Marine Toxins Detection by Biosensors Based on Aptamers
Abstract
:1. Introduction
2. The Toxicity of Representative Marine Toxins
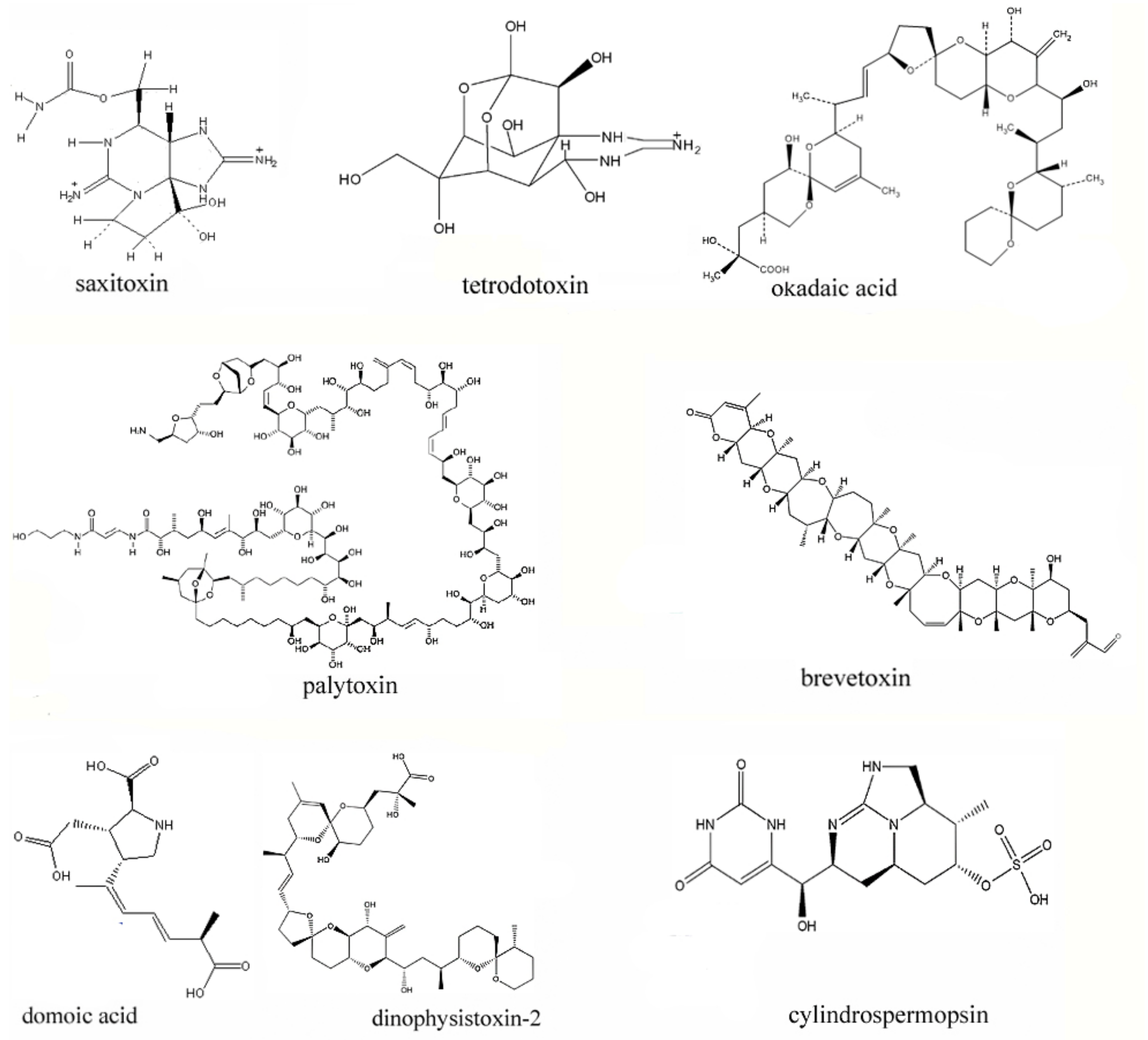
2.1. The Representative Marine Toxins
2.2. The Toxicity of Representative Marine Toxins
2.3. The Development of Approaches for Marine Toxins Detection
2.3.1. The Classical Detection Methods for Marine Toxins
2.3.2. New Detection Methods Based on Marine Toxins Receptor
3. The Development of Biosensors Based on Aptamers
3.1. The Features of Aptamers
3.2. The Development of Biosensors
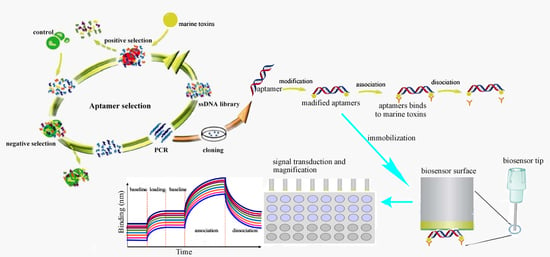
3.3. The Calssification and Procedure of Aptamer-Based Biosensors
3.3.1. The Classification of Aptasensors
3.3.2. The Procedure for the Detection of Marine Toxins by Aptasensors
3.3.3. The Preparation and Optimization of Apatamer-Based Biosensors
4. The Detection of Marine Toxins by Aptamer-Based Biosensors
4.1. The Detection of Representative Marine Toxins by Aptasensors
4.2. Recent Advances in Detecting Toxins by Aptasensors
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AgPs | silver nanoparticles |
| AuPs | gold nanoparticles |
| BLI | biolayer Interferometry |
| BTX | brevetoxin |
| cAMP | cyclic adenosine monophosphate |
| CYN | cylindrospermopsin |
| DA | domoic acid |
| DTX | dinophysistoxin |
| EDC | 1-Ethyl-3-(3′-dimethylaminopropyl)carbodiimide |
| ELAA | enzyme-linked ligand analysis |
| ELISA | enzyme-linked immunosorbent assay |
| FAM | carboxyfluorescein |
| GC | gas chromatography |
| GO | graphene oxide |
| HPLC | high-performance liquid chromatography |
| LC-MS/MS | liquid chromatography-mass spectrometry |
| LD50 | mild toxic dose |
| LOAD | lab-on-a-disc |
| LOC | lab-on-a-chip |
| LOD | limit of detection |
| LRD | linear range of detection |
| MBA | mouse biosassay |
| MC-LR | microrocystin LR containing a leucine substituent |
| MRE | molecular recognition element |
| MRGO | magnetic reduced graphene oxide |
| MTT | 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide |
| NDR | nodularin |
| NHS | N-Hydroxysuccinimide |
| OA | okadaic acid |
| OTA | Ochration A |
| PTX | palytoxin |
| QDs | quantum dots |
| QGRS | q uadruplex-forming G-Rich Sequences |
| RSD | relative standard deviation |
| SELEX | systematic evolution of ligands by exponential enrichment |
| SERS | surface enhaced micro-Raman scattering |
| SPR | surface plasmon resonance |
| STX | saxitoxin |
| UV | ultraviolet |
References
- Yasumoto, T.; Murata, M. Marine toxins. Chem. Rev. 1993, 93, 1897–1909. [Google Scholar]
- Yasumoto, T. The chemistry and biological function of natural marine toxins. Chem. Rec. 2001, 1, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Gran, L.E.; Turner, J.T. Ecology of Harmful Algae; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Ciminiello, P.; Forino, M.; Dell’aversano, C. Seafood toxins: Classes, sources, and toxicology. In Handbook of Marine Natural Products; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1345–1387. [Google Scholar]
- Whittle, K.; Gallacher, S. Marine toxins. Br. Med. Bull. 2000, 56, 236–253. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, T. Chemistry, etiology and food chain dynamics of marine toxins. Proc. Jpn. Acad. B Phys. 2005, 81, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Badeno, D.G. Marine food-borne dinoflagellate toxins. Int. Rev. Cytol. 1983, 82, 99–150. [Google Scholar]
- Sobel, J.; Painter, J. Illnesses caused by marine toxins. Clin. Infect. Dis. 2005, 41, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, N.; Hallegraeff, G.; Ruth, P.; Ginkel, R.; McNabb, P.; Hay, B.; O’Connor, W.; Kiermeier, A.; Deveney, M.; McLeod, C. Uptake, distribution and depuration of paralytic shellfish toxins from Alexandrium minutum in Australian greenlip abalone, Haliotis laevigata. Toxicon 2011, 58, 101–111. [Google Scholar] [CrossRef]
