MazF Endoribonucleolytic Toxin Conserved in Nitrospira Specifically Cleaves the AACU, AACG, and AAUU Motifs
Abstract
:1. Introduction
2. Results
2.1. Identifying the TA System in Nitrospira Strain ND1
2.2. Estimating MazF-nd1 Cleavage Sites
2.3. MazF-nd1 Recognizes RNA at AACU, AACG, and AAUU Sequences
2.4. Analysis of Intracellular Targets of MazF-nd1
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plasmids, RNA, and Oligonucleotides
5.2. In Vivo Toxicity of MazF-nd1
5.3. Protein Expression
5.4. Protein Purification
5.5. Enzymatic Activity of MazF-nd1 and MazE-nd1
5.6. Cleavage Sequence Identification
5.7. Fluorometric Detection of MazF-nd1 Activity
5.8. Analyzing the Frequency of MazF-nd1 Cleavage Sites in Nitrospira Strain ND1 Genome
5.9. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yamaguchi, Y.; Park, J.-H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, A.M.; Gollan, B.; Helaine, S. Toxin-antitoxin systems: Reversible toxicity. Curr. Opin. Microbiol. 2017, 36, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012, 9, 1488–1490. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, K.; Christensen, S.K.; Løbner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef]
- Goeders, N.; Chai, R.; Chen, B.; Day, A.; Salmond, G.P. Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins 2016, 8, 282. [Google Scholar] [CrossRef] [Green Version]
- Masuda, H.; Tan, Q.; Awano, N.; Wu, K.P.; Inouye, M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 2012, 84, 979–989. [Google Scholar] [CrossRef]
- Masuda, H.; Tan, Q.; Awano, N.; Yamaguchi, Y.; Inouye, M. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol. Lett. 2012, 328, 174–181. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lord, D.M.; Cheng, H.Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell 2013, 52, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, J.; Hoeflich, K.; Ikura, M.; Qing, G.; Inouye, M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 2003, 12, 913–923. [Google Scholar] [CrossRef]
- Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5’-bispyrophosphate: A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 1996, 93, 6059–6063. [Google Scholar] [CrossRef] [Green Version]
- Hazan, R.; Sat, B.; Engelberg-Kulka, H. Escherichia colimazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004, 186, 3663–3669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amitai, S.; Kolodkin-Gal, I.; Hananya-Meltabashi, M.; Sacher, A.; Engelberg-Kulka, H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 2009, 5, e1000390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vesper, O.; Amitai, S.; Belitsky, M.; Byrgazov, K.; Kaberdina, A.C.; Engelberg-Kulka, H.; Moll, I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 2011, 147, 147–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schifano, J.M.; Woychik, N.A. Cloaked dagger: tRNA slicing by an unlikely culprit. RNA Biol. 2017, 14, 15–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, Y.; Nariya, H.; Park, J.; Inouye, M. Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat. Commun. 2012, 3, 607. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, T.; Yokota, A.; Tsuneda, S.; Noda, N. AAU-specific RNA cleavage mediated by MazF toxin endoribonuclease conserved in Nitrosomonas europaea. Toxins 2016, 8, 174. [Google Scholar] [CrossRef]
- Miyamoto, T.; Yokota, A.; Ota, Y.; Tsuruga, M.; Aoi, R.; Tsuneda, S.; Noda, N. Nitrosomonas europaea MazF specifically recognises the UGG motif and promotes selective RNA degradation. Front. Microbiol. 2018, 9, 2386. [Google Scholar] [CrossRef]
- Daims, H.; Lücker, S.; Wagner, M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef]
- Lücker, S.; Wagner, M.; Maixner, F.; Pelletier, E.; Koch, H.; Vacherie, B.; Rattei, T.; Sinninghe Damsté, J.S.; Spieck, E.; Le Paslier, D.; et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 13479–13484. [Google Scholar]
- Koch, H.; Lücker, S.; Albertsen, M.; Kitzinger, K.; Herbold, C.; Spieck, E.; Nielsen, P.H.; Wagner, M.; Daims, H. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. USA 2015, 112, 11371–11376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujitani, H.; Ushiki, N.; Tsuneda, S.; Aoi, Y. Isolation of sublineage I Nitrospira by a novel cultivation strategy. Environ. Microbiol. 2014, 16, 3030–3040. [Google Scholar] [CrossRef] [PubMed]
- Ushiki, N.; Fujitani, H.; Shimada, Y.; Morohoshi, T.; Sekiguchi, Y.; Tsuneda, S. Genomic analysis of two phylogenetically distinct Nitrospira species reveals their genomic plasticity and functional diversity. Front. Microbiol. 2018, 8, 2637. [Google Scholar] [CrossRef] [Green Version]
- Sevin, E.; Barloy-Hubler, F. RASTA-Bacteria: A web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007, 8, R155. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, T.; Kato, Y.; Sekiguchi, Y.; Tsuneda, S.; Noda, N. Characterization of MazF-mediated sequence-specific RNA cleavage in Pseudomonas putida using massive parallel sequencing. PLoS ONE 2016, 11, e0149494. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Rothenbacher, F.P.; Suzuki, M.; Hurley, J.M.; Montville, T.J.; Kim, T.J.; Ouyang, M.; Woychik, N.A. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 2012, 194, 3464–3474. [Google Scholar] [CrossRef] [Green Version]
