Looking for the X Factor in Bacterial Pathogenesis: Association of orfX-p47 Gene Clusters with Toxin Genes in Clostridial and Non-Clostridial Bacterial Species
Abstract
:1. Introduction
2. Results and Discussion
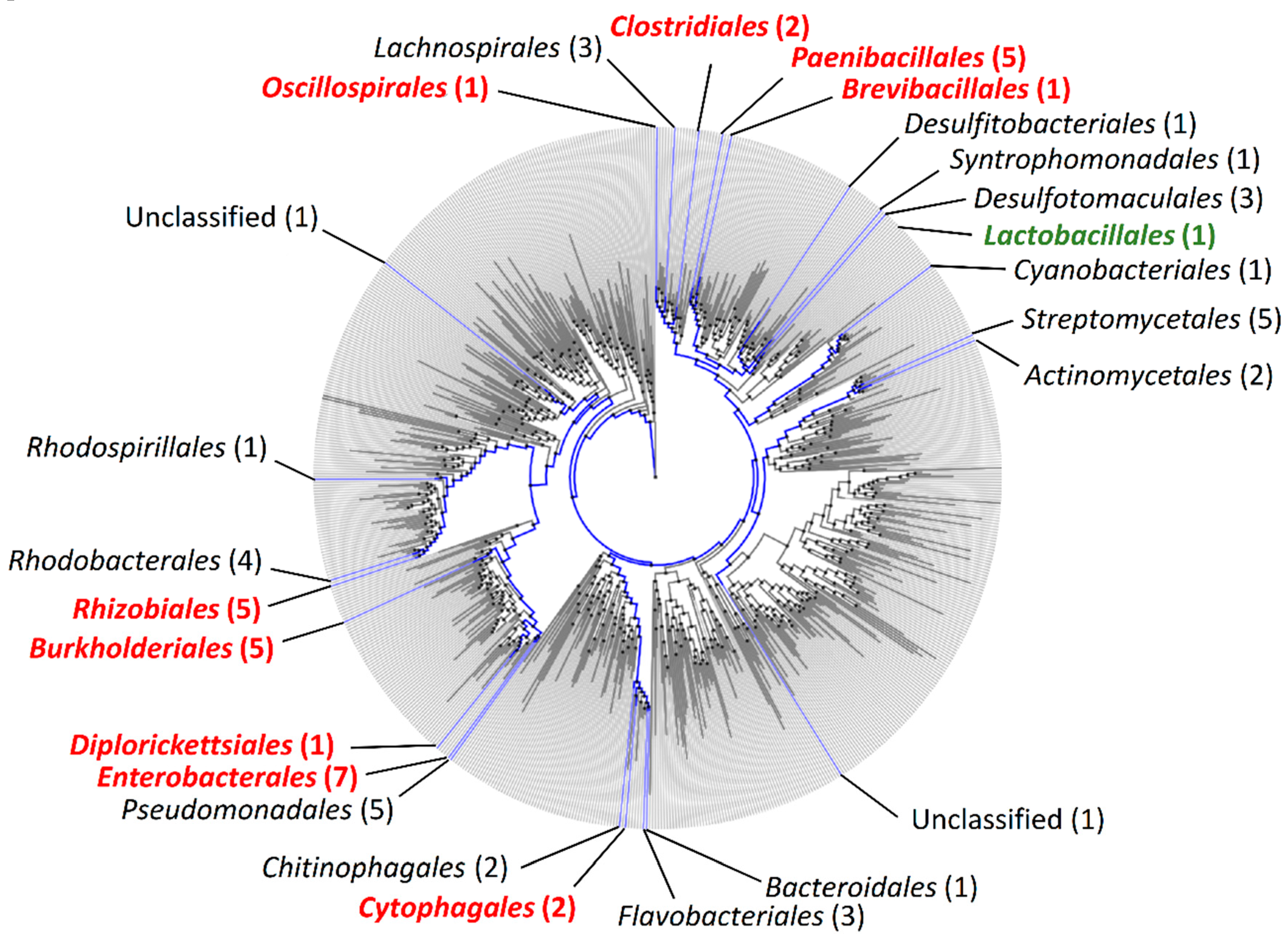
2.1. Phylogenetic Distribution of orfX1, orfX2, orfX3, and p47 in Bacteria
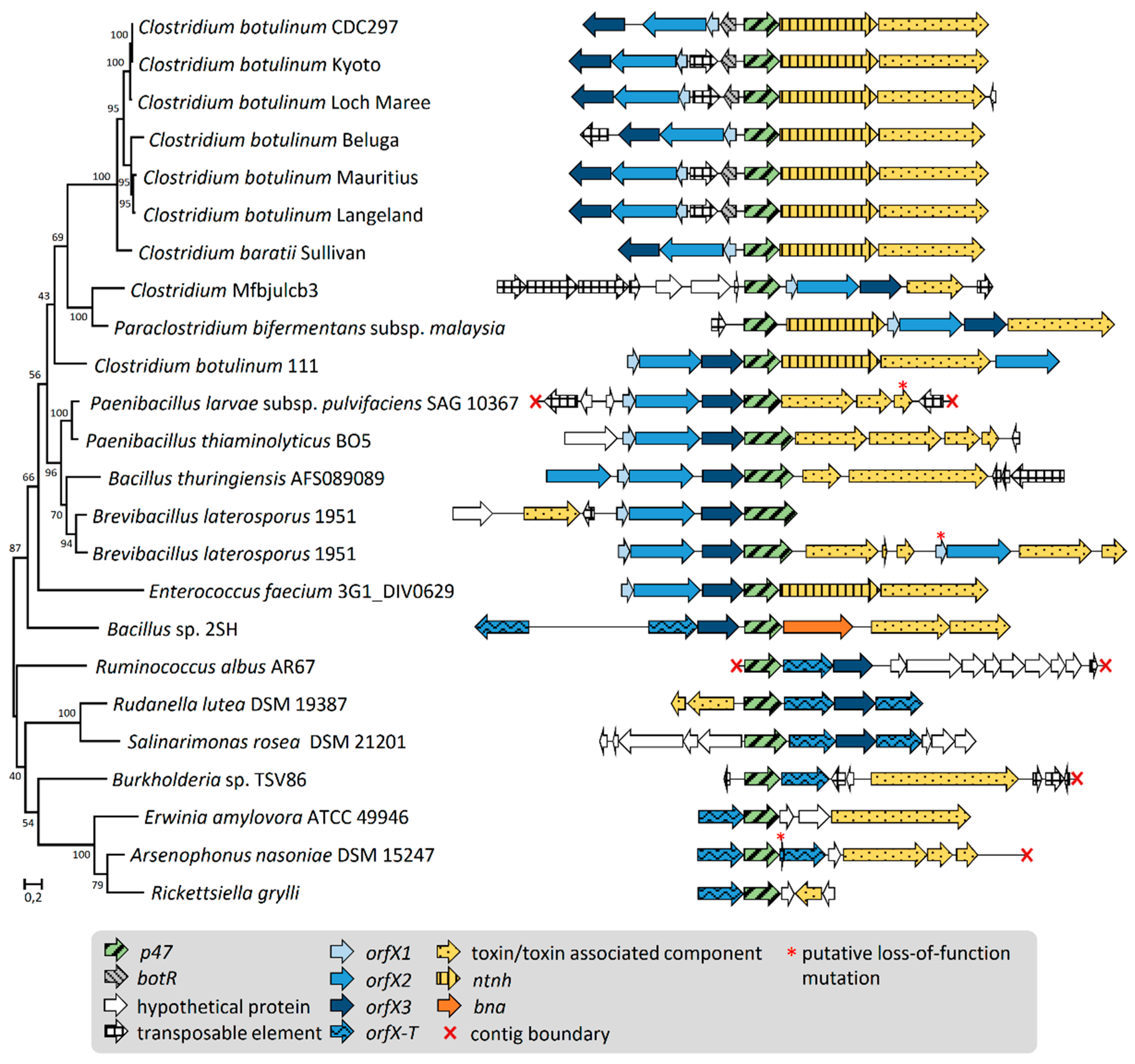
2.2. Gene Organization and Arrangement of orfX1, orfX2, orfX3, and p47
2.3. P47, OrfX1, OrfX2, and OrfX3 Have a Common Origin
2.4. Association of orfX1, orfX2, orfX3, and p47 with Toxin Genes
2.5. Identification of BoNT/NTNH-Like Protein in Bacillus sp. 2SH
3. Conclusions
4. Materials and Methods
4.1. Sequence Database Mining and Conserved Domain Analysis
4.2. Comparative Sequence Analysis, Motif-Based Sequence Analysis, and Phylogenetic Tree Analysis
4.3. Sequence Analysis of the Putative Toxin Gene Cluster in Bacillus sp. 2SH and Structural Modeling
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sobel, J. Botulism. Clin. Infect. Dis. 2005, 41, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.; Kiviniemi, K.; Korkeala, H. Hazard and control of group II (non-proteolytic) Clostridium botulinum in modem food processing. Int. J. Food Microbiol. 2006, 108, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.; Fredriksson-Ahomaa, M.; Korkeala, H. Molecular epidemiology of group I and II Clostridium botulinum. In Clostridia: Molecular Biology in the Post-Genomic Era; Holger Brüggemann, G.G., Ed.; Caister Academic Press: Poole, UK, 2009; pp. 103–130. [Google Scholar]
- Montecucco, C.; Molgo, J. Botulinal neurotoxins: Revival of an old killer. Curr. Opin. Pharmacol. 2005, 5, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.K.; Xie, G.; Foley, B.T.; Smith, T.J.; Munk, A.C.; Bruce, D.; Smith, L.A.; Brettin, T.S.; Detter, J.C. Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol. 2009, 7, 66. [Google Scholar] [CrossRef]
- Hill, K.K.; Smith, T.J. Genetic diversity within Clostridium botulinum serotypes, botulinum neurotoxin gene clusters and toxin subtypes. Curr. Top. Microbiol. 2013, 364, 1–20. [Google Scholar]
- Gu, S.; Rumpel, S.; Zhou, J.; Strotmeier, J.; Bigalke, H.; Perry, K.; Shoemaker, C.B.; Rummel, A.; Jin, R. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science 2012, 335, 977–981. [Google Scholar] [CrossRef] [Green Version]
- Ito, H.; Sagane, Y.; Miyata, K.; Inui, K.; Matsuo, T.; Horiuchi, R.; Ikeda, T.; Suzuki, T.; Hasegawa, K.; Kouguchi, H.; et al. HA-33 facilitates transport of the serotype D botulinum toxin across a rat intestinal epithelial cell monolayer. FEMS Immunol. Med. Microbiol. 2011, 61, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Jin, R. Assembly and function of the botulinum neurotoxin progenitor complex. Curr. Top. Microbiol. 2013, 364, 21–44. [Google Scholar]
- Fujinaga, Y.; Inoue, K.; Watanabe, S.; Yokota, K.; Hirai, Y.; Nagamachi, E.; Oguma, K. The haemagglutinin of Clostridium botulinum type C progenitor toxin plays an essential role in binding of toxin to the epithelial cells of guinea pig small intestine, leading to the efficient absorption of the toxin. Microbiology 1997, 143, 3841–3847. [Google Scholar] [CrossRef] [Green Version]