- Joseph, H.; Natalie, H. The evolution of cultural adaptations: Fijian food taboos protect against dangerous marine toxins. Proc. Biol. Sci. 2010, 277, 3715–3724. [Google Scholar]
- Pravda, M.K.; MarK, P.G.; George, G. Analysis of important freshwater and marine toxins. Anal. Let. 2002, 1, 1–15. [Google Scholar] [CrossRef]
- Arjen, G.; Irene, E.P.; Marnix, P.; Patrick, P.J.M.; Hester, V.D.T.; Jacob, D.B. Marine Toxins: Chemistry, Toxcity, Occurrence and Detection, with Special Reference to the Dutch Situation. Toxins 2010, 2, 878–904. [Google Scholar]
- Wang, Y.; Gan, N.; Zhou, Y.; Li, T.; Cao, Y.; Chen, Y. Novel single-stranded DNA binding protein-assisted fluorescence aptamer switch based on FRET for homogeneous detection of antibiotics. Biosens. Bioelectron. 2017, 87, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Khoshfetrat, S.M.; Mehrgardi, M.A. Amplified detection of leukemia cancer cells using an aptamer-conjugated gold-coated magnetic nanoparticles on a nitrogen-doped graphene modified electrode. Bioelectrochemistry 2017, 114, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Li, B.; Yao, R.; Li, Z.; Wang, X.; Dong, X.; Qu, H.; Li, Q.; Li, N.; Chi, H.; et al. Intuitive Label-Free SERS Detection of Bacteria Using Aptamer-Based in Situ Silver Nanoparticles Synthesis. Anal. Chem. 2017, 89, 9836–9842. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, B.; Shen, J.; Xiong, X.L.; Deng, S.X. Aptamer based bare eye detection of kanamycin by using a liquid crystal film on a glass support. Microchim. Acta 2017, 184, 3765–3771. [Google Scholar] [CrossRef]
- Handy, S.M.; Yakes, B.J.; DeGrasse, J.A.; Campbell, K.; Elliott, C.T.; Kanyuck, K.M.; DeGrasse, S.L. First report of the use of a saxitoxin-protein conjugate to develop a DNA aptamer to a small molecule toxin. Toxicon 2013, 61, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Oikawa, H.; Fujita, T.; Saito, K.; Watabe, S.; Satomi, M.; Yano, Y. Comparison of paralytic shellfish poisoning toxin between carnivorous crabs (Telmessus acutidens and Charybdis japonica) and their prey mussel (Mytilus galloprovincialis) in an inshore food chain. Toxicon 2004, 43, 713–719. [Google Scholar] [CrossRef]
- Hiroshi, O.; Tsuneo, F.; Ken, S.; Massataka, S.; Yutaka, Y. Difference in the level of paralytic shellfish poisoning toxin accumulation between the crabs Telmessus acutidens and Charybdis japonica collected in Onahama, Fukushima Prefecture. Fish. Sci. 2007, 73, 395–403. [Google Scholar]
- Jester, R.J.; Baugh, K.A.; Lefebvre, K.A. Presence of Alexandrium catenella and paralytic shellfish toxins in finfish, shellfish and rock crabs in Monterey Bay, California, USA. Mar. Biol. 2009, 156, 493–504. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, T.; Miyazawa, K.; Daigo, K.; Arakawa, O. Paralytic shellfish poisoning (PSP) toxin- and/or tetrodotoxin-contaminated crabs and food poisoning by them. Toxin Rev. 2011, 30, 91–102. [Google Scholar] [CrossRef]
- Vilariño, N.; Louzao, M.C.; Vieytes, M.R.; Botana, L.M. Biological methods for marine toxin detection. Anal. Bioanal. Chem. 2010, 397, 1673–1681. [Google Scholar] [CrossRef]