- Schifano, J.M.; Vvedenskaya, I.O.; Knoblauch, J.G.; Ouyang, M.; Nickels, B.E.; Woychik, N.A. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014, 5, 3538. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.R.; Hergenrother, P.J. A continuous fluorometric assay for the assessment of MazF ribonuclease activity. Anal. Biochem. 2007, 371, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Inoue, K.; Yoshizumi, S.; Kobayashi, H.; Zhang, Y.; Ouyang, M.; Kato, F.; Sugai, M.; Inouye, M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009, 191, 3248–3255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, Y.; Yumoto, N.; Tokushige, M.; Fukui, K.; Ohya-Nishiguchi, H. Purification and characterization of two types of fumarases from Escherichia coli. J. Biochem. 1991, 109, 728–733. [Google Scholar]
- Park, S.; Gunsalus, R. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: Role of the arcA, fnr, and soxR gene products. J. Bacteriol. 1995, 177, 6255–6262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, J.; Hara, H.; Kato, I.; Inouye, M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2004, 280, 3143–3150. [Google Scholar] [CrossRef] [Green Version]
- Chopra, N.; Saumitra, P.A.; Bhatnagar, R.; Bhatnagar, S. Linkage, mobility, and selfishness in the MazF family of bacterial toxins: A snapshot of bacterial evolution. Genome Biol. Evol. 2013, 5, 2268–2284. [Google Scholar]
- Park, J.H.; Yamaguchi, Y.; Inouye, M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 2011, 585, 2526–2532. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, T.; Ota, Y.; Yokota, A.; Suyama, T.; Tsuneda, S.; Noda, N. Characterization of a Deinococcus radiodurans MazF: A UACA-specific RNA endoribonuclease. MicrobiologyOpen 2017, 6, e501. [Google Scholar] [CrossRef] [Green Version]
- Shaku, M.; Park, J.H.; Inouye, M.; Yamaguchi, Y. Identification of MazF homologue in Legionella pneumophila which cleaves RNA at the AACU sequence. J. Mol. Microbiol. Biotechnol. 2018, 28, 269–280. [Google Scholar] [CrossRef]
- Barth, V.C.; Woychik, N.A. The sole Mycobacterium smegmatis MazF toxin targets tRNALys to impart highly selective, codon-dependent proteome reprogramming. Front. Genet. 2019, 10, 1356. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, Y.; Teh, J.S.; Zhang, J.; Connell, N.; Rubin, H.; Inouye, M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 18638–18643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schifano, J.M.; Edifor, R.; Sharp, J.D.; Ouyang, M.; Konkimalla, A.; Husson, R.N.; Woychik, N.A. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. USA 2013, 110, 8501–8506. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Phadtare, S.; Nariya, H.; Ouyang, M.; Husson, R.N.; Inouye, M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 2008, 69, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Schifano, J.M.; Cruz, J.W.; Vvedenskaya, I.O.; Edifor, R.; Ouyang, M.; Husson, R.N.; Nickels, B.E.; Woychik, N.A. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 2016, 44, 1256–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, C.F.; Park, J.H.; Prax, M.; Herbig, A.; Nieselt, K.; Rosenstein, R.; Inouye, M.; Bertram, R. Characterization of a mazEF toxin-antitoxin homologue from Staphylococcus equorum. J. Bacteriol. 2013, 195, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]