- Fujinaga, Y.; Inoue, K.; Watarai, S.; Sakaguchi, Y.; Arimitsu, H.; Lee, J.C.; Jin, Y.; Matsumura, T.; Kabumoto, Y.; Watanabe, T.; et al. Molecular characterization of binding subcomponents of Clostridium botulinum type C progenitor toxin for intestinal epithelial cells and erythrocytes. Microbiology 2004, 150, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Niwa, K.; Koyama, K.; Inoue, S.; Suzuki, T.; Hasegawa, K.; Watanabe, T.; Ikeda, T.; Ohyama, T. Role of nontoxic components of serotype D botulinum toxin complex in permeation through a Caco-2 cell monolayer, a model for intestinal epithelium. FEMS Immunol. Med. Microbiol. 2007, 49, 346–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafsson, R.; Berntsson, R.P.; Martinez-Carranza, M.; El Tekle, G.; Odegrip, R.; Johnson, E.A.; Stenmark, P. Crystal structures of OrfX2 and P47 from a botulinum neurotoxin OrfX-type gene cluster. FEBS Lett. 2017, 591, 3781–3792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, K.H.; Qi, R.; Liu, S.; Kroh, A.; Yao, G.; Perry, K.; Rummel, A.; Jin, R. The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein. Toxicon 2018, 147, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Kalb, S.R.; Baudys, J.; Smith, T.J.; Smith, L.A.; Barr, J.R. Characterization of hemagglutinin negative botulinum progenitor toxins. Toxins 2017, 9, 193. [Google Scholar] [CrossRef] [Green Version]
- Marvaud, J.C.; Gibert, M.; Inoue, K.; Fujinaga, Y.; Oguma, K.; Popoff, M.R. BotR/A is a positive regulator of botulinum neurotoxin and associated non-toxin protein genes in Clostridium botulinum A. Mol. Microbiol. 1998, 29, 1009–1018. [Google Scholar] [CrossRef]
- Couesnon, A.; Raffestin, S.; Popoff, M.R. Expression of botulinum neurotoxins A and E, and associated non-toxin genes, during the transition phase and stability at high temperature: Analysis by quantitative reverse transcription-PCR. Microbiology 2006, 152, 759–770. [Google Scholar] [CrossRef] [Green Version]
- Williamson, C.H.; Sahl, J.W.; Smith, T.J.; Xie, G.; Foley, B.T.; Smith, L.A.; Fernandez, R.A.; Lindström, M.; Korkeala, H.; Keim, P.; et al. Comparative genomic analyses reveal broad diversity in botulinum-toxin-producing clostridia. BMC Genom. 2016, 17, 180. [Google Scholar] [CrossRef] [Green Version]
- Campbell, K.; Collins, M.D.; East, A.K. Nucleotide sequence of the gene coding for Clostridium botulinum (Clostridium argentinense) type G neurotoxin: Genealogical comparison with other clostridial neurotoxins. Biochim. Biophys. Acta 1993, 1216, 487–491. [Google Scholar] [CrossRef]
- Zhang, S.; Masuyer, G.; Zhang, J.; Shen, Y.; Lundin, D.; Henriksson, L.; Miyashita, S.I.; Martinez-Carranza, M.; Dong, M.; Stenmark, P. Identification and characterization of a novel botulinum neurotoxin. Nat. Commun. 2017, 8, 14130. [Google Scholar] [CrossRef]
- Brunt, J.; Carter, A.T.; Stringer, S.C.; Peck, M.W. Identification of a novel botulinum neurotoxin gene cluster in Enterococcus. FEBS Lett. 2018, 592, 310–317. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, M.J.; Wentz, T.G.; Zhang, S.; Lee, E.J.; Dong, M.; Sharma, S.K.; Doxey, A.C. Bioinformatic discovery of a toxin family in Chryseobacterium piperi with sequence similarity to botulinum neurotoxins. Sci. Rep. 2019, 9, 1634. [Google Scholar] [CrossRef] [PubMed]