- Ling, S.; Xiao, S.; Xie, C.; Wang, R.; Zeng, L.; Wang, K.; Zhang, D.; Li, X.; Wang, S. Preparation of Monoclonal Antibody for Brevetoxin 1 and Development of Ic-ELISA and Colloidal Gold Strip to Detect Brevetoxin 1. Toxins 2018, 10, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naar, J.; Bourdelais, A.; Tomas, C.; Kubanek, J.; Whitney, P.L.; Flewelling, L.; Steidinger, K.; Lancaster, J.; Baden, D.G. A competitive ELISA to detect brevetoxins from Karenia brevis (formerly Gymnodinium breve) in seawater, shellfish, and mammalian body fluid. Environ. Health Perspect. 2002, 110, 179–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campora, C.E.; Hokama, Y.; Ebesu, J.S. Comparative analysis of purified Pacific and Caribbean ciguatoxin congeners and related marine toxins using a modified ELISA technique. J. Clin. Lab. Anal. 2006, 20, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Gerssen, A.; Mulder, P.P.; McElhinney, M.A.; Boer, J. Liquid chromatography-tandem mass spectrometry method for the detection of marine lipophilic toxins under alkaline conditions. J. Chromatogr. A 2009, 1216, 1421–1430. [Google Scholar] [CrossRef]
- Elqarch, A.; Vale, P.; Rifai, S.; Fassouane, A. Detection of diarrheic shellfish poisoning and azaspiracid toxins in Moroccan mussels: Comparison of the LC-MS method with the commercial immunoassay kit. Mar. Drugs 2008, 6, 587–594. [Google Scholar] [CrossRef]
- Bernd, K.; Julia, A.B.; Urban, T.; Francisco, G.C.; Asterio, S.M.; Juan, J.G.R.; Lorenzo, L.R.; Karl, B.A.; Margarita, F.T.; Matthias, W.; et al. LC-MS/MS Detection of Karlotoxins Reveals New Variants in Strains of the Marine Dinoflagellate Karlodinium veneficum from the Ebro Delta (NW Mediterranean). Mar. Drugs 2017, 15, 391. [Google Scholar]
- Chandran, K.; Raju, R.; Kalpana, B. Introduction to biosensors. Biosens. Bioelctron. 2015, 1–68. [Google Scholar] [CrossRef]
- Shi, J.; McLamore, E.S.; Marshall, P.D. Nanomaterial based self-referencing microbiosensors for cell and tissue physiology research. Biosens. Bioelctron. 2013, 40, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Asphahani, F.; Zhang, M. Cellular impedance biosensors for drug screening and toxin detection. Analyst 2007, 132, 835–841. [Google Scholar] [CrossRef] [Green Version]
- Maria, A.A.L.; Olga, D.R. Screen-printed biosensors in drug analysis. Curr. Pharm. Anal. 2017, 13, 169–174. [Google Scholar]
- Lv, M.; Liu, Y.; Geng, J.; Kou, X.; Xin, Z.; Yang, D. Engineering nanomaterials-based biosensors for food safety detection. Biosens. Bioelectron. 2018, 106, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Hanif, K.; Bahram, N. A novel biosensor nano material for ultraselective and ultrasensitive electrochemical diagnosis of breast cancer-related BRCA1 gene. Anal. Methods 2016, 8, 3069–3074. [Google Scholar]
- Bunka, D.H.; Stockley, P.G. Aptamers come of age-at last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Szpechcinski, A.; Grzanka, A. Aptamers in clinical diagnostics. Postepy Biochem. 2006, 52, 260–270. [Google Scholar]
- Famulok, M.; Mayer, G. Aptamers and SELEX in chemistry & biology. Chem. Biol. 2014, 21, 1055–1058. [Google Scholar]
- Yüce, M.; Ullah, N.; Budak, H. Trends in aptamer selection methods and applications. Analyst 2015, 140, 5379–5399. [Google Scholar] [CrossRef]
- Duan, N.; Wu, S.; Dai, S.; Gu, H.; Hao, L.; Ye, H.; Wang, Z. Advances in aptasensors for the detection of food contaminants. Analyst 2016, 141, 3942–3961. [Google Scholar] [CrossRef]
- Lan, L.; Yao, Y.; Ping, J.; Ying, Y. Recent Progress in Nanomaterial-based optical aptamer assay for the detection of food chemical contaminants. ACS Appl. Mater. Interfaces 2017, 9, 23287–23301. [Google Scholar] [CrossRef]
- Isbister, G.K.; Kiernan, M.C. Neurotoxic marine poisoning. Lancet Neurol. 2005, 4, 219–228. [Google Scholar]
- Wiese, M.; D’Agostino, P.M.; Mihali, T.K.; Moffitt, M.C.; Neilan, B.A. Neurotoxic alkaloids: Saxitoxin and its analogs. Mar. Drugs 2010, 8, 2185–2211. [Google Scholar] [CrossRef] [Green Version]