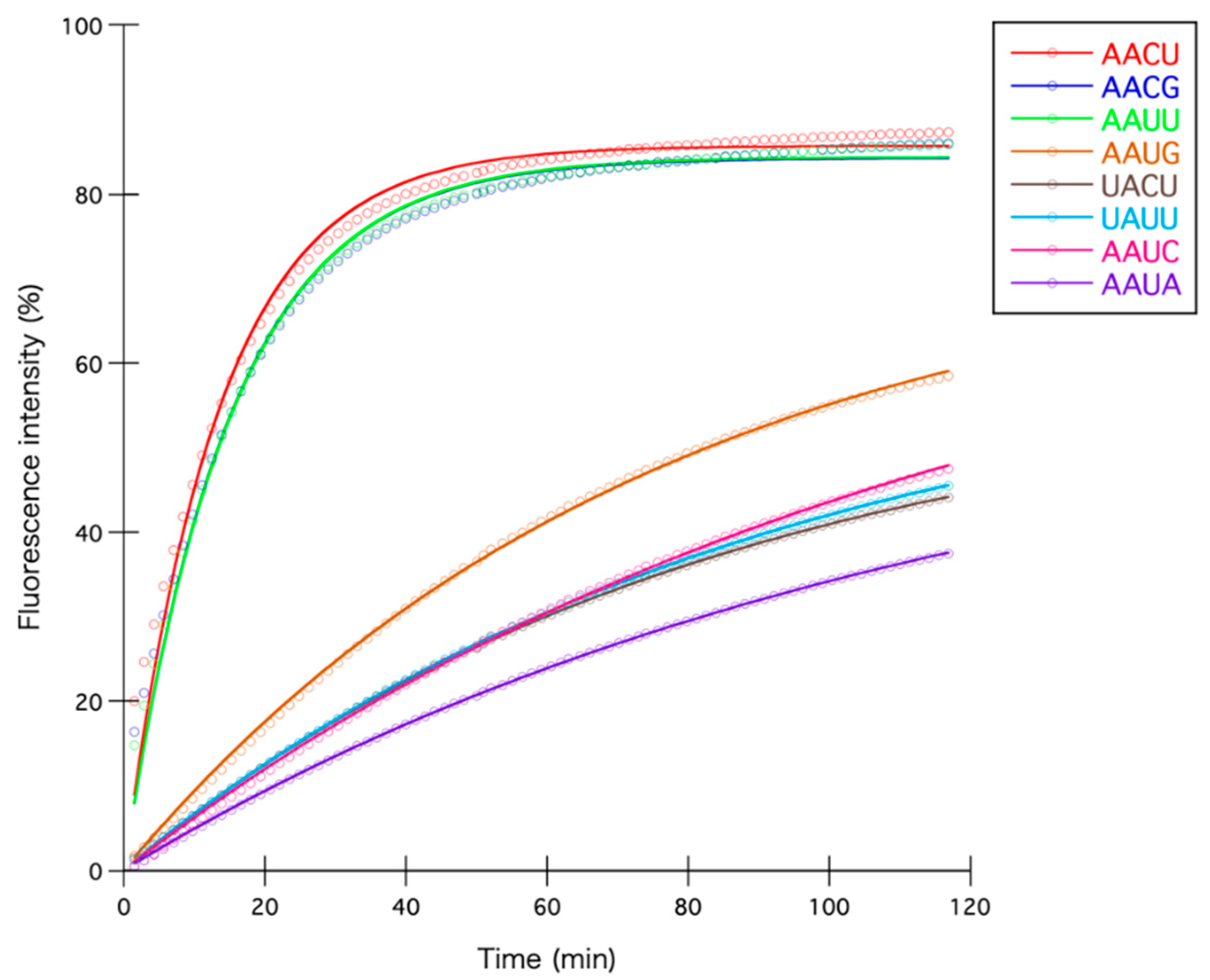
| Sequence 1 | Occurrence in Substrate RNA 2 | Initial Reaction Velocity 3 | Fmax3 | k3 |
|---|---|---|---|---|
| AACU | 17 | 6.421 | 85.67 | 7.50 × 10−2 |
| AACG | 24 | 5.669 | 84.23 | 6.73 × 10−2 |
| AAUU | 12 | 5.652 | 84.36 | 6.70 × 10−2 |
| AAUG | 21 | 0.998 | 74.66 | 1.34 × 10−2 |
| UACU | 32 | 0.701 | 58.59 | 1.20 × 10−2 |
| UAUU | 11 | 0.697 | 62.44 | 1.12 × 10−2 |
| AAUC | 28 | 0.649 | 75.43 | 8.61 × 10−3 |
| AAUA | 20 | 0.509 | 59.29 | 8.58 × 10−3 |
| AACA | 21 | 0.427 | 53.82 | 7.93 × 10−3 |
| CAUU | 20 | 0.423 | 53.20 | 7.95 × 10−3 |
| GACU | 15 | 0.421 | 65.96 | 6.38 × 10−3 |
| AACC | 28 | 0.410 | 49.96 | 8.21 × 10−3 |
| CACU | 17 | 0.398 | 58.92 | 6.75 × 10−3 |
| GAUU | 24 | 0.390 | 63.03 | 6.19 × 10−3 |
| Rank | Locus Tag | Product | Length (bp) | K1 | E2 | P3 |
|---|---|---|---|---|---|---|
| 1 | 60124 | Protein of unknown function | 1980 | 38 | 15.66 | 1.11 × 10−6 |
| 2 | 50240 | Conserved protein of unknown function | 3540 | 44 | 22.66 | 4.30 × 10−5 |
| 3 | 50464 | FumC: fumarate hydratase (fumarase C), aerobic class II | 1455 | 29 | 12.96 | 7.75 × 10−5 |
| 4 | 62020 | Conserved protein of unknown function | 3270 | 43 | 23.02 | 1.20 × 10−4 |
| 5 | 60419 | IspU: undecaprenyl pyrophosphate synthase | 786 | 16 | 5.99 | 4.71 × 10−4 |
| 6 | 63509 | Conserved protein of unknown function | 5319 | 53 | 32.37 | 5.16 × 10−4 |
| 7 | 50535 | Sensory response regulator with diguanylate cyclase domain | 2127 | 32 | 16.78 | 5.75 × 10−4 |
| 8 | 61480 | Conserved protein of unknown function | 2316 | 37 | 20.91 | 8.77 × 10−4 |
| 9 | 63396 | YaeT: putative outer membrane protein assembly factor | 2325 | 36 | 20.29 | 9.74 × 10−4 |
| 10 | 62925 | Sigma-54 dependent transcriptional regulator (Modular protein) | 2601 | 33 | 18.10 | 1.00 × 10−3 |
| 11 | 61386 | TonB-dependent siderophore receptor | 2334 | 34 | 19.27 | 1.45 × 10−3 |
| 12 | 62179 | Conserved exported protein of unknown function | 507 | 11 | 3.69 | 1.45 × 10−3 |
| 13 | 50325 | FlgG: flagellar component of cell-distal portion of basal-body rod | 792 | 16 | 6.71 | 1.50 × 10−3 |
| 14 | 50347 | Protein of unknown function | 5361 | 59 | 38.96 | 1.59 × 10−3 |
| 15 | 63512 | Conserved protein of unknown function | 3252 | 31 | 17.19 | 1.67 × 10−3 |
| 16 | 60586 | Exported protein of unknown function | 1371 | 23 | 11.52 | 1.77 × 10−3 |
| 17 | 61395 | Putative TonB-dependent receptor | 2262 | 35 | 20.30 | 1.80 × 10−3 |