- Contreras, E.; Masuyer, G.; Qureshi, N.; Chawla, S.; Dhillon, H.S.; Lee, H.L.; Chen, J.; Stenmark, P.; Gill, S.S. A neurotoxin that specifically targets Anopheles mosquitoes. Nat. Commun. 2019, 10, 2869. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Hill, K.K.; Foley, B.T.; Detter, J.C.; Munk, A.C.; Bruce, D.C.; Doggett, N.A.; Smith, L.A.; Marks, J.D.; Xie, G.; et al. Analysis of the neurotoxin complex genes in Clostridium botulinum A1-A4 and B1 strains: BoNT/A3,/Ba4 and/B1 clusters are located within plasmids. PLoS ONE 2007, 2, e1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, T.J.; Hill, K.K.; Xie, G.; Foley, B.T.; Williamson, C.H.D.; Foster, J.T.; Johnson, S.L.; Chertkov, O.; Teshima, H.; Gibbons, H.S.; et al. Genomic sequences of six botulinum neurotoxin-producing strains representing three clostridial species illustrate the mobility and diversity of botulinum neurotoxin genes. Infect. Genet. Evol. 2015, 30, 102–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosomi, K.; Sakaguchi, Y.; Kohda, T.; Gotoh, K.; Motooka, D.; Nakamura, S.; Umeda, K.; Iida, T.; Kozaki, S.; Mukamoto, M. Complete nucleotide sequence of a plasmid containing the botulinum neurotoxin gene in Clostridium botulinum type B strain 111 isolated from an infant patient in Japan. Mol. Genet. Genom. 2014, 289, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Pedron, R.; Esposito, A.; Bianconi, I.; Pasolli, E.; Tett, A.; Asnicar, F.; Cristofolini, M.; Segata, N.; Jousson, O. Genomic and metagenomic insights into the microbial community of a thermal spring. Microbiome 2019, 7, 8. [Google Scholar] [CrossRef]
- Glare, T.R.; Durrant, A.; Berry, C.; Palma, L.; Ormskirk, M.M.; Cox, M.P. Phylogenetic determinants of toxin gene distribution in genomes of Brevibacillus laterosporus. Genomics 2019. [Google Scholar] [CrossRef]
- Sebaihia, M.; Bocsanczy, A.M.; Biehl, B.S.; Quail, M.A.; Perna, N.T.; Glasner, J.D.; DeClerck, G.A.; Cartinhour, S.; Schneider, D.J.; Bentley, S.D.; et al. Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J. Bacteriol. 2010, 192, 2020–2021. [Google Scholar] [CrossRef] [Green Version]
- Dingman, D.W. Four complete Paenibacillus larvae genome sequences. Genome Announc. 2017, 5, e00407–e00417. [Google Scholar] [CrossRef] [Green Version]
- Grubbs, K.J.; Bleich, R.M.; Santa Maria, K.C.; Allen, S.E.; Farag, S.; Shank, E.A.; Bowers, A.A. Large-scale bioinformatics analysis of Bacillus genomes uncovers conserved roles of natural products in bacterial physiology. mSystems 2017, 2, e00040-17. [Google Scholar] [CrossRef] [Green Version]
- Hazes, B. The (QxW)3 domain: A flexible lectin scaffold. Protein Sci. 1996, 5, 1490–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arndt, J.W.; Gu, J.; Jaroszewski, L.; Schwarzenbacher, R.; Hanson, M.A.; Lebeda, F.J.; Stevens, R.C. The structure of the neurotoxin-associated protein HA33/A from Clostridium botulinum suggests a reoccurring beta-trefoil fold in the progenitor toxin complex. J. Mol. Biol. 2005, 346, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Paaventhan, P.; Joseph, J.S.; Seow, S.V.; Vaday, S.; Robinson, H.; Chua, K.Y.; Kolatkar, P.R. A 1.7A structure of Fve, a member of the new fungal immunomodulatory protein family. J. Mol. Biol. 2003, 332, 461–470. [Google Scholar] [CrossRef]