- Lipkind, G.M.; Fozzard, H.A. A structural model of the tetrodotoxin and saxitoxin binding site of the Na+ channel. Biophy. J. 1994, 66, 1–13. [Google Scholar] [CrossRef]
- Bricelj, V.M.; Connell, L.; Konoki, K.; Macquarrie, S.P.; Scheuer, T.; Catterall, W.A.; Trainer, V.L. Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature 2005, 434, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Benton, B.J.; Keller, S.A.; Spriggs, D.L.; Capacio, B.R.; Chang, F.C. Recovery from the lethal effects of saxitoxin: A therapeutic window for 4-aminopyridine (4-AP). Toxicon 1998, 36, 571–588. [Google Scholar] [CrossRef]
- Kazuo, T.; Paul, J.S. Okadaic acid, a cytotoxic polyether from two marine sponges of the genus Halichondria. J. Am. Chem. Soc. 1981, 103, 2469–2471. [Google Scholar]
- Aune, T.; Larsen, S.; Aasen, J.A.; Rehmann, N.; Satake, M.; Hess, P. Relative toxicity of dinophysistoxin-2 (DTX-2) compared with okadaic acid, based on acute intraperitoneal toxicity in mice. Toxicon 2007, 49, 1–7. [Google Scholar] [CrossRef]
- Garcia, L.; Garcia, F.; Llorens, F.; Unzeta, M.; Itarte, E.; Gómez, N. PP1/PP2A phosphatases inhibitors okadaic acid and calyculin A block ERK5 activation by growth factors and oxidative stress. FEBS Lett. 2002, 523, 90–94. [Google Scholar] [CrossRef] [Green Version]
- Lago, J.; Rodriguez, L.P.; Blanco, L.; Vieites, J.M.; Cabado, A.G. Tetrodotoxin, an extremely potent marine neurotoxin: Distribution, toxicity, origin and therapeutical uses. Mar. Drugs 2015, 13, 6384–6406. [Google Scholar] [CrossRef]
- Abal, P.; Louzao, M.C.; Antelo, A.; Alvarez, M.; Caqide, E.; Vilarino, N.; Vieytes, M.R.; Botana, L.M. Acute oral toxicity of tetrodotoxin in mice: Determination of lethal dose 50 (LD50) and no observed adverse effect levelb (NOAEL). Toxins 2017, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Habermann, E.; Ahnert-Hilger, G.; Chhatwal, G.S.; Beress, L. Delayed haemolytic action of palytoxin. General characteristics. BBA Biomembr. 1981, 649, 481–486. [Google Scholar] [CrossRef]
- Wiles, J.S.; Vick, J.A.; Christensen, M.K. Toxicological evaluation of palytoxin in several animal species. Toxicon 1974, 12, 427–433. [Google Scholar] [CrossRef]
- Valdiglesias, V.; Prego-Faraldo, M.; Pásaro, E.; Mendez, J.; Laffon, B. Okadaic Acid: More than a Diarrheic Toxin. Mar. Drugs 2013, 11, 4328–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamat, P.K.; Rai, S.; Swarnkar, S.; Nath, C. Molecular and cellular mechanism of okadaic acid (OKA)-induced neurotoxicity: A novel tool for Alzheimer’s disease therapeutic application. Mol. Neurobiol. 2014, 50, 852–865. [Google Scholar] [CrossRef] [PubMed]
- Gold, E.P.; Jacocks, H.M.; Bourdelais, A.J.; Baden, D.G. Brevenal, a brevetoxin antagonist from Karenia brevis, binds to a previously unreported site on mammalian sodium channels. Harmful Algae 2013, 26, 12–19. [Google Scholar] [PubMed] [Green Version]
- Zaias, J.; Fleming, L.E.; Baden, D.G.; Abraham, W.M. Repeated exposure to aerosolized brevetoxin-3 induces prolonged airway hyperresponsiveness and lung inflammation in sheep. Inhal. Toxicol. 2011, 23, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Froscio, S.M.; Humpage, A.R.; Burcham, P.C.; Falconer, I.R. Cylindrospermopsin-induced protein synthesis inhibition and its dissociation from acute toxicity in mouse hepatocytes. Environ. Toxicol. 2003, 18, 234–251. [Google Scholar] [CrossRef] [PubMed]
- Chiamulera, C.; Costa, S.; Valerio, E.; Reqqiani, A. Domoic acid toxicity in rats and mice after intracerebroventricular administration: Comparison with excitatory amino acid agonists. Pharmacol. Toxicol. 1992, 70, 115–120. [Google Scholar] [CrossRef]
