and immunoreactivities of members of a gene family of sphingomyelinases from Loxosceles venom glands. Mol. Immunol. 2004, 41, 831–840. [Google Scholar] [CrossRef]
- de Almeida, D.M.; Fernandes-Pedrosa, M.D.; de Andrade, R.M.G.; Marcelino, J.R.; Gondo-Higashi, H.; de Azevedo, I.; Ho, P.L.; van den Berg, C.; Tambourgi, D.V. A new anti-loxoscelic serum produced against recombinant sphingomyelinase D: Results of preclinical trials. Am. J. Trop. Med. Hyg. 2008, 79, 463–470. [Google Scholar] [CrossRef]
- Guimarães, G.; Dias-Lopes, C.; Duarte, C.G.; Felicori, L.; Machado de Avila, R.A.; Figueiredo, L.F.M.; de Moura, J.; Faleiro, B.T.; Barro, J.; Flores, K.; et al. Biochemical and immunological characteristics of Peruvian Loxosceles laeta spider venom: Neutralization of its toxic effects by anti-loxoscelic antivenoms. Toxicon 2013, 70, 90–97. [Google Scholar] [CrossRef]
- Duarte, C.G.; Bonilla, C.; Guimarães, G.; Machado de Avila, R.A.; Mendes, T.M.; Silva, W.; Tintaya, B.; Yarleque, A.; Chávez-Olórtegui, C. Anti-loxoscelic horse serum produced against a recombinant dermonecrotic protein of Brazilian Loxosceles intermedia spider neutralize lethal effects of Loxosceles laeta venom from Peru. Toxicon 2015, 93, 37–40. [Google Scholar] [CrossRef]
- Felicori, L.; Fernandes, P.B.; Giusta, M.S.; Duarte, C.G.; Kalapothakis, E.; Nguyen, C.; Molina, F.; Granier, C.; Chávez-Olórtegui, C. An in vivo protective response against toxic effects of the dermonecrotic protein from Loxosceles intermedia spider venom elicited by synthetic epitopes. Vaccine 2009, 27, 4201–4208. [Google Scholar] [CrossRef]
- Souza, N.A.; Dias-Lopes, C.; Matoso, Í.H.G.; de Oliveira, C.F.B.; CháVez-Olortegui, C.D.; Minozzo, J.C.; Felicori, L.F. Immunoprotection elicited in rabbit by a chimeric protein containing B-cell epitopes of Sphingomyelinases D from Loxosceles spp. spiders. Vaccine 2018, 36, 7324–7330. [Google Scholar] [CrossRef]
- Fernandes-Pedrosa, M.d.F.; Junqueira-de-Azevedo, I.d.L.M.; Gonçalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D.V. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genom. 2008, 9, 279. [Google Scholar] [CrossRef]
- Williamson, A.L.; Lustigman, S.; Oksov, Y.; Deumic, V.; Plieskatt, J.; Mendez, S.; Zhan, B.; Bottazzi, M.E.; Hotez, P.J.; Loukas, A. Ancylostoma caninum MTP-1, an Astacin- Like Metalloprotease Secreted by Infective Hookworm Larvae, Is Involved in Tissue Migration. Infect. Immun. 2006, 74, 961. [Google Scholar] [CrossRef]
- Feitosa, L.; Gremski, W.; Veiga, S.S.; Elias, M.C.; Graner, E.; Mangili, O.C.; Brentani, R.R. Detection and characterization of metalloproteinases with gelatinolytic, fibronectinolytic and fibrinogenolytic activities in brown spider (Loxosceles intermedia) venom. Toxicon 1998, 36, 1039. [Google Scholar] [CrossRef]
- Veiga, S.S.; Zanetti, V.; Braz, A.; Mangili, O.C.; Gremski, W. Extracellular matrix molecules as targets for brown spider venom toxins. Braz. J. Med. Biol. Res. 2001, 34, 843–850. [Google Scholar] [CrossRef]
- Young, A.R.; Pincus, S.J. Comparison of enzymatic activity from three species of necrotising arachnids in Australia: Loxosceles rufescens, Badumna insignis and Lampona cylindrata. Toxicon 2001, 39, 391–400. [Google Scholar] [CrossRef]