- Chen, W.J.; Hsieh, F.C.; Hsu, F.C.; Tasy, Y.F.; Liu, J.R.; Shih, M.C. Characterization of an insecticidal toxin and pathogenicity of Pseudomonas taiwanensis against insects. PLoS Pathog. 2014, 10, e1004288. [Google Scholar] [CrossRef]
- Waterfield, N.R.; Bowen, D.J.; Fetherston, J.D.; Perry, R.D.; ffrench-Constant, R.H. The tc genes of Photorhabdus: A growing family. Trends Microbiol. 2001, 9, 185–191. [Google Scholar] [CrossRef]
- Reyes, A.G.; Anne, J.; Mejia, A. Ribosome-inactivating proteins with an emphasis on bacterial RIPs and their potential medical applications. Future Microbiol. 2012, 7, 705–717. [Google Scholar] [CrossRef]
- Sadana, P.; Geyer, R.; Pezoldt, J.; Helmsing, S.; Huehn, J.; Hust, M.; Dersch, P.; Scrima, A. The invasin D protein from Yersinia pseudotuberculosis selectively binds the Fab region of host antibodies and affects colonization of the intestine. J. Biol. Chem. 2018, 293, 8672–8690. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, M.J.; Adams, J.B.; Doxey, A.C. Botulinum neurotoxin homologs in non-Clostridium species. FEBS Lett. 2015, 589, 342–348. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Infante, N.; Moreno-Fierros, L. An overview of the safety and biological effects of Bacillus thuringiensis Cry toxins in mammals. J. Appl. Toxicol. 2016, 36, 630–648. [Google Scholar] [CrossRef]
- Leuber, M.; Orlik, F.; Schiffler, B.; Sickmann, A.; Benz, R. Vegetative insecticidal protein (Vip1Ac) of Bacillus thuringiensis HD201: Evidence for oligomer and channel formation. Biochemistry 2006, 45, 283–288. [Google Scholar] [CrossRef]
- Han, S.; Craig, J.A.; Putnam, C.D.; Carozzi, N.B.; Tainer, J.A. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Struct. Biol. 1999, 6, 932–936. [Google Scholar] [PubMed]
- Barth, H.; Hofmann, F.; Olenik, C.; Just, I.; Aktories, K. The N-terminal part of the enzyme component (C2I) of the binary Clostridium botulinum C2 toxin interacts with the binding component C2II and functions as a carrier system for a Rho ADP-ribosylating C3-like fusion toxin. Infect. Immun. 1998, 66, 1364–1369. [Google Scholar] [PubMed]
- Wilkes, T.E.; Darby, A.C.; Choi, J.H.; Colbourne, J.K.; Werren, J.H.; Hurst, G.D.D. The draft genome sequence of Arsenophonus nasoniae, son-killer bacterium of Nasonia vitripennis, reveals genes associated with virulence and symbiosis. Insect Mol. Biol. 2010, 19, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Binz, T.; Bade, S.; Rummel, A.; Kollewe, A.; Alves, J. Arg (362) and Tyr (365) of the botulinum neurotoxin type a light chain are involved in transition state stabilization. Biochemistry 2002, 41, 1717–1723. [Google Scholar] [CrossRef] [PubMed]
- Dover, N.; Barash, J.R.; Hill, K.K.; Xie, G.; Arnon, S.S. Molecular characterization of a novel botulinum neurotoxin type H gene. J. Infect. Dis. 2014, 209, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Montal, M. Crucial role of the disulfide bridge between botulinum neurotoxin light and heavy chains in protease translocation across membranes. J. Biol. Chem. 2007, 282, 29604–29611. [Google Scholar] [CrossRef] [Green Version]
- Depaiva, A.; Poulain, B.; Lawrence, G.W.; Shone, C.C.; Tauc, L.; Dolly, J.O. A role for the interchain disulfide or its participating thiols in the internalization of botulinum neurotoxin a revealed by a toxin derivative that binds to ecto-acceptors and inhibits transmitter release intracellularly. J. Biol. Chem. 1993, 268, 20838–20844. [Google Scholar]