- Xi, D.; Ramsdell, J.S. Glutamate receptors and calcium entry mechanisms for domoic acid in hippocampal neurons. Neuroreport 1996, 7, 115–120. [Google Scholar] [CrossRef]
- Larsen, K.; Petersen, D.; Wilkins, A.L.; Samdal, I.A.; Sandvik, M.; Rundberget, T.; Goldstone, D.; Arcus, V.; Hovgaard, P.; Rise, F.; et al. Clarification of the c-35 stereochemistries of dinophysistoxin-1 and dinophysistoxin-2 and its consequences for binding to protein phosphatase. Chem. Res. Toxicol. 2007, 20, 868–875. [Google Scholar] [CrossRef]
- Stabell, O.; Steffenak, I.; Aune, T. An evaluation of the mouse bioassay applied to extracts of‘diarrhoetic’shellfish toxins. Food Chem. Toxicol. 1992, 30, 139–144. [Google Scholar] [CrossRef]
- Tubaro, A.; Florio, C.; Luxich, E.; Vertua, R.; Della, L.R.; Yasumoto, T. Suitability of the MTT-based cytotoxicity assay to detect okadaic acid contamination of mussels. Toxicon 1996, 34, 965–974. [Google Scholar] [CrossRef]
- Hamada-Sato, N.; Minamitani, N.; Inaba, Y. Development of amperometric sensor system for measurement of diarrheic shellfish poisoning (DSP) toxin, okadaic acid (OA). Sens. Mater. 2004, 16, 99–107. [Google Scholar]
- Cheun, B.S.; Loughran, M.; Hayashi, T.; Nagashima, Y.; Watanabe, E. Use of a channel biosensor for the assay of paralytic shellfish toxins. Toxicon 1998, 36, 1371–1381. [Google Scholar] [CrossRef]
- Marquette, C.A.; Coulet, P.R.; Blum, L.C.J. Semi-automated membrane based chemiluminescent immunosensor for flow injection analysis of okadaic acid in mussels. Anal. Chim. Acta 1999, 398, 173–182. [Google Scholar] [CrossRef]
- Pinzaru, S.C.; Müller, C.; Ujevi, I.; Venter, M.M.; Chis, V.; Glamuzina, B. Lipophilic marine biotoxins sers sensing in solutions and in mussel tissue. Talanta 2018, 187, 47–58. [Google Scholar] [CrossRef]
- Hong, K.L.; Sooter, L.J. Single-Stranded DNA Aptamers against Pathogens and Toxins: Identification and Biosensing Applications. Biomed. Res. Int. 2015, 2015, 419318. [Google Scholar] [CrossRef]
- Feng, C.; Dai, S.; Wang, L. Optical aptasensors for quantitative detection of small biomolecules: A review. Biosens. Bioelectron. 2014, 59, 64–74. [Google Scholar] [CrossRef]
- Kyo, M.; Ohtsuka, K.; Okamoto, E.; Inamori, K. High-throughput SPR biosensor. Methods Mol. Biol. 2009, 577, 227–234. [Google Scholar]
- Petersen, R.L. Strategies using Bio-Layer interferometry biosensor technology for vaccine research and development. Biosensors 2017, 7, 49. [Google Scholar] [CrossRef] [Green Version]
- Gu, H.J.; Duan, N.; Wu, S.J.; Hao, L.L.; Xia, Y.; Ma, X.Y.; Wang, Z.P. Graphene oxide-assisted non-immobilized SELEX of okdaic acid aptamer and the analytical application of aptasensor. Sci. Rep. 2016, 6, 21665. [Google Scholar] [CrossRef] [Green Version]
- He, J.L.; Wu, Z.S.; Zhou, H.; Wang, H.Q.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Fluorescence aptameric sensor for strand displacement amplification detection of cocaine. Anal. Chem. 2010, 82, 1358–1364. [Google Scholar] [CrossRef]
- Park, J.W.; Tatavarty, R.; Kim, D.W.; Jung, H.T.; Gu, M.B. Immobilization-free screening of aptamers assisted by graphene oxide. Chem. Commun. 2012, 48, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Guo, L.; He, J.; Xu, H.; Xie, J. Stepping library-based post-SELEX strategy approaching to the minimized aptamer in SPR. Anal. Chem. 2017, 89, 6559–6566. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.X.; Zheng, X.; Jiao, B.H.; Wang, L.H. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.X.; Hu, B.; Zheng, X.; Cao, Y.; Liu, D.J.; Sun, M.J.; Jiao, B.H.; Wang, L.H. Gonyautoxin 1/4 aptamers with high-affinity and high-specificity: From efficient selection to aptasensor application. Biosens. Bioelectron. 