Association of Nrf2, SOD2 and GPX1 Polymorphisms with Biomarkers of Oxidative Distress and Survival in End-Stage Renal Disease Patients
Abstract
:1. Introduction
2. Results
2.1. Positive Association Between the Presence of SOD2, GPX1 and Nrf2 Polymorphisms and the Risk of ESRD
2.2. Association of SOD2, GPX, and Nrf2 Polymorphisms with Biomarkers of Oxidative Damage
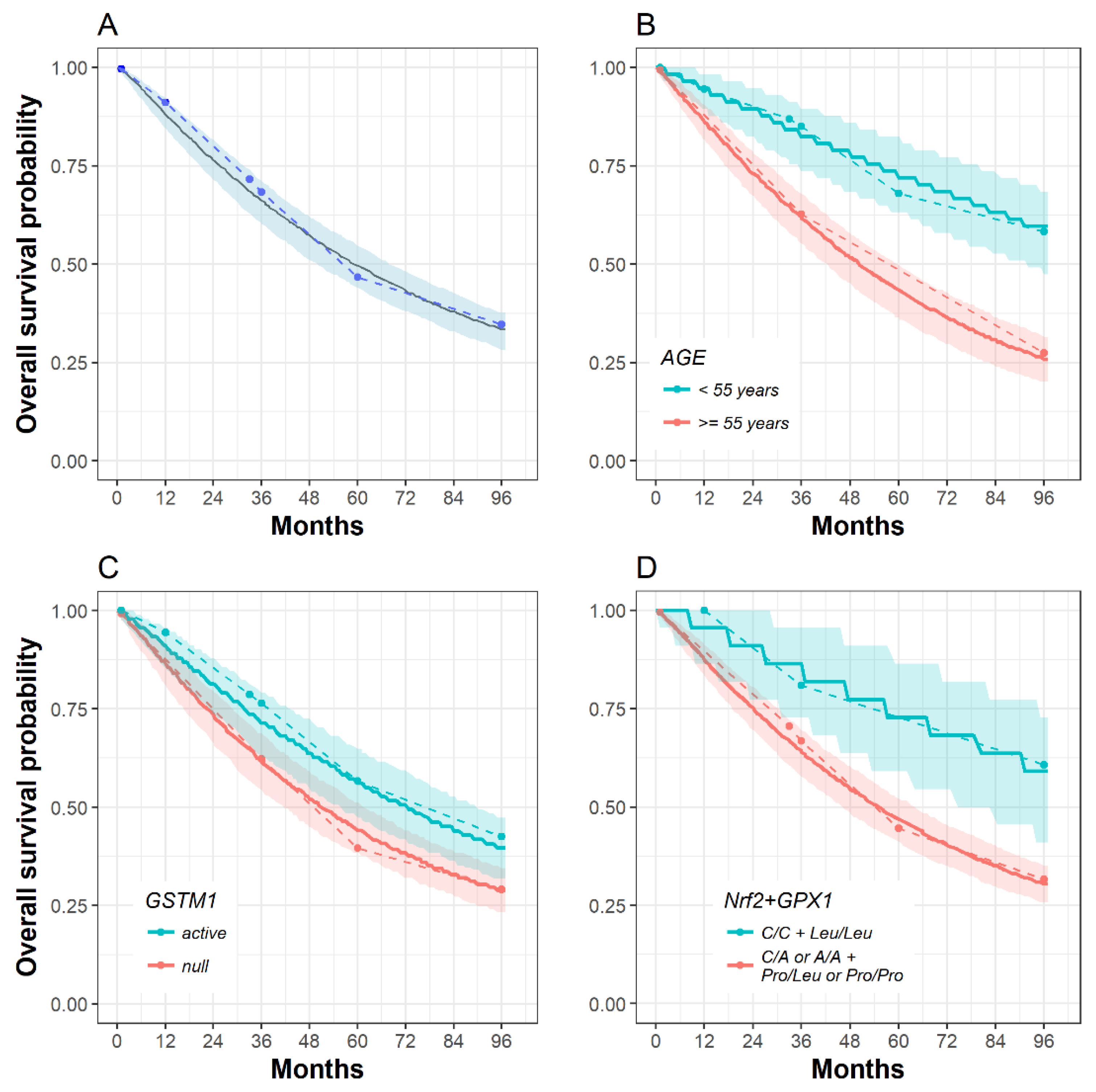
2.3. The Influence of SOD2, GPX1, Nrf2 and GSTM1 Polymorphisms on Patient Survival
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Study Population
5.2. Analysis of the SOD2, GPX1, Nrf2, and GSTM1 Genotypes
5.3. Oxidative Stress Byproducts Measurement in Plasma of ESRD Patients
5.4. Data Analysis
5.5. Time-to-Event Modelling
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Köttgen, A.; Glazer, N.L.; Dehghan, A.; Hwang, S.-J.; Katz, R.; Li, M.; Yang, Q.; Gudnason, V.; Launer, L.J.; Harris, T.B.; et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 2009, 41, 712. [Google Scholar] [CrossRef] [PubMed]
- Ju, W.; Eichinger, F.; Bitzer, M.; Oh, J.; McWeeney, S.; Berthier, C.C.; Shedden, K.; Cohen, C.D.; Henger, A.; Krick, S.; et al. Renal gene and protein expression signatures for prediction of kidney disease progression. Am. J. Pathol. 2009, 174, 2073–2085. [Google Scholar] [CrossRef] [PubMed]
- Martini, S.; Nair, V.; Keller, B.J.; Eichinger, F.; Hawkins, J.J.; Randolph, A.; Böger, C.A.; Gadegbeku, C.A.; Fox, C.S.; Cohen, C.D.; et al. Integrative biology identifies shared transcriptional networks in CKD. J. Am. Soc. Nephrol. 2014, 25, 2559–2572. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, M.; Yamamoto, M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 2006, 46, 113–140. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, Y.; Mitsuda, Y.; Tsuruta, Y.; Hamajima, N.; Niwa, T. Polymorphism of Nrf2, an antioxidative gene, is associated with blood pressure and cardiovascular mortality in hemodialysis patients. Int. J. Med. Sci. 2014, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Nomiyama, T.; Tanaka, Y.; Piao, L.; Nagasaka, K.; Sakai, K.; Ogihara, T.; Nakajima, K.; Watada, H.; Kawamori, R. The polymorphism of manganese superoxide dismutase is associated with diabetic nephropathy in Japanese type 2 diabetic patients. J. Hum. Genet. 2003, 48, 138. [Google Scholar]