- Schiavo, G.; Rossetto, O.; Santucci, A.; Dasgupta, B.R.; Montecucco, C. Botulinum neurotoxins are zinc proteins. J. Biol. Chem. 1992, 267, 23479–23483. [Google Scholar]
- Collins, M.D.; East, A.K. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J. Appl. Microbiol. 1998, 84, 5–17. [Google Scholar] [CrossRef]
- Doxey, A.C.; Lynch, M.D.J.; Muller, K.M.; Meiering, E.M.; McConkey, B.J. Insights into the evolutionary origins of clostridial neurotoxins from analysis of the Clostridium botulinum strain a neurotoxin gene cluster. BMC Evol. Biol. 2008, 8, 316. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; de Souza, R.F.; Anantharaman, V.; Iyer, L.M.; Aravind, L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowen, D.; Rocheleau, T.A.; Blackburn, M.; Andreev, O.; Golubeva, E.; Bhartia, R.; ffrench-Constant, R.H. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 1998, 280, 2129–2132. [Google Scholar] [CrossRef] [PubMed]
- Waterfield, N.; Hares, M.; Yang, G.; Dowling, A.; ffrench-Constant, R.H. Potentiation and cellular phenotypes of the insecticidal toxin complexes of Photorhabdus bacteria. Cell. Microbiol. 2005, 7, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Hoch, D.H.; Romero-Mira, M.; Ehrlich, B.E.; Finkelstein, A.; DasGupta, B.R.; Simpson, L.L. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: Relevance to translocation of proteins across membranes. Proc. Natl. Acad. Sci. USA 1985, 82, 1692–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roderer, D.; Raunser, S. Tc toxin complexes: Assembly, membrane permeation, and protein translocation. Annu. Rev. Microbiol. 2019, 73, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Owen-Hughes, T. Snf2-family proteins: Chromatin remodellers for any occasion. Curr. Opin. Chem. Biol. 2011, 15, 649–656. [Google Scholar] [CrossRef] [Green Version]
- Sergeant, M.; Jarrett, P.; Ousley, M.; Morgan, J.A. Interactions of insecticidal toxin gene products from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 2003, 69, 3344–3349. [Google Scholar] [CrossRef] [Green Version]
- Busby, J.N.; Panjikar, S.; Landsberg, M.J.; Hurst, M.R.; Lott, J.S. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 2013, 501, 547–550. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Mendler, K.; Chen, H.; Parks, D.H.; Lobb, B.; Hug, L.A.; Doxey, A.C. AnnoTree: Visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 2019, 47, 4442–4448. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
| Bacterial Species | Strain Name | Taxonomy (Class) | Isolation Source | Accession Number | Reference |
|---|---|---|---|---|---|
| Clostridium botulinum | Beluga | Clostridia | Fermented whale flippers, Canada | NZ_ACSC01000002 | Direct submission |
| Clostridium botulinum | Kyoto-F | Clostridia | Infant feces, Japan | CP001581 | [24] |
| Clostridium botulinum | CDC_297 | Clostridia | Liver paste, USA | CP006907 | [25] |
| Clostridium botulinum | 111 | Clostridia | Infant feces, Japan | AP014696 | [26] |
| Clostridium botulinum | Loch Maree | Clostridia | Duck liver paste, Scotland | CP000962 | [24] |
| Clostridium botulinum | Mauritius | Clostridia | Fish, Mauritius | NZ_LFPL01000000 | [18] |