2016, 79, 938–944. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, B.; Gao, S.X.; Liu, D.J.; Sun, M.J.; Jiao, B.H.; Wang, L.H. A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 2015, 101, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Gu, H.J. Study on Non-Immobilized Screening of Aptamers against Four Marine Biotoxins and the Application in Bioassay; Jiangnan University: Jiangsu, China, 2018. [Google Scholar]
- Eissa, S.; Ng, A.; Siaj, M.; Tavares, A.C.; Zourob, M. Selection and identification of DNA aptamers against okadaic acid for biosensing application. Anal. Chem. 2013, 85, 11794–11801. [Google Scholar] [CrossRef]
- Shao, B.Y.; Chen, B.; Chen, W.B.; Yang, F.; Miao, T.Y.; Peng, J. Preparation and application of tetrodotoxin DNA aptamer. Food. Sci. 2014, 35, 205–208. [Google Scholar]
- Gao, S.X.; Zheng, X.; Hu, B.; Sun, M.J.; Wu, J.; Jiao, B.H.; Wang, L. Enzyme-linked, aptamer-based, competitive biolayer interferometry biosensor for palytoxin. Biosens. Bioelectron. 2017, 89, 952–958. [Google Scholar] [CrossRef]
- Tang, D.; Zhang, B.; Tang, J.; Hou, L.; Chen, G. Displacement-type quartz crystal microbalance immunosensing platform for ultrasensitive monitoring of small molecular toxins. Anal. Chem. 2013, 85, 6958–6966. [Google Scholar] [CrossRef]
- Su, B.L.; Tang, J.; Yang, H.H.; Chen, G.N.; Huang, J.X.; Tang, D.P. A graphene platform for sensitive electrochemical immunoassay of carcinoembryoninc antigen based on gold-nanoflower-biolabels. Electroanalysis 2011, 23, 832–841. [Google Scholar] [CrossRef]
- Tang, J.; Hou, L.; Tang, D.; Zhou, J.; Wang, Z.; Li, J.; Chen, G. Magneto-controlled electrochemical immunoassay of brevetoxin B in seafood based on guanine-functionalized graphene nanoribbons. Biosens. Bioelectron. 2012, 38, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Siaj, M.; Zourob, M. Aptamer-based competitive electrochemical biosensor for brevetoxin-2. Biosens. Bioelectron. 2015, 69, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Elshafey, R.; Siaj, M.; Zourob, M. In vitro selection, characterization, and biosensing application of high-affinity cylindrospermopsin-targeting aptamers. Anal. Chem. 2014, 86, 9196–9203. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Hu, B.; Gao, S.X.; Zheng, X.; Ouyang, S.Q.; Li, Z.G.; Zhou, R.; Li, Z.; Jiao, B.H. Screening, verification and application of specific aptamers for marine biotoxins. In Proceedings of the 12th Congress of the Chinese Society of Biochemistry and Molecular Biology and the 2018 National academic Conference, Chongqing City, China, 25–28 October 2018. [Google Scholar]
- Guo, M.M.; Tan, Z.J.; Wu, H.Y.; Li, Z.X.; Zhai, Y.X. Simultaneous determination of Okadaic Acid, dinophysistoxin, saxitoxin and yessotoxin in shellfish by liquid chromatography-tandem mass spectrometry. Chromatography 2012, 3, 256–261. [Google Scholar]
- Eissa, S.; Ng, A.; Siaj, M.; Zourob, M. Selection, characterization, and application of high affinity microcystin-targeting aptamers in a graphene-based biosensing platform. J. Phys. Chem. A 2015, 40, 178–186. [Google Scholar]
- Chikashi, N.; Kobayashi, T.; Miyake, M.; Shirai, M.; Miyakea, J. Usage of a DNA aptamer as a ligand targeting microcystin. Mol. Cryst. Liq. Cryst. 2001, 371, 369–374. [Google Scholar]
- He, F.; Liang, L.; Zhou, S.; Xie, W.; He, S.; Wang, Y.; Tlili, C.; Tong, S.; Wang, D. Label-free sensitive detection of microcystin-LR via aptamer-conjugated gold nanoparticles based on solid-state nanopores. Langmuir 2018, 34, 14825–14833. [Google Scholar] [CrossRef]
