Label-Free Direct Detection of Saxitoxin Based on a Localized Surface Plasmon Resonance Aptasensor
Abstract
1. Introduction
2. Results
2.1. Development of a New STX Aptamer
2.1.1. GO-SELEX for STX Aptamer
2.1.2. Characterization of the STX Aptamer
2.2. Application to LSPR Aptasensor
2.2.1. Optimization of the Oligonucleotide Molar Ratio
2.2.2. Screening of Working Buffer Components
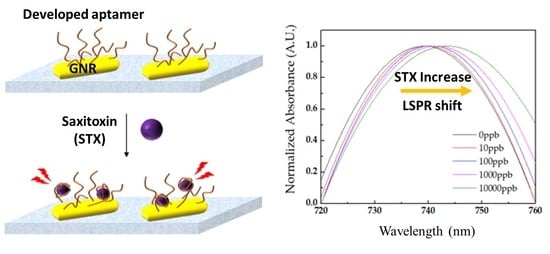
2.3. STX Detection Using the LSPR Aptasensor
2.3.1. Quantitative and Selectivity Analysis of STX
2.3.2. Selectivity Test
2.3.3. Spiking Test for Real Sample Analysis
3. Conclusions
4. Discussion
5. Materials and Methods
5.1. Materials and Reagents
5.2. In vitro Selection of the DNA Aptamer
5.2.1. Graphene Oxide-SELEX Process
5.2.2. Cloning and Sequencing of Selected DNA
5.3. Characterization of the Developed Aptamer
5.3.1. CD Spectrum Measurement
5.3.2. Fluorescence Assay
5.4. Dvelopment of LSPR Aptasensor Chip
5.4.1. Synthesis of GNRs
5.4.2. Fabrication of the GNR Substrate
5.4.3. Functionalization of GNR Substrate with Aptamers
5.5. Determination of the Sensitivity and Specificity of the LSPR Aptasensor
5.6. Preparation of STX-Spiked Real Samples
5.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Deeds, J.; Landsberg, J.; Etheridge, S.; Pitcher, G.; Longan, S. Non-traditional vectors for paralytic shellfish poisoning. Mar. Drugs 2008, 6, 308–348. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.; Jo, H.; Kim, S.H.; Kang, G.J. Exposure assessment to paralytic shellfish toxins through the shellfish consumption in Korea. Food Res. Int. 2018, 108, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, J.; Hoogenboom, R.L.; Hendriksen, P.J.; Bodero, M.; Bovee, T.F.; Rietjens, I.M.; Gerssen, A. Marine biotoxins and associated outbreaks following seafood consumption: Prevention and surveillance in the 21st century. Glob. Food Secur. 2017, 15, 11–21. [Google Scholar] [CrossRef]
- Cusick, K.D.; Sayler, G.S. An overview on the marine neurotoxin, saxitoxin: Genetics, molecular targets, methods of detection and ecological functions. Mar. Drugs 2013, 11, 991–1018. [Google Scholar] [CrossRef] [PubMed]
- Wiese, M.; D’agostino, P.M.; Mihali, T.K.; Moffitt, M.C.; Neilan, B.A. Neurotoxic alkaloids: Saxitoxin and its analogs. Mar. Drugs 2010, 8, 2185–2211. [Google Scholar] [CrossRef]
- Humpage, A.R.; Ledreux, A.; Fanok, S.; Bernard, C.; Briand, J.F.; Eaglesham, G.; Papageorgiou, J.; Nicholson, B.; Steffensen, D. Application of the neuroblastoma assay for paralytic shellfish poisons to neurotoxic freshwater cyanobacteria: Interlaboratory calibration and comparison with other methods of analysis. Environ. Toxicol. Chem. 2007, 26, 1512–1519. [Google Scholar] [CrossRef]
- Hanke, W.; Boheim, G.; Barhanin, J.; Pauron, D.; Lazdunski, M. Reconstitution of highly purified saxitoxin-sensitive Na+-channels into planar lipid bilayers. EMBO J. 1984, 3, 509–515. [Google Scholar] [CrossRef]
- Jin, X.; Chen, J.; Zeng, X.; Xu, L.; Wu, Y.; Fu, F. A signal-on magnetic electrochemical immunosensor for ultra-sensitive detection of saxitoxin using palladium-doped graphitic carbon nitride-based non-competitive strategy. Biosens. Bioelectron. 2019, 128, 45–51. [Google Scholar] [CrossRef]
- Wekell, J.C.; Hurst, J.; Lefebvre, K.A. The origin of the regulatory limits for PSP and ASP toxins in shellfish. J. Shellfish Res. 2004, 23, 927–930. [Google Scholar]