- Zanetti, V.C.; da Silveira, R.B.; Dreyfuss, J.L.; Haoach, J.; Mangili, O.C.; Veiga, S.S.; Gremski, W. Morphological and biochemical evidence of blood vessel damage and fibrinogenolysis triggered by brown spider venom. Blood Coagul. Fibrinolysis 2002, 13, 135–148. [Google Scholar] [CrossRef]
- Da Silveira, R.B.; Wille, A.C.M.; Chaim, O.M.; Appel, M.H.; Silva, D.T.; Franco, C.R.C.; Toma, L.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; et al. Identification, cloning, expression and functional characterization of an astacin-like metalloprotease toxin from Loxosceles intermedia (brown spider) venom. Biochem. J. 2007, 406, 355–363. [Google Scholar] [CrossRef]
- Hopp, T.P.; Woods, K.R. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 1981, 78, 3824. [Google Scholar] [CrossRef]
- de Giuseppe, P.O.; Ullah, A.; Silva, D.T.; Gremski, L.H.; Wille, A.C.M.; Moreira, D.C.; Ribeiro, A.S.; Chaim, O.M.; Murakami, M.T.; Veiga, S.S.; et al. Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulphide bridge. Biochem. Biophys. Res. Commun. 2011, 409, 622–627. [Google Scholar] [CrossRef]
- Guevara, T.; Yiallouros, I.; Kappelhoff, R.; Bissdorf, S.; Stöcker, W.; Gomis-Rüth, F.X. Proenzyme structure and activation of astacin metallopeptidase. J. Biol. Chem. 2010, 285, 13958. [Google Scholar] [CrossRef]
- Nicholson, G.M.; Graudins, A. Antivenoms for the Treatment of Spider Envenomation. J. Toxicol. Toxin Rev. 2003, 22, 35–59. [Google Scholar] [CrossRef]
- Isbister, G.K.; Fan, H.W. Spider bite. Lancet 2011, 378, 2039–2047. [Google Scholar] [CrossRef]
- Hubiche, T.; Delaunay, P.; del Giudice, P. A case of loxoscelism in southern France. Am. J. Trop. Med. Hyg. 2013, 88, 807–808. [Google Scholar] [CrossRef]
- Coutinho, I.; Rocha, S.; Ferreira, M.E.; Vieira, R.; Cordeiro, M.R.; Reis, J.P. Cutaneous loxoscelism in Portugal: A rare cause of dermonecrosis. Acta Med. Port. 2014, 27, 654. [Google Scholar] [CrossRef]
- Morales-Moreno, H.J.; Carranza-Rodriguez, C.; Borrego, L. Cutaneous loxoscelism due to Loxosceles rufescens. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 1431–1432. [Google Scholar] [CrossRef]
- Olvera, A.; Ramos-Cerrillo, B.; Estévez, J.; Clement, H.; de Roodt, A.; Paniagua-Solís, J.; Vázquez, H.; Zavaleta, A.; Salas Arruz, M.; Stock, R.P.; et al. North and South American Loxosceles spiders: Development of a polyvalent antivenom with recombinant sphingomyelinases D as antigens. Toxicon 2006, 48, 64–74. [Google Scholar] [CrossRef]
- Fernandes Pedrosa, M.d.F.; Junqueira de Azevedo, I.d.L.M.; Gonçalves-de-Andrade, R.M.; van Den Berg, C.W.; Ramos, C.R.R.; Lee Ho, P.; Tambourgi, D.V. Molecular cloning and expression of a functional dermonecrotic and haemolytic factor from Loxosceles laeta venom. Biochem. Biophys. Res. Commun. 2002, 298, 638–645. [Google Scholar] [CrossRef]
- De Castro, C.S.; Silvestre, F.G.; Araujo, S.C.; Gabriel de, M.Y.; Mangili, O.C.; Cruz, I.; Chavez-Olortegui, C.; Kalapothakis, E. Identification and molecular cloning of insecticidal toxins from the venom of the brown spider Loxosceles intermedia. Toxicon. 2004, 44, 3–273. [Google Scholar] [CrossRef]
- Arlian, L.G. Arthropod allergens and human health. Ann. Rev. Entomol. 2002, 47, 395–433. [Google Scholar] [CrossRef]