- Möllsten, A.; Marklund, S.L.; Wessman, M.; Svensson, M.; Forsblom, C.; Parkkonen, M.; Brismar, K.; Groop, P.-H.; Dahlquist, G. A functional polymorphism in the manganese superoxide dismutase gene and diabetic nephropathy. Diabetes 2007, 56, 265–269. [Google Scholar] [CrossRef]
- Möllsten, A.; Jorsal, A.; Lajer, M.; Vionnet, N.; Tarnow, L. The V16A polymorphism in SOD2 is associated with increased risk of diabetic nephropathy and cardiovascular disease in type 1 diabetes. Diabetologia 2009, 52, 2590–2593. [Google Scholar] [CrossRef] [Green Version]
- Crawford, A.; Fassett, R.G.; Coombes, J.S.; Kunde, D.A.; Ahuja, K.D.K.; Robertson, I.K.; Ball, M.J.; Geraghty, D.P. Glutathione peroxidase, superoxide dismutase and catalase genotypes and activities and the progression of chronic kidney disease. Nephrol. Dial. Transplant. 2011, 26, 2806–2813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mimić-Oka, J.; Simić, T.; Djukanović, L.; Reljić, Z.; Davicević, Z. Alteration in plasma antioxidant capacity in various degrees of chronic renal failure. Clin. Nephrol. 1999, 51, 233–241. [Google Scholar] [PubMed]
- Suvakov, S.; Damjanovic, T.; Stefanovic, A.; Pekmezovic, T.; Savic-Radojevic, A.; Pljesa-Ercegovac, M.; Matic, M.; Djukic, T.; Coric, V.; Jakovljevic, J.; et al. Glutathione S-transferase A1, M1, P1 and T1 null or low-activity genotypes are associated with enhanced oxidative damage among haemodialysis patients. Nephrol. Dial. Transplant. 2013, 28, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Suvakov, S.; Jerotic, D.; Damjanovic, T.; Milic, N.; Pekmezovic, T.; Djukic, T.; Jelic-Ivanovic, Z.; Radojevic, A.S.; Pljesa-Ercegovac, M.; Matic, M.; et al. Markers of Oxidative Stress and Endothelial Dysfunction Predict Haemodialysis Patients Survival. Am. J. Nephrol. 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mohammedi, K.; Bellili-Muñoz, N.; Driss, F.; Roussel, R.; Seta, N.; Fumeron, F.; Hadjadj, S.; Marre, M.; Velho, G. Manganese superoxide dismutase (SOD2) polymorphisms, plasma advanced oxidation protein products (AOPP) concentration and risk of kidney complications in subjects with type 1 diabetes. PLoS ONE 2014, 9, e96916. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.; Fassett, R.G.; Coombes, J.S.; Kunde, D.A.; Ahuja, K.D.K.; Robertson, I.K.; Ball, M.J.; Geraghty, D.P. Relationship between antioxidant enzyme genotype and activity and kidney function: a case-control study. Clin. Nephrol. 