- Yang, X.; Zhou, L.; Tan, Y.; Shi, X.; Zhao, Z.; Nie, D.; Zhou, C.; Liu, H. Development and validation of a liquid chromatography-tandem mass spectrometry method coupled with dispersive solid-phase extraction for simultaneous quantification of eight paralytic shellfish poisoning toxins in shellfish. Toxins 2017, 9, 206. [Google Scholar] [CrossRef]
- Dell’Aversano, C.; Hess, P.; Quilliam, M.A. Hydrophilic interaction liquid chromatography–mass spectrometry for the analysis of paralytic shellfish poisoning (PSP) toxins. J. Chromatogr. A 2005, 1081, 190–201. [Google Scholar] [CrossRef]
- Rey, V.; Botana, A.M.; Alvarez, M.; Antelo, A.; Botana, L.M. Liquid chromatography with a fluorimetric detection method for analysis of paralytic shellfish toxins and tetrodotoxin based on a porous graphitic carbon column. Toxins 2016, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.F.; Niedzwiadek, B.; Menard, C. Quantitative determination of paralytic shellfish poisoning toxins in shellfish using prechromatographic oxidation and liquid chromatography with fluorescence detection: Collaborative study. J. AOAC Int. 2005, 88, 1714–1732. [Google Scholar] [PubMed]
- Bragg, W.A.; Lemire, S.W.; Coleman, R.M.; Hamelin, E.I.; Johnson, R.C. Detection of human exposure to saxitoxin and neosaxitoxin in urine by online-solid phase extraction-liquid chromatography–tandem mass spectrometry. Toxicon 2015, 99, 118–124. [Google Scholar] [CrossRef]
- Turrell, E.; Stobo, L.; Lacaze, J.P.; Piletsky, S.; Piletska, E. Optimization of hydrophilic interaction liquid chromatography/mass spectrometry and development of solid-phase extraction for the determination of paralytic shellfish poisoning toxins. J. AOAC Int. 2008, 91, 1372–1386. [Google Scholar] [PubMed]
- Yu, E.; Choi, S.J. Development of an improved stationary liquid-phase lab-on-a-chip for the field monitoring of paralytic shellfish toxins. BioChip J. 2017, 11, 30–38. [Google Scholar] [CrossRef]
- Haughey, S.A.; Campbell, K.; Yakes, B.J.; Prezioso, S.M.; DeGrasse, S.L.; Kawatsu, K.; Elliott, C.T. Comparison of biosensor platforms for surface plasmon resonance based detection of paralytic shellfish toxins. Talanta 2011, 85, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zheng, X.; Wu, J. A biolayer interferometry-based competitive biosensor for rapid and sensitive detection of saxitoxin. Sensors Actuators B: Chem. 2017, 246, 169–174. [Google Scholar] [CrossRef]
- Hou, L.; Jiang, L.; Song, Y.; Ding, Y.; Zhang, J.; Wu, X.; Tang, D. Amperometric aptasensor for saxitoxin using a gold electrode modified with carbon nanotubes on a self-assembled monolayer, and methylene blue as an electrochemical indicator probe. Microchim. Acta 2016, 183, 1971–1980. [Google Scholar] [CrossRef]
- Sun, A.; Chai, J.; Xiao, T.; Shi, X.; Li, X.; Zhao, Q.; Li, D.; Chen, J. Development of a selective fluorescence nanosensor based on molecularly imprinted-quantum dot optosensing materials for saxitoxin detection in shellfish samples. Sensors Actuators B: Chem. 2018, 258, 408–414. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, G.; Mao, X.; Yang, S.; De Ruyck, K.; Wu, Y. High sensitivity immunoassays for small molecule compounds detection–Novel noncompetitive immunoassay designs. TrAC Trends Anal. Chem. 2018, 103, 198–208. [Google Scholar] [CrossRef]