- Sade, Y.B.; Boia-Ferreira, M.; Gremski, L.H.; da Silveira, R.B.; Gremski, W.; Senff-Ribeiro, A.; Chaim, O.M.; Veiga, S.S. Molecular cloning, heterologous expression and functional characterization of a novel translationally-controlled tumor protein (TCTP) family member from Loxosceles intermedia (brown spider) venom. Int. J. Biochem. Cell Biol. 2012, 44, 170–177. [Google Scholar] [CrossRef]
- Veiga, S.S.; Feitosa, L.; dos Santos, V.L.; de Souza, G.A.; Ribeiro, A.S.; Mangili, O.C.; Porcionatto, M.A.; Nader, H.B.; Dietrich, C.P.; Brentani, R.R.; et al. Effect of brown spider venom on basement membrane structures. Histochem. J. 2000, 32, 7–397. [Google Scholar] [CrossRef]
- Da Silveira, R.B.; Chaim, O.M.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Hyaluronidases in Loxosceles intermedia (Brown spider) venom are endo-β-N-acetyl-d-hexosaminidases hydrolases. Toxicon 2007, 49, 758–768. [Google Scholar] [CrossRef]
- Hiroshi, Y.; Masaya, M. Refolding Techniques for Recovering Biologically Active Recombinant Proteins from Inclusion Bodies. Biomolecules 2014, 4, 235–251. [Google Scholar] [CrossRef]
- Trevisan-Silva, D.; Gremski, L.H.; Chaim, O.M.; Da Silveira, R.B.; Meissner, G.O.; Mangili, O.C.; Barbaro, K.C.; Gremski, W.; Veiga, S.S.; Senff-Ribeiro, A. Astacin- like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 2010, 92, 21. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Herman, C.; Murmu, A.; Singh, A.; Panda, A.K. Kinetics of Inclusion Body Formation and Its Correlation with the Characteristics of Protein Aggregates in Escherichia coli. PLoS ONE 2012, 7, e33951. [Google Scholar] [CrossRef]
- Novo, J.; Oliveira, M.; Magalhães, G.; Morganti, L.; Raw, I.; Ho, P. Generation of Polyclonal Antibodies Against Recombinant Human Glucocerebrosidase Produced in Escherichia coli. Mol. Biotechnol. 2010, 46, 279–286. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, T.; Xu, K.; Lei, J.; Wang, L.; Li, Z.; Zhang, Z. A novel and convenient method to immunize animals: Inclusion bodies from recombinant bacteria as antigen to directly immunize animals. Afr. J. Biotechnol. 2011, 10, 8146–8150. [Google Scholar] [CrossRef]
- Lorch, M.S.; Collado, M.S.; Argüelles, M.H.; Rota, R.P.; Spinsanti, L.I.; Lozano, M.E.; Goñi, S.E. Production of recombinant NS1 protein and its possible use in encephalitic flavivirus differential diagnosis. Protein Expr. Purif. 2019, 153, 18–25. [Google Scholar] [CrossRef]
- Jiang, X.; Xia, S.; He, X.; Ma, H.; Feng, Y.; Liu, Z.; Wang, W.; Tian, M.; Chen, H.; Peng, F.; et al. Targeting peptide-enhanced antibody and CD11c(+)dendritic cells to inclusion bodies expressing protective antigen against ETEC in mice. FASEB J. 2019, 33, 2836–2847. [Google Scholar] [CrossRef]
- Kesik, M.; Saczyńska, V.; Szewczyk, B.; Płucienniczak, A. Inclusion bodies from recombinant bacteria as a novel system for delivery of vaccine antigen by the oral route. Immunol. Lett. 2004, 91, 197–204. [Google Scholar] [CrossRef]
- Wedrychowicz, H.; Kesik, M.; Kaliniak, M.; Kozak-Cieszczyk, M.; Jedlina-Panasiuk, L.; Jaros, S.; Plucienniczak, A. Vaccine potential of inclusion bodies containing cysteine proteinase of Fasciola hepatica in calves and lambs experimentally challenged with metacercariae of the fluke. Vet. Parasitol. 2007, 147, 77–88. [Google Scholar] [CrossRef]