| Clostridium botulinum | Langeland | Clostridia | Liver paste, Denmark | CP000728 | Direct submission |
| Clostridium botulinum | Mfbjulcb3 | Clostridia | Retail fish market, India | CP027780 | Direct submission |
| Clostridium baratii | Sullivan | Clostridia | Adult human feces, USA | CP006905 | [25] |
| Enterococcus faecium | 3G1_DIV0629 | Bacilli | Cow feces, USA | NGLI00000000 | Direct submission |
| Paraclostridium bifermentans subsp. malaysia | Pbm | Clostridia | Swamp soil, Malaysia | CM017269 | [23] |
| Arsenophonus nasoniae | DSM 15247 | Gammaproteobacteria | Son-killer of Nasonia vitripennis, USA | AUCC00000000 | Direct submission |
| Bacillus sp. | 2SH | Bacilli | Alpine fresh spring, Italy | SCNA01000023 | [27] |
| Brevibacillus laterosporus | 1951 | Bacilli | Forage rape seed, New Zealand | RHPK00000000 | [28] |
| Burkholderia sp. | TSV86 | Betaproteobacteria | Water, Australia | GCA_001522865 | Direct submission |
| Erwinia amylovora | ATCC 49946 | Gammaproteobacteria | Infected apple tree, USA | FN666575 | [29] |
| Paenibacillus larvae subsp. pulvifaciens | SAG 10367 | Bacilli | Apis mellifera (honeybee), Chile | NZ_CP020557 | [30] |
| Paenibacillus thiaminolyticus | BO5 | Bacilli | Soil, Russia | GCA_003591545.1 | Direct submission |
| Rickettsiella grylli | - | Gammaproteobacteria | Pill bugs, USA | NZ_AAQJ00000000 | Direct Submission |
| Rudanella lutea | DSM 19387 | Cytophagia | Air sample, South Korea | NZ_ARPG00000000 | Direct submission |
| Ruminococcus albus | AR67 | Clostridia | Sheep rumen, New Zealand | GCA_900112155 | Direct submission |
| Salinarimonas rosea | DSM 21201 | Alphaproteobacteria | Salt mine sediment, China | NZ_AUBC00000000 | Direct submission |
| Bacillus thuringiensis | AFS089089 | Bacilli | Grainbin dust, USA | NVNL01000046 | [31] |
| Bacterial Strain | OrfX-assisting Gene | Protein ID | Predicted Annotation | NCBI Conserved Domain Search Output (Accession Number) | E-score |
|---|---|---|---|---|---|
| Clostridium botulinum Beluga | bont/E | EES49627.1 | BoNT type E | Clostridial neurotoxin, translocation domain (cl06820) | 3.07 × 10−93 |
| Clostridial neurotoxin zinc protease (cl15546) | 1.10 × 10−79 | ||||
| Clostridial neurotoxin, N-terminal receptor binding (PF07953) | 9.90 × 10−68 | ||||
| Clostridial neurotoxin, C-terminal receptor binding (cl08467) | 3.90 × 10−22 | ||||
| ntnh/E | EES49602.1 | NTNH protein | Clostridial neurotoxin zinc protease (cl15546) | 1.15 × 10−83 | |
| Laminin G domain (cl22861) | 1.79 × 10−47 | ||||
| Non-toxic non-hemagglutinin C-terminal (cl07187) | 2.43 × 10−40 | ||||
| Clostridium Mfbjulcb3 | C7M59_04110 | AVQ52086.1 | Crystal insecticidal protein (Cry)/insecticidal delta-endotoxin | Delta-endotoxin, C-terminal domain (cd04085) | 5.38 × 10−39 |
| Delta-endotoxin (cl15971) | 5.32 × 10−14 | ||||
| Delta-endotoxin, N-terminal domain (cl04339) | 2.11 × 10−13 | ||||
| Arsenophonus nasoniae DSM 15247 | NNa | WP_026823093.1 | RHS repeat protein | RHS Repeat (PF05593) | 1.19 × 10−5 |
| Uncharacterized conserved protein RhaS (COG3209) | 3.84 × 10−3 | ||||
| NN | WP_026823094.1 | RHS repeat protein | - | - | |
| NN | WP_081700660.1 | RHS repeat protein | RHS repeat-associated core domain (cl37315) | 2.68 × 10−12 | |
| Beta-eliminating lyase (cl18945) | 2.98 × 10−3 | ||||