- Park, J.H.; Byun, J.Y.; Mun, H.; Shim, W.B.; Shin, Y.B.; Li, T.; Kim, M.G. A regeneratable, label-free, localized surface plasmon resonance (LSPR) aptasensor for the detection of ochratoxin A. Biosens. Bioelectron. 2014, 59, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Byun, J.Y.; Shim, W.B.; Kim, S.U.; Kim, M.G. High-sensitivity detection of ATP using a localized surface plasmon resonance (LSPR) sensor and split aptamers. Biosens. Bioelectron. 2015, 73, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Park, J.H.; Byun, J.Y.; Kim, J.H.; Kim, M.G. An optical fiber-based LSPR aptasensor for simple and rapid in-situ detection of ochratoxin A. Biosens. Bioelectron. 2018, 102, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Byun, J.Y.; Jang, H.; Hong, D.; Kim, M.G. A highly sensitive and widely adaptable plasmonic aptasensor using berberine for small-molecule detection. Biosens. Bioelectron. 2017, 97, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Willets, K.A.; Van Duyne, R.P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Lee, K.S.; Ahn, J.; Lee, J.J.; Kim, M.G.; Shin, Y.B. Highly sensitive biosensing using arrays of plasmonic Au nanodisks realized by nanoimprint lithography. ACS Nano 2011, 5, 897–904. [Google Scholar] [CrossRef]
- Miller, M.M.; Lazarides, A.A. Sensitivity of metal nanoparticle surface plasmon resonance to the dielectric environment. J. Phys. Chem. B 2005, 109, 21556–21565. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, K.; Micheletto, R.; Oyama, M.; Umar, A.A.; Kawai, S.; Kawakami, Y. An original planar multireflection system for sensing using the local surface plasmon resonance of gold nanospheres. J. Opt. A: Pure Appl. Opt. 2006, 8. [Google Scholar] [CrossRef]
- Gu, H.; Duan, N.; Xia, Y.; Hun, X.; Wang, H.; Wang, Z. Magnetic Separation-Based Multiple SELEX for Effectively Selecting Aptamers against Saxitoxin, Domoic Acid, and Tetrodotoxin. J. Agric. Food Chem. 2018, 66, 9801–9809. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Zheng, B.; Yao, D.; Kuai, S.; Tian, J.; Liang, H.; Ding, Y. Study of the binding way between saxitoxin and its aptamer and a fluorescent aptasensor for detection of saxitoxin. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2018, 204, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Tatavarty, R.; Kim, D.W.; Jung, H.T.; Gu, M.B. Immobilization-free screening of aptamers assisted by graphene oxide. Chem. Commun. 2012, 48, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Gao, S.; Hu, B.; Zheng, X.; Cao, Y.; Liu, D.; Sun, M.; Jiao, B.; Wang, L. Gonyautoxin 1/4 aptamers with high-affinity and high-specificity: From efficient selection to aptasensor application. Biosens. Bioelectron. 2016, 79, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Kejnovská, I.; Bednářová, K.; Renčiuk, D.; Kypr, J. Circular dichroism spectroscopy of DNA: From duplexes to quadruplexes. Chirality 2012, 24, 691–698. [Google Scholar] [CrossRef]
- Handy, S.M.; Yakes, B.J.; DeGrasse, J.A.; Campbell, K.; Elliott, C.T.; Kanyuck, K.M.; DeGrasse, S.L. First report of the use of a saxitoxin–protein conjugate to develop a DNA aptamer to a small molecule toxin. Toxicon 2013, 61, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Hu, B.; Gao, S.X.; Liu, D.J.; Sun, M.J.; Jiao, B.H.; Wang, L.H. A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 2015, 101, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Kankia, B. Monomolecular tetrahelix of polyguanine with a strictly defined folding pattern. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Kankia, B.I.; Marky, L.A. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: Stability, heat, and hydration. J. Am. Chem. Soc. 2001, 123, 10799–10804. [Google Scholar] [CrossRef]
- Cai, S.; Yan, J.; Xiong, H.; Liu, Y.; Peng, D.; Liu, Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 2018, 143, 5317–5338. [Google Scholar] [CrossRef] [PubMed]