2012, 78, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Shimoda-Matsubayashi, S.; Matsumine, H.; Kobayashi, T.; Nakagawa-Hattori, Y.; Shimizu, Y.; Mizuno, Y. Structural dimorphism in the mitochondrial targeting sequence in the human manganese superoxide dismutase gene: a predictive evidence for conformational change to influence mitochondrial transport and a study of allelic association in Parkinson’s disease. Biochem. Biophys. Res. Commun. 1996, 226, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Mohammedi, K.; Patente, T.A.; Bellili-Muñoz, N.; Driss, F.; Le Nagard, H.; Fumeron, F.; Roussel, R.; Hadjadj, S.; Corrêa-Giannella, M.L.; Marre, M.; et al. Glutathione peroxidase-1 gene (GPX1) variants, oxidative stress and risk of kidney complications in people with type 1 diabetes. Metabolism 2016, 65, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Aminzadeh, M.A.; Nicholas, S.B.; Norris, K.C.; Vaziri, N.D. Role of impaired Nrf2 activation in the pathogenesis of oxidative stress and inflammation in chronic tubulo-interstitial nephropathy. Nephrol. Dial. Transplant. 2013, 28, 2038–2045. [Google Scholar] [CrossRef] [PubMed]
- Pergola, P.E.; Raskin, P.; Toto, R.D.; Meyer, C.J.; Huff, J.W.; Grossman, E.B.; Krauth, M.; Ruiz, S.; Audhya, P.; Christ-Schmidt, H. Bardoxolone methyl and kidney function in CKD with type 2 diabetes. N. Engl. J. Med. 2011, 365, 327–336. [Google Scholar] [CrossRef]
- Soerensen, M.; Christensen, K.; Stevnsner, T.; Christiansen, L. The Mn-superoxide dismutase single nucleotide polymorphism rs4880 and the glutathione peroxidase 1 single nucleotide polymorphism rs1050450 are associated with aging and longevity in the oldest old. Mech. Ageing Dev. 2009, 130, 308–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Droge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Nikic, P.; Dragicevic, D.; Savic-Radojevic, A.; Pljesa-Ercegovac, M.; Coric, V.; Jovanovic, D.; Bumbasirevic, U.; Pekmezovic, T.; Simic, T.; Dzamic, Z. Association between GPX1 and SOD2 genetic polymorphisms and overall survival in patients with metastatic urothelial bladder cancer: a single-center study in Serbia. J. BUON 2018, 23, 1130–1135. [Google Scholar] [PubMed]
- Jocelyn, P.C. Spectrophotometric Assay of Thiols. Methods Enzymol. 1987, 143, 44–67. [Google Scholar]
- Witko-Sarsat, V.; Friedlander, M.; Capeillère-Blandin, C.; Nguyen-Khoa, T.; Nguyen, A.T.; Zingraff, J.; Jungers, P.; Descamps-Latscha, B. Advanced oxidation protein products as a novel marker of oxidative stress in uremia. Kidney Int. 1996, 49, 1304–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dousset, J.; Trouilh, M.; Foglietti, M.-J. Plasma malonaldehyde levels during myocardial infarction. Clin. Chim. Acta 1983, 129, 319–322. [Google Scholar] [CrossRef]