- Such, D.R.; Souza, F.N.; Meissner, G.O.; Morgon, A.M.; Gremski, L.H.; Ferrer, V.P.; Trevisan-Silva, D.; Matsubara, F.H.; Boia-Ferreira, M.; Sade, Y.B.; et al. Brown spider (Loxosceles genus) venom toxins: Evaluation of biological conservation by immune cross-reactivity. Toxicon 2015, 108, 154–166. [Google Scholar] [CrossRef]
- Lima, S.d.A.; Guerra-Duarte, C.; Costal-Oliveira, F.; Mendes, T.M.; Figueiredo, L.F.M.; Oliveira, D.; Machado de Avila, R.A.; Ferrer, V.P.; Trevisan-Silva, D.; Veiga, S.S.; et al. Recombinant Protein Containing B-Cell Epitopes of Different Spider Toxins Generates Neutralizing Antibodies in Immunized Rabbits. Front. Immunol. 2018, 9, 653. [Google Scholar] [CrossRef]
- Tavares, F.L.; Peichoto, M.E.; Rangel, D.D.; Barbaro, K.C.; Cirillo, M.C.; Santoro, M.L.; Sano-Martins, I.S. Loxosceles gaucho spider venom and its sphingomyelinase fraction trigger the main functions of human and rabbit platelets. Hum. Exp. Toxicol. 2011, 30, 1567–1574. [Google Scholar] [CrossRef]
- Appel, M.H.; Da Silveira, R.B.; Chaim, O.M.; Paludo, K.S.; Silva, D.T.; Chaves, D.M.; Da Silva, P.H.; Mangili, O.C.; Senff-Ribeiro, A.; Gremski, W.; et al. Identification, cloning and functional characterization of a novel dermonecrotic toxin (phospholipase D) from brown spider (Loxosceles intermedia) venom. Biochim. Biophys. Acta 2008, 1780, 167–178. [Google Scholar] [CrossRef]
- Pezzi, M.; Giglio, A.M.; Scozzafava, A.; Filippelli, O.; Serafino, G.; Verre, M. Spider Bite: A Rare Case of Acute Necrotic Arachnidism with Rapid and Fatal Evolution. Case Rep. Emerg. Med. 2016, 2016. [Google Scholar] [CrossRef]
- Mariutti, R.B.; Chaves-Moreira, D.; Vuitika, L.; Caruso, Í.P.; Coronado, M.A.; Azevedo, V.A.; Murakami, M.T.; Veiga, S.S.; Arni, R.K. Bacterial and Arachnid Sphingomyelinases D: Comparison of Biophysical and Pathological Activities. J. Cell. Biochem. 2017, 118, 2053–2063. [Google Scholar] [CrossRef]
- Chaves-Moreira, D.; De Moraes, F.R.; Caruso, Í.P.; Chaim, O.M.; Senff-Ribeiro, A.; Ullah, A.; Da Silva, L.S.; Chahine, J.; Arni, R.K.; Veiga, S.S. Potential Implications for Designing Drugs Against the Brown Spider Venom Phospholipase-D. J. Cell. Biochem. 2017, 118, 726–738. [Google Scholar] [CrossRef]
- Mendes, T.M.; Oliveira, D.; Figueiredo, L.F.M.; Machado-de-Avila, R.A.; Duarte, C.G.; Dias-Lopes, C.; Guimaraes, G.; Felicori, L.; Minozzo, J.C.; Chavez-Olortegui, C. Generation and characterization of a recombinant chimeric protein (rCpLi) consisting of B-cell epitopes of a dermonecrotic protein from Loxosceles intermedia spider venom. Vaccine 2013, 31, 2749–2755. [Google Scholar] [CrossRef]
- Ribeiro, M.F.; Oliveira, F.L.; Monteiro-Machado, M.; Cardoso, P.F.; Guilarducci-Ferraz, V.V.C.; Melo, P.A.; Souza, C.M.V.; Calil-Elias, S. Pattern of inflammatory response to Loxosceles intermedia venom in distinct mouse strains: A key element to understand skin lesions and dermonecrosis by poisoning. Toxicon 2015, 96, 10–23. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- ImageJ, U.S. Available online: imagej.nih.gov/ij/ (downloaded on 15 December 2018)(ImageJ bundled with 64-bit Java 1.8.0_112 windows version).