- Valdiglesias, V.; Prego-Faraldo, M.; Pásaro, E.; Méndez, J.; Laffon, B. Okadaic acid: More than a diarrheic toxin. Mar. Drugs 2013, 11, 4328–4349. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.; Yin, Y.; Fu, W.; Qiu, B.; Lin, Z.; Yang, Y.; Zheng, L.; Li, J.; Chen, G. Determination of paralytic shellfish poisoning toxins by HILIC–MS/MS coupled with dispersive solid phase extraction. Food Chem. 2013, 137, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Wharton, R.E.; Feyereisen, M.C.; Gonzalez, A.L.; Abbott, N.L.; Hamelin, E.I.; Johnson, R.C. Quantification of saxitoxin in human blood by ELISA. Toxicon 2017, 133, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Nikoobakht, B.; El-Sayed, M.A. Preparation and growth mechanism of gold nanorods (NRs) using seed-mediated growth method. Chem. Mater. 2003, 15, 1957–1962. [Google Scholar] [CrossRef]
- Orendorff, C.J.; Murphy, C.J. Quantitation of metal content in the silver-assisted growth of gold nanorods. J. Phys. Chem. B 2006, 110, 3990–3994. [Google Scholar] [CrossRef] [PubMed]
- Barbera-Sánchez, L.; Soler, J.F.; Rojas de Astudillo, L.; Chang-Yen, I. Paralytic Shellfish Poisoning (PSP) in Margarita Island, Venezuela. Rev. Biol. Trop. 2004, 52, 89–98. [Google Scholar] [PubMed]





| Spiked STX Concentration (μg/L) | LSPR Shift by Real Sample (△nm) | LSPR Shift by Standard STX (△nm) | Found STX Concentration (μg/L) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| 10 | 0.63 | 0.67 | 9.40 | 98.43 | 1.40 |
| 100 | 1.59 | 1.37 | 116.05 | 116.05 | 2.39 |
| 2000 | 2.24 | 2.33 | 1922.74 | 96.13 | 3.44 |
| Method | LOD (μg/L) | Linear Range (μg/L) | Recovery (%) | Measurement Steps | Ref |
|---|---|---|---|---|---|
| HILIC-MS/MS | 1.7 (buffer) | 8.1–225.5 | 86.6–91.9 | 1) Automatic toxin sepration by instrument (20 min) 2) Measurement | [42] |
| Competitive biosensor | 0.5 (buffer) | 100–800 | 101.4–107.3 | 1) Baseline (2min) 2) Loading (5min) 3) Washing (2min) 4) Association (3min) 5) Dissociation (3min) | [18] |
| Fluorescence nanosensor | 0.3 (shellfish) | 20–100 | 89.4–102.4 | 1) Incubation (6min) 2) Measurement | [20] |
| Direct ELISA | - | 0.02–0.8 (human whole blood) | 109–110 | 1) Sample fixation 2) Antibody/enzyme conjugate (1h) 3) Washing 4) Substrate solution adding & incubation (1min) 5) Stop solution 6) Measurment | [43] |
| Fluorescence switch sensor | 1.8 (buffer) | 0–24 | 105.7–111.2 | 1) Incubation (30min) 2) Heating at 61°C (10min) 3) Measurement | [31] |
| Electrochemical aptasensor | 0.11 (buffer) | 0.27–9 | 63–121 | 1) Sample deposition (30min) 2) Washing 3) Measurement | [19] |
| LSPR aptasensor | 2.46 (buffer) | 5–10,000 | 96.13–116.05 | 1) Incubation (30min) 2) Measurement | This work |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, S.-J.; Park, J.-H.; Lee, B.; Kim, M.-G. Label-Free Direct Detection of Saxitoxin Based on a Localized Surface Plasmon Resonance Aptasensor. Toxins 2019, 11, 274. https://doi.org/10.3390/toxins11050274
Ha S-J, Park J-H, Lee B, Kim M-G. Label-Free Direct Detection of Saxitoxin Based on a Localized Surface Plasmon Resonance Aptasensor. Toxins. 2019; 11(5):274. https://doi.org/10.3390/toxins11050274
Chicago/Turabian StyleHa, Su-Ji, Jin-Ho Park, Bobin Lee, and Min-Gon Kim. 2019. "Label-Free Direct Detection of Saxitoxin Based on a Localized Surface Plasmon Resonance Aptasensor" Toxins 11, no. 5: 274. https://doi.org/10.3390/toxins11050274
APA StyleHa, S.-J., Park, J.-H., Lee, B., & Kim, M.-G. (2019). Label-Free Direct Detection of Saxitoxin Based on a Localized Surface Plasmon Resonance Aptasensor. Toxins, 11(5), 274. https://doi.org/10.3390/toxins11050274