- Erel, O. A new automated colorimetric method for measuring total oxidant status. Clin. Biochem. 2005, 38, 1103–1111. [Google Scholar] [CrossRef]
- Alamdari, D.H.; Paletas, K.; Pegiou, T.; Sarigianni, M.; Befani, C.; Koliakos, G. A novel assay for the evaluation of the prooxidant–antioxidant balance, before and after antioxidant vitamin administration in type II diabetes patients. Clin. Biochem. 2007, 40, 248–254. [Google Scholar] [CrossRef]
- Lavielle, M. Mixed Effects Models for the Population Approach, 1st ed.; Chapman and Hall/CRC: New York, NY, USA, 2018; ISBN 1482226510. [Google Scholar]
- Wählby, U.; Jonsson, E.N.; Karlsson, M.O. Comparison of stepwise covariate model building strategies in population pharmacokinetic-pharmacodynamic analysis. AAPS PharmSci 2002, 4, 68–79. [Google Scholar] [CrossRef]
- Jonsson, E.N.; Karlsson, M.O. Automated covariate model building within NONMEM. Pharm. Res. 1998, 15, 1463–1468. [Google Scholar] [CrossRef]
- Wählby, U.; Jonsson, E.N.; Karlsson, M.O. Assessment of actual significance levels for covariate effects in NONMEM. J. Pharmacokinet. Pharmacodyn. 2001, 28, 231–252. [Google Scholar] [CrossRef] [PubMed]

| Variable | Controls | Patients | p |
|---|---|---|---|
| Age (years) | 61.09 ± 10.78 | 62.41 ± 11.91 | 0.155 |
| Gender, n (%) | |||
| Male | 199 (53) | 147 (57) | |
| Female | 175 (47) | 109 (43) | 0.297 |
| Smoking, n (%) a | |||
| Never | 189 (51) | 167 (74) | |
| Ever | 181 (49) | 58 (26) | <0.001 |
| Hypertension, n (%) a | |||
| No | 245 (69) | 42 (20) | |
| Yes | 109 (31) | 171 (80) | <0.001 |
| Diabetes, n (%) a | |||
| No | 281 (100) | 188 (87) | |
| Yes | 0 (0) | 27 (13) | <0.001 |
| BMI (kg/m2) a | 26.17 ± 4.27 | 24.57 ± 4.07 | <0.001 |
| Biochemical serum parameters a | |||
| Urea (mmol/L) | 5.36 ± 1.99 | 23.92 ± 5.01 | <0.001 |
| Creatinine (µmol/L) | 82.09 ± 15.04 | 856.93 ± 233.63 | <0.001 |
| Albumin (g/L) | 43.93 ± 3.79 | 38.61 ± 4.40 | <0.001 |
| Total cholesterol (mmol/L) | 4.34 ± 0.99 | 4.63 ± 1.14 | <0.003 |
| TAG (mmol/L) | 1.61 ± 0.59 | 2.06 ± 1.34 | <0.001 |
| Haemoglobin (g/L) | 142.14 ± 17.23 | 105.17 ± 14.76 | <0.001 |
| Haematocrit (%) | 41.00 ± 6.36 | 31.55 ± 4.56 | <0.001 |
| Serum iron (µmol/L) | 19.00 ± 4.32 | 11.29 ± 5.97 | <0.001 |
| Ferritin (ng/mL) | 56.44 ± 28.26 | 377.15 ± 258.81 | <0.001 |