- Theakston, R.D.G.; Reid, H.A. Enzyme-linked Immunosorbent Assay (ELISA) in assessing antivenom potency. Toxicon 1979, 17, 511–515. [Google Scholar] [CrossRef]
- Santoro, M.L.; Sousa-e-Silva, M.C.C.; Goncalves, L.R.C.; Almeida-Santos, S.M.; Cardoso, D.F.; Laporta-Ferreira, I.L.; Saiki, M.; Peres, C.A.; Sano-Martins, I.S. Comparison of the biological activities in venoms from three subspecies of the South American rattlesnake (Crotalus durissus terrificus, C. durissus cascavella and C. durissus collilineatus). Comp. Biochem. Physiol. C: Pharmacol. Toxicol. Endocrinol. 1999, 123, 293. [Google Scholar] [CrossRef]






| L. gaucho | L. intermedia | L. laeta |
|---|---|---|
| PLD LgRec1 | PLD | PLD |
| hydrophilic peptides | LiRecDT1 | Smase I |
| SNSIETDVSFDKQ | 78.6% * | 50.0% |
| KFNDFLKGLRKVTTPGDSK | 78.9% | 63.1% |
| KLITGFKETLKNEGHEELLEKVGTDFSGNDDISDVQKTYNKAG | 62.7% | 55.8% |
| LLRGLTRVKAAVANRDSGSG | 75.0% | 40.0% |
| DKRQSTRDTLDAN | 69.2% | 38.4% |
| PDITVEILNEAAYKKKFRIATYEDNPWET | 68.9% | 51.7% |
| L. gaucho | L. intermedia | L. laeta |
|---|---|---|
| Metalloprotease LgALP1 | Metalloprotease | Metalloprotease |
| hydrophilic peptides | LALP2 | LLAE0237C |
| ALFPGDIKKAMRHIEENTCIKFKSRKNEEGYVKIYKGKKES | 90.4% * | 48.7% |
| HEHTRPDRDLYITVHEDNIRPSSKRNYKKT | 90.3% | 46.6% |
| LTSARYKDSLTDLDIKKINTLYN | 86.9% | 47.8% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calabria, P.A.L.; Shimokawa-Falcão, L.H.A.L.; Colombini, M.; Moura-da-Silva, A.M.; Barbaro, K.C.; Faquim-Mauro, E.L.; Magalhaes, G.S. Design and Production of a Recombinant Hybrid Toxin to Raise Protective Antibodies against Loxosceles Spider Venom. Toxins 2019, 11, 108. https://doi.org/10.3390/toxins11020108
Calabria PAL, Shimokawa-Falcão LHAL, Colombini M, Moura-da-Silva AM, Barbaro KC, Faquim-Mauro EL, Magalhaes GS. Design and Production of a Recombinant Hybrid Toxin to Raise Protective Antibodies against Loxosceles Spider Venom. Toxins. 2019; 11(2):108. https://doi.org/10.3390/toxins11020108
Chicago/Turabian StyleCalabria, Paula A. L., Lhiri Hanna A. L. Shimokawa-Falcão, Monica Colombini, Ana M. Moura-da-Silva, Katia C. Barbaro, Eliana L. Faquim-Mauro, and Geraldo S. Magalhaes. 2019. "Design and Production of a Recombinant Hybrid Toxin to Raise Protective Antibodies against Loxosceles Spider Venom" Toxins 11, no. 2: 108. https://doi.org/10.3390/toxins11020108
APA StyleCalabria, P. A. L., Shimokawa-Falcão, L. H. A. L., Colombini, M., Moura-da-Silva, A. M., Barbaro, K. C., Faquim-Mauro, E. L., & Magalhaes, G. S. (2019). Design and Production of a Recombinant Hybrid Toxin to Raise Protective Antibodies against Loxosceles Spider Venom. Toxins, 11(2), 108. https://doi.org/10.3390/toxins11020108