| Bacillus sp. 2SH | BNAb | WP_137842862.1 | BoNT/NTNH-like A component (BNA) | Clostridial neurotoxin zinc protease (cl15546) | 3.55 × 10−34 |
| Clostridial neurotoxin, translocation domain (cl06820) | 1.14 × 10−26 | ||||
| NN | WP_137842861.1 | RHS repeat protein | - | - | |
| NN | WP_137842860.1 | RHS repeat protein | RHS repeat-associated core domain (TIGR03696) | 2.88 × 10−26 | |
| Bacterial SNF2 helicase associated domain (cl07173) | 1.81 × 10−3 | ||||
| Brevibacillus laterosporus 1951 | EEL31_08340 (cluster I) | TPG68525.1 | Crystal insecticidal protein (Cry)/insecticidal delta-endotoxin | Delta-endotoxin, C-terminal domain (cd04085) | 1.86 × 10−22 |
| Delta-endotoxin (cl15971) | 1.13 × 10−13 | ||||
| Delta-endotoxin, N-terminal domain (cl04339) | 1.03 × 10−11 | ||||
| EEL31_17680 (cluster II) | TPG70133.1 | Binary toxin/vegetative insecticidal protein (VIP1) | Clostridial binary toxin B/anthrax toxin PA domain 2 (cl38748) | 2.52 × 10−39 | |
| Clostridial binary toxin B/anthrax toxin PA Ca-binding domain (cl09551) | 2.12 × 10−14 | ||||
| Clostridial binary toxin B/anthrax toxin PA domain 3 (cl38749) | 6.89 × 10−12 | ||||
| EEL31_17670 (cluster II) | TPG71603.1 | Vegetative insecticidal protein (VIP2) | VIP2, ADP-ribosyltransferase exoenzyme (cl00173) | 4.71 × 10−51 | |
| Clostridial binary toxin B/anthrax toxin PA domain 2 (cl38748) | 1.60 × 10−39 | ||||
| EEL31_17650 (cluster II) | TPG70130.1 | Binary toxin/vegetative insecticidal protein (VIP1) | Clostridial binary toxin B/anthrax toxin PA Ca-binding domain (cl09551) | 1.44 × 10−14 | |
| Clostridial binary toxin B/anthrax toxin PA domain 3 (cl38749) | 3.11 × 10−8 | ||||
| EEL31_17645 (cluster II) | TPG70129.1 | Vegetative insecticidal protein (VIP2) | VIP2, ADP-ribosyltransferase exoenzyme (cl00173) | 3.92 × 10−66 | |
| Burkholderia sp. TSV86 | WS68_18250 | WP_059573479.1 | Autotransporter protein | Outer membrane autotransporter barrel domain (cl36898) | 5.76 × 10−52 |
| Autotransport protein MisL (cl36477) | 1.07 × 10−19 | ||||
| Large exoprotein involved in heme utilization or adhesion (COG3210) | 2.08 × 10−6 | ||||
| Extended signal peptide of type V secretion system (PF13018) | 4.04 × 10−5 | ||||
| Erwinia amylovora ATCC 49946 | EAM_RS01885 | WP_004160289.1 | RHS repeat protein | RHS repeat-associated core domain (TIGR03696) | 1.12 × 10−23 |
| Uncharacterized conserved protein RhaS (COG3209) | 8.37 × 10−8 | ||||
| Paenibacillus larvae subsp. pulvifaciens SAG 10367 | B7C51_09885 | ARF68072.1 | Binary toxin/vegetative insecticidal protein (VIP1) | Clostridial binary toxin B/anthrax toxin PA domain 2 (cl38748) | 4.05 × 10−40 |
| Clostridial binary toxin B/anthrax toxin PA Ca-binding domain (cl09551) | 1.82 × 10−10 | ||||
| Clostridial binary toxin B/anthrax toxin PA domain 3 (cl38749) | 6.37 × 10−7 | ||||
| PA14 domain (cl08459) | 3.38 × 10−5 | ||||
| B7C51_09880 | ARF68071.1 | Vegetative insecticidal protein (VIP2) | VIP2, ADP-ribosyltransferase exoenzyme (cl00173) | 1.86 × 10−8 | |
| Anthrax toxin lethal factor (cl08465) | 2.48 × 10−3 | ||||
| B7C51_09875 | NN | Anthrax toxin lethal factor/vegetative insecticidal protein (VIP2) | VIP2, ADP-ribosyltransferase exoenzyme (PF03496) | 3.59 × 10−62 | |
| Paenibacillus thiaminolyticus BO5 | DQX05_07030 | WP_119792154.1 | Binary toxin/vegetative insecticidal protein (VIP1) | Clostridial binary toxin B/anthrax toxin PA domain 2 (cl38748) | 1.14 × 10−39 |