| Genotypes | Controls, n (%) | Patients, n (%) | OR (95% CI) | p |
|---|---|---|---|---|
| SOD2 rs4880 | ||||
| Ala/Ala | 113 (32) | 56 (23) | 1.0 a | |
| Ala/Val | 167 (46) | 111 (45) | 1.31 (0.88–1.97) | 0.180 |
| Val/Val | 79 (22) | 77 (32) | 2.01 (1.28–3.16) | 0.002 |
| GPX1 rs1050450 | ||||
| Pro/Pro | 158 (42) | 101 (40) | 1.0 a | |
| Pro/Leu | 164 (45) | 122 (48) | 1.22 (0.86–1.72) | 0.271 |
| Leu/Leu | 43 (12) | 32 (12) | 1.17 (0.69–1.98) | 0.558 |
| Nrf2 rs6721961 | ||||
| C/C | 241 (71) | 185 (73) | 1.0 a | |
| C/A | 94 (27) | 64 (25) | 0.87 (0.59–1.26) | 0.461 |
| A/A | 7 (2) | 4 (2) | 0.75 (0.21–2.61) | 0.649 |
| SOD2 and GPX1 | ||||
| (Ala/Ala+Alal/Val)/(Pro/Pro+Pro/Leu) | 230 (68) | 151 (62) | 1.0 a | |
| (Ala/Ala+Ala/Val)/(Leu/Leu) | 36 (10) | 16 (7) | 0.69 (0.37–1.28) | 0.239 |
| (Val/Val)/(Pro/Pro+Pro/Leu) | 67 (20) | 63 (26) | 1.49 (0.99–2.24) | 0.051 |
| (Val/Val)/(Leu/Leu) | 6 (2) | 13 (5) | 3.27(1.12–8.25) | 0.019 * |
| Nrf2 and SOD2 | ||||
| (C/C)/(Ala/Ala + Ala/Val) | 183 (55) | 122 (50) | 1.0 a | |
| (C/C)/(Val/Val) | 49 (15) | 56 (23) | 1.80 (1.14–2.82) | 0.011 * |
| (C/A+A/A)/(Ala/Ala + Ala/Val) | 76 (23) | 44 (18) | 0.85 (0.55–1.32) | 0.465 |
| (C/A+A/A)/(Val/Val) | 22 (7) | 21 (9) | 1.46 (0.76–2.80) | 0.256 |
| Nr2 and GPX1 | ||||
| (C/C)/(Pro/Pro+Pro/Leu) | 202 (60) | 158 (63) | 1.0 a | |
| (C/C)/(Leu/Leu) | 34 (10) | 26 (10) | 0.96 (0.55–1.67) | 0.876 |
| (C/A+A/A)/(Pro/Pro+Pro/Leu) | 94 (28) | 64 (25) | 0.84 (0.57–1.24) | 0.391 |
| (C/A+A/A)/(Leu/Leu) | 5 (2) | 4 (2) | 1.04 (0.27–3.97) | 0.951 |
| Genotypes | Protein oxidative byproducts | Lipid oxidative byproducts | TOS and PAB | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PSH mmol/l | Carbonyls nmol/g | AOPP µmol/l | Nitrotyrosine nmol/l | MDA mmol/l | MDAadd pmol/l | TOS nmol/g | PAB mmol/l | ||
| SOD2 | Ala/Ala a | 6.9 [5.4–8.9] 100% | 2.14 ± 0.13 100% | 64.3 [56.7–69.1] 100% | 64.5 [46.1–91.2] 100% | 2.17 ± 0.78 100% | 40.28 ± 8.04 100% | 18.6 [13.7–34.1] 100% | 142.2 [71.8–184.4] 100% |
| Ala/Val | 6.7 [5.5–8.3] 97% | 2.30 ± 0.23 107% | 61.1 [46.8–74.3] 95% | 59.3 [45.2–87.5] 95% | 2.37 ± 0.72 109% | 38.93 ± 9.42 97% | 18.2 [12.8–50.8] 98% | 113.9 [61.8–218.3] 80% | |
| Val/Val | 6.1 [5.3–7.3] 88% | 2.32 ± 0.26 * 108% | 64.6 [48.1–80.8] 100% | 72.3 [46.1–101.2] 112% | 2.57 ± 0.79 * 118% | 43.17 ± 10.15 107% | 24.5 [12.8–56.3] 131% | 160.4 [105.1–251.5] 113% | |