- Ng, A.; Chinnappan, R.; Eissa, S.; Liu, H.; Tlili, C.; Zourob, M. Selection, characterization, and biosensing application of high affinity congener-specific microcystin-targeting aptamers. Environ. Sci. Technol. 2012, 46, 10697–10703. [Google Scholar] [CrossRef]
- Wu, S.J.; Li, Q.; Duan, N.; Ma, H.L.; Wang, Z.P. DNA aptamer selection and aptamer-based fluorometric displacement assay for the hepatotoxin microcystin-RR. Microchim. Acta 2016, 183, 2555–2562. [Google Scholar] [CrossRef]
- Maguire, I.; Fitzgerald, J.; Heery, B.; Nwankire, C.; Regan, F. Novel microfluidic analytical sensing platform for the simultaneous detection of three algal toxins in water. Acs Omega 2018, 3, 6624–6634. [Google Scholar] [CrossRef]
- Sabet, F.S.; Hosseini, M.; Khabbaz, H.; Dadmehr, M.; Ganjali, M.R. FRET-based aptamer biosensor for selective and sensitive detection of aflatoxin B1 in peanut and rice. Food Chem. 2017, 220, 527–532. [Google Scholar] [CrossRef] [PubMed]
- AI-Rubaye, A.G.; Nabok, A.; Catanante, G.; Marty, J.L.; Takacs, E.; Szekacs, A. Label-free optical detection of mycotoxins using specific aptamers immobilized on gold nanostructures. Toxins 2018, 10, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Zhu, Z.; Li, B.; Liu, Z.; Jia, L.; Zuo, L.; Chen, L.; Zhu, Z.; Shan, G.; Luo, S.Z. A direct determination of AFBs in vinegar by aptamer-based surface plasmon resonance biosensor. Toxincon 2018, 146, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.P.; Li, C.; Wu, S.Q.; Zhang, Q.C. Label-free aptamer-based detection of microcystin-LR using a microcantilever array biosensor. Sens. Actuators B Chem. 2018, 260, 42–47. [Google Scholar] [CrossRef]
- Wu, K.; Ma, C.; Zhao, H.; He, H.; Chen, H. Label-free G-quadruplex aptamer fluorescence assay for ochratoxin A using a thioflavin T probe. Toxins 2018, 10, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frohnmeyer, E.; Tuschel, N.; Sitz, T.; Hermann, C.; Dahl, G.T.; Schulz, F.; Baeumner, A.J.; Fischer, M. Aptamer lateral flow assays for rapid and sensitive detection of cholera toxin. Analyst 2019, 144, 1840–1849. [Google Scholar] [CrossRef]
- Jeddi, I.; Saiz, L. Three-dimensional modeling of single stranded DNA hairpins for aptamer-based biosensors. Sci. Rep. 2017, 7, 1178. [Google Scholar] [CrossRef] [Green Version]
- Frohnmeyer, E.; Frisch, F.; Falke, S.; Betzel, C.; Fischer, M. Highly affine and selective aptamers against cholera toxin as capture elements in magnetic bead-based sandwich ELAA. J. Biotechnol. 2018, 269, 35–42. [Google Scholar] [CrossRef]
- Zhang, Z.; Tao, C.; Yin, J.; Wang, Y.; Li, Y. Enhancing the response rate of strand displacement-based electrochemical aptamer sensors using bivalent binding aptamer-cDNA probes. Biosens. Bioelectron. 2018, 103, 39–44. [Google Scholar] [CrossRef]
- Alizadeh, N.; Memar, M.Y.; Mehramuz, B.; Abibiqlou, S.S.; Hemmati, F.; Samadi, K.H. Current advances in aptamer-assisted technologies for detecting bacterial and fungal toxins. J. Appl. Microbiol. 2018, 124, 644–651. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.H.; Huang, Y.F.; Dong, Y.Y.; Han, X.T.; Wang, S.; Liang, X.G. Aptamers and aptasensors for highly specific recognition and sensitive detection of marine biotoxins: Recent advances and perspectives. Toxins 2018, 10, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogiazi, V.; Cruz, A.; Mishra, S.; Shanov, V.; Heineman, W.R.; Dionysiou, D. A comprehensive review: Development of electrochemical biosensors for detection of cyanotoxins in freshwater. ACS Sens. 2019, 4, 1151–1173. [Google Scholar] [CrossRef] [PubMed]