| Clostridial binary toxin B/anthrax toxin PA Ca-binding domain (cl09551) | 5.76 × 10−15 | ||||
| Clostridial binary toxin B/anthrax toxin PA domain 3 (cl38749) | 1.63 × 10−7 | ||||
| PA14 domain (cl08459) | 1.49 × 10−4 | ||||
| Ricin-type beta-trefoil lectin domain-like (PF14200) | 4.93 × 10−3 | ||||
| DQX05_07025 | WP_119792152.1 | Binary toxin/vegetative insecticidal protein (VIP1) | Clostridial binary toxin B/anthrax toxin PA domain 2 (cl38748) | 2.27 × 10−41 | |
| Clostridial binary toxin B/anthrax toxin PA Ca-binding domain (cl09551) | 1.32 × 10−15 | ||||
| Clostridial binary toxin B/anthrax toxin PA domain 3 (cl38749) | 2.02 × 10−6 | ||||
| DQX05_07020 | WP_119792150 | Vegetative insecticidal protein (VIP2) | VIP2, ADP-ribosyltransferase exoenzyme (cl00173) | 1.20 × 10−6 | |
| DQX05_07015 | WP_119792149 | Anthrax toxin lethal factor/vegetative insecticidal protein (VIP2) | Anthrax toxin lethal factor (cl08465) | 6.60 × 10−4 | |
| VIP2, ADP-ribosyltransferase exoenzyme (cl00173) | 5.05 × 10−63 | ||||
| Rickettsiella grylli | RICGR_0720 | WP_081441678.1 | Shiga toxin A-chain (rRNA N-glycosidase) | Ribosome inactivating protein (cl08249) | 7.84 × 10−20 |
| Rudanella lutea DSM 19387 | NN | WP_019988042.1 | BIG-5 domain containing protein | Bacterial Ig-like domain, BIG5 (PF13205) | 1.62 × 10−16 |
| NN | WP_019988043.1 | Low affinity iron permease | Low affinity iron permease (PF04120) | 1.20 × 10−70 | |
| Ruminococcus albus AR67 | SAMN02910406 _03599 | WP_074963339.1 | Starch-binding protein | Uncharacterized conserved protein YjdB, contains Ig-like domain (COG5492) | 1.24 × 10−10 |
| Starch-binding module 26 (PF16738) | 1.92 × 10−8 | ||||
| Bacillus thuringiensis AFS089089 | CON71_23765 | WP_098902378.1 | Crystal insecticidal protein (Cry) | Insecticidal crystal toxin, P42 (cl05149) | 5.68 × 10−11 |
| CON71_23770 | WP_098902379.1 | RHS repeat protein | RHS repeat-associated core domain (TIGR03696) | 7.79 × 10−25 | |
| Uncharacterized conserved protein RhaS (COG3209) | 7.58 × 10−7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowakowska, M.B.; Douillard, F.P.; Lindström, M. Looking for the X Factor in Bacterial Pathogenesis: Association of orfX-p47 Gene Clusters with Toxin Genes in Clostridial and Non-Clostridial Bacterial Species. Toxins 2020, 12, 19. https://doi.org/10.3390/toxins12010019
Nowakowska MB, Douillard FP, Lindström M. Looking for the X Factor in Bacterial Pathogenesis: Association of orfX-p47 Gene Clusters with Toxin Genes in Clostridial and Non-Clostridial Bacterial Species. Toxins. 2020; 12(1):19. https://doi.org/10.3390/toxins12010019
Chicago/Turabian StyleNowakowska, Maria B., François P. Douillard, and Miia Lindström. 2020. "Looking for the X Factor in Bacterial Pathogenesis: Association of orfX-p47 Gene Clusters with Toxin Genes in Clostridial and Non-Clostridial Bacterial Species" Toxins 12, no. 1: 19. https://doi.org/10.3390/toxins12010019
APA StyleNowakowska, M. B., Douillard, F. P., & Lindström, M. (2020). Looking for the X Factor in Bacterial Pathogenesis: Association of orfX-p47 Gene Clusters with Toxin Genes in Clostridial and Non-Clostridial Bacterial Species. Toxins, 12(1), 19. https://doi.org/10.3390/toxins12010019