| Ala/Ala+ Ala/Val a | 6.7 [5.5–8.5] 100% | 2.23 ± 0.21 100% | 62.7 [48.4–73.3] 100% | 60.3 [46.1–87.5] 100% | 2.29 ± 0.75 100% | 39.49 ± 8.84 100% | 18.4 [13.3–48.3] 100% | 126.6 [67.7–211.0] 100% | |
| Val/Val | 6.1 [5.3–7.3] * 91% | 2.32 ± 0.26 104% | 64.6 [48.1–80.8] 103% | 72.3 [46.1–101.2] 119% | 2.57 ± 0.79 * 112% | 43.17 ± 10.15 * 109% | 24.5 [12.8–56.3] 133% | 160.4 [105.1–251.5] * 127% | |
| GPX1 | Pro/Pro a | 6.1 [5.1–8.0] 100% | 2.27 ± 0.23 100% | 63.8 [46.5–73.7] 100% | 64.5 [50.1–93.1] 100% | 2.33 ± 0.79 100% | 40.17 ± 7.63 100% | 17.8 [14.0–41.0] 100% | 153.8 [83.8–233.0] 100% |
| Pro/Leu | 6.5 [5.5–8.3] 106% | 2.29 ± 0.24 101% | 63.1 [49.3–74.4] 99% | 64.5 [46.1–93.1] 100% | 2.34 ± 0.74 100% | 41.82 ± 11.00 104% | 19.1 [12.8–53.2] 107% | 143.0 [71.7–226.0] 93% | |
| Leu/Leu | 6.6 [5.5–7.3] 108% | 2.19 ± 0.19 96% | 67.5 [48.2–85.1] 106% | 60.2 [44.7–78.7] 93% | 2.65 ± 0.81 114% | 38.91 ± 8.19 97% | 27.2 [16.8–47.5] 152% | 128.6 [68.4–191.8] 84% | |
| Pro/Pro+Pro/Leu | 6.3 [5.4–8.1] 100% | 2.28 ± 0.23 100% | 63.5 [48.6–74.3] 100% | 64.5 [46.1–93.1] 100% | 2.33 ± 0.77 100% | 41.04 ± 9.54 100% | 18.2 [13.2–48.4] 100% | 146.8 [78.2–229.9] 100% | |
| Leu/Leu | 6.6 [5.5–7.3] 105% | 2.19 ± 0.19 96% | 67.5 [48.2–85.1] 106% | 60.3 [44.7–78.7] 93% | 2.65 ± 0.81 113% | 38.91 ± 8.19 97% | 27.2 [16.8–47.5] 145% | 128.6 [68.4–191.8] 88% | |
| NRF2 | C/C a | 6.3 [5.5–8.0] 100% | 2.24 ± 0.22 100% | 65.7 [48.8–75.7] 100% | 64.5 [50.1–93.1] 100% | 2.39 ± 0.78 100% | 40.17 ± 9.12 100% | 22.0 [13.3–53.6] 100% | 142.6 [83.0–216.9] 100% |
| C/A+A/A | 6.4 [5.5–8.2] 102% | 2.34 ± 0.25 104% | 56.7 [44.6–70.5] 86% | 52.3 [38.5–87.5] * 81% | 2.33 ± 0.75 97% | 41.88 ± 9.83 104% | 16.2 [13.2–28.3] 73% | 159.4 [72.8–252.6] 112% | |
| Patient’s Characteristics | Probability (%) to Survive at Least | ||||
|---|---|---|---|---|---|
| Age (years) | GSTM1 Genotype | Nrf2+GPX1 Genotype | 3 Years | 5 Years | 8 Years |
| 40 | active | C/C + Leu/Leu | 95.91 | 93.10 | 88.73 |
| C/A or A/A + Pro/Leu or Pro/Pro | 91.02 | 84.59 | 74.25 | ||
| null | C/C + Leu/Leu | 94.11 | 90.00 | 83.52 | |
| C/A or A/A + Pro/Leu or Pro/Pro | 86.92 | 77.28 | 61.40 | ||
| 55 | active | C/C + Leu/Leu | 91.40 | 85.27 | 75.43 |
| C/A or A/A + Pro/Leu or Pro/Pro | 80.62 | 65.75 | 40.80 | ||
| null | C/C + Leu/Leu | 87.49 | 78.31 | 63.23 | |
| C/A or A/A + Pro/Leu or Pro/Pro | 71.23 | 48.16 | 12.48 | ||
| 70 | active | C/C + Leu/Leu | 84.68 | 73.22 | 54.14 |
| C/A or A/A + Pro/Leu or Pro/Pro | 64.29 | 35.21 | 1.14 | ||