- Cunha, I.; Biltes, R.; Sales, M.G.F.; Vasconcelos, V. Aptamer-based biosensors to detect aquatic phycotoxins and cyanotoxins. Sensors 2018, 18, 2367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farzin, L.; Shamsipur, M.; Sheibani, S. A review: Aptamer-based analytical strategies using the nanomaterials for environmental and human monitoring of toxic heavy metals. Talanta 2017, 174, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wan, Z.; Zhong, L.; Li, X.; Wu, Q.; Wang, J.; Wang, P. Label-free okadaic acid detection using growth of gold nanoparticles in sensor gaps as a conductive tag. Biomed. Microdevices. 2017, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Ghows, N.; Shaegh, S.A.M.; Abnous, K. A novel fluorescent aptasensor for ultrasensitive detection of microcystin-LR based on single-walled carbon nanotubes and dapoxyl. Talanta 2017, 16, 187–192. [Google Scholar] [CrossRef]


| Marine Toxins | Toxicity (LD or LD50) Towards Mice | Toxic Mechanism | Aptasensor Type | LOD | LRD | Aptamer Sequences | The Immobilization Method | Reference |
|---|---|---|---|---|---|---|---|---|
| STX | LD50 = 10 μg/kg | binds to sodium channel proteins | graphene quantum dots | 0.1 ng/μL | 0.1–100 ng/μL | CTTTTTACAAAATTCTCTTTTTACCTATATTATGAACAGA | Physical absorption of MRGO | [42,43,44,45,73,77,78] |
| TTX | LD50 = 10 μg/kg | blocks nerve conduction | aptamer fluorochrome (EvaGreen) | 1 μM | TCAAATTTTCGTCTACTCAATCTTTCTGTCTTATC | ___ | [49,50,78,80] | |
| OA | LD50 = 166 μg/kg | activating cAMP mediator system, inhibiting PP1A, PP2A | Fluorescence combined with rolling cycle amplification | 10 pg/mL | 0.01–100 ng/mL | GGTCACCAACAACAGGGAGCGCTACGCGAAGGGTCAATGTGACGTCATGCGGATGTGTGG | The binding of biotin-labeled aptamer to streptavidin-catalase complex | [46,47,71,78,79] |
| PTX | LD = 2.3-31.5 μg/kg | activating cells to release potassium ions rapidly | Biolayer Interferometry (AR2G biosensor) | 0.04 pg/mL | 200–700 pg/mL | ACCGACCGTGCTGGACTCAGGAGGTGGTGGGGACTTTGCTTGTACTGGGCGCCCGGTTGAAACTATGAGCGAGCCTGGCG | EDC/NHS method | [51,52,81] |
| BTX | LD50 = 55.36 mg/kg | open and activate sodium channel | label-free impedimetric biosensor (electrochemical biosensor) | 106 pg/mL | GGCCACCAAACCACACCGTCGCAACCGCGAGAACCGAAGTAGTGATCATGTCCCTGCGTG | The BTX was immobilized on cysteamine-modified gold electrodes | [55,56,85] | |
| DA | LD50 = 10 mg/kg | bind with the receptor of glutamic neurotransmitter | ___ | ___ | ___ | [53,54,58,59,78] | ||
| DTX-2 | LD50 = 338 μg/kg | inhibiting PP2A | ___ | ___ | ___ | ___ | [60,88] | |
| CYN | LD50 = 200 μg/kg | Inhibit the synthesis of protein, glutathione and P450 activity | Label free impedimetric biosensor (Graphene-Based biosensor) | 1.9 pM | GGCATCAGGCAACAACCGATGGTCCGGCCACCCTAACAACCAGCCCACCCACCACCCCGCCG | utilizs an unlabeled aptamer noncovalently assembled on a graphene electrode | [85,86,87] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, W.; Liu, T.; Zhang, W.; Zhu, M.; Liu, Z.; Kong, Y.; Liu, S. Marine Toxins Detection by Biosensors Based on Aptamers. Toxins 2020, 12, 1. https://doi.org/10.3390/toxins12010001
Ye W, Liu T, Zhang W, Zhu M, Liu Z, Kong Y, Liu S. Marine Toxins Detection by Biosensors Based on Aptamers. Toxins. 2020; 12(1):1. https://doi.org/10.3390/toxins12010001
Chicago/Turabian StyleYe, Wei, Taomei Liu, Weimin Zhang, Muzi Zhu, Zhaoming Liu, Yali Kong, and Shan Liu. 2020. "Marine Toxins Detection by Biosensors Based on Aptamers" Toxins 12, no. 1: 1. https://doi.org/10.3390/toxins12010001
APA StyleYe, W., Liu, T., Zhang, W., Zhu, M., Liu, Z., Kong, Y., & Liu, S. (2020). Marine Toxins Detection by Biosensors Based on Aptamers. Toxins, 12(1), 1. https://doi.org/10.3390/toxins12010001