| null | C/C + Leu/Leu | 77.42 | 59.79 | 30.42 | |
| C/A or A/A + Pro/Leu or Pro/Pro | 45.88 | 6.71 | ≈0 | ||
| Patient’s Characteristics | Probability (%) to Survive at Least | ||||
|---|---|---|---|---|---|
| Age (years) | GSTM1 Genotype | GPX1 Genotype | 3 Years | 5 Years | 8 Years |
| 40 | active | Leu/Leu | 99.60 | 99.07 | 98.00 |
| Pro/Leu Pro/Pro | 98.39 | 96.31 | 92.20 | ||
| null | Leu/Leu | 98.87 | 97.40 | 94.47 | |
| Pro/Leu Pro/Pro | 95.52 | 89.95 | 79.54 | ||
| 55 | active | Leu/Leu | 98.43 | 96.42 | 92.42 |
| Pro/Leu Pro/Pro | 93.85 | 86.35 | 72.81 | ||
| null | Leu/Leu | 95.65 | 90.23 | 80.07 | |
| Pro/Leu Pro/Pro | 83.60 | 66.11 | 40.88 | ||
| 70 | active | Leu/Leu | 95.67 | 90.27 | 80.14 |
| Pro/Leu Pro/Pro | 83.67 | 66.22 | 41.03 | ||
| null | Leu/Leu | 88.26 | 74.92 | 53.58 | |
| Pro/Leu Pro/Pro | 60.49 | 31.29 | 8.11 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jerotic, D.; Matic, M.; Suvakov, S.; Vucicevic, K.; Damjanovic, T.; Savic-Radojevic, A.; Pljesa-Ercegovac, M.; Coric, V.; Stefanovic, A.; Ivanisevic, J.; et al. Association of Nrf2, SOD2 and GPX1 Polymorphisms with Biomarkers of Oxidative Distress and Survival in End-Stage Renal Disease Patients. Toxins 2019, 11, 431. https://doi.org/10.3390/toxins11070431
Jerotic D, Matic M, Suvakov S, Vucicevic K, Damjanovic T, Savic-Radojevic A, Pljesa-Ercegovac M, Coric V, Stefanovic A, Ivanisevic J, et al. Association of Nrf2, SOD2 and GPX1 Polymorphisms with Biomarkers of Oxidative Distress and Survival in End-Stage Renal Disease Patients. Toxins. 2019; 11(7):431. https://doi.org/10.3390/toxins11070431
Chicago/Turabian StyleJerotic, Djurdja, Marija Matic, Sonja Suvakov, Katarina Vucicevic, Tatjana Damjanovic, Ana Savic-Radojevic, Marija Pljesa-Ercegovac, Vesna Coric, Aleksandra Stefanovic, Jasmina Ivanisevic, and et al. 2019. "Association of Nrf2, SOD2 and GPX1 Polymorphisms with Biomarkers of Oxidative Distress and Survival in End-Stage Renal Disease Patients" Toxins 11, no. 7: 431. https://doi.org/10.3390/toxins11070431
APA StyleJerotic, D., Matic, M., Suvakov, S., Vucicevic, K., Damjanovic, T., Savic-Radojevic, A., Pljesa-Ercegovac, M., Coric, V., Stefanovic, A., Ivanisevic, J., Jelic-Ivanovic, Z., McClements, L., Dimkovic, N., & Simic, T. (2019). Association of Nrf2, SOD2 and GPX1 Polymorphisms with Biomarkers of Oxidative Distress and Survival in End-Stage Renal Disease Patients. Toxins, 11(7), 431. https://doi.org/10.3390/toxins11070431