| 18 | 60858 | ThiI: putative tRNA sulfurtransferase | 1179 | 21 | 10.20 | 1.90 × 10−3 |
| 19 | 50308 | FlgE: flagellar hook protein | 1224 | 22 | 10.98 | 2.09 × 10−3 |
| 20 | 61921 | Putative regulatory protein, MerR family, cobalamin B12-binding | 918 | 16 | 6.9 | 2.10 × 10−3 |
| 21 | 63111 | Efflux transporter, outer membrane factor lipoprotein | 1521 | 23 | 11.69 | 2.12 × 10−3 |
| 22 | 63116 | HyfR: hydrogenase-4 transcriptional activator | 2043 | 29 | 16.12 | 2.33 × 10−3 |
| 23 | 62910 | Putative di-haem cytochrome c | 2076 | 28 | 15.46 | 2.50 × 10−3 |
| 24 | 63252 | RhlE: ATP-dependent RNA helicase | 1353 | 18 | 8.49 | 2.86 × 10−3 |
| 25 | 61398 | Putative TonB-dependent receptor | 2325 | 35 | 20.92 | 2.89 × 10−3 |
| Locus Tag | Product | Length (bp) |
|---|---|---|
| 50353 | FlaG: flagellar protein | 372 |
| 50758 | Putative nucleotidyltransferase (fragment) | 183 |
| 50957 | Insertion element ISR1 uncharacterized 10 kDa protein A3 (fragment) | 159 |
| 60210 | TadA: tRNA-specific adenosine deaminase | 489 |
| 60360 | RcnA: putative nickel/cobalt efflux system | 708 |
| 60698 | AcyP: acylphosphatase | 348 |
| 60976 | Bfr: bacterioferritin, iron storage and detoxification protein | 480 |
| 60984 | MreD: putative cell shape-determining protein | 501 |
| 61420 | Nqo: NADH-quinone oxidoreductase chain 10 | 513 |
| 61675 | MerT: mercuric transport protein | 381 |
| 61748 | Putative membrane protein insertion efficiency factor (modular protein) | 264 |
| 62137 | K+-transporting ATPase, F subunit (fragment) | 90 |
| 62252 | RbfA: ribosome-binding factor A | 399 |
| 62630 | Cytochrome bd-type quinol oxidase, subunit 2 | 1017 |
| 62661 | AtpE: ATP synthase subunit c | 231 |
| 62937 | YitW: MIP18 family protein | 324 |
| 63168 | TatA: Sec-independent protein translocase protein | 294 |
| 63277 | Sulfate-binding protein (fragment) | 492 |
| Name | Sequence 1 |
|---|---|
| DR-14-AAUU | aaaaaAAUUaaaaa |
| DR-14-UAUU | aaaaaUAUUaaaaa |
| DR-14-GAUU | aaaaaGAUUaaaaa |
| DR-14-CAUU | aaaaaCAUUaaaaa |
| DR-14-AAUA | aaaaaAAUAaaaaa |
| DR-14-AAUG | aaaaaAAUGaaaaa |
| DR-14-AAUC | aaaaaAAUCaaaaa |
| DR-14-AACU | aaaaaAACUaaaaa |
| DR-14-UACU | aaaaaUACUaaaaa |
| DR-14-GACU | aaaaaGACUaaaaa |
| DR-14-CACU | aaaaaCACUaaaaa |
| DR-14-AACA | aaaaaAACAaaaaa |
| DR-14-AACG | aaaaaAACGaaaaa |
| DR-14-AACC | aaaaaAACCaaaaa |
| D-13-AAA | aaaaaaaaaaaaa |
| R-13- GUUGU | GUUGUCAUGCCGG |
| R-13- UCUCG | UCUCGGUGCGUUG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aoi, R.; Miyamoto, T.; Yokota, A.; Ota, Y.; Fujitani, H.; Tsuneda, S.; Noda, N. MazF Endoribonucleolytic Toxin Conserved in Nitrospira Specifically Cleaves the AACU, AACG, and AAUU Motifs. Toxins 2020, 12, 287. https://doi.org/10.3390/toxins12050287
Aoi R, Miyamoto T, Yokota A, Ota Y, Fujitani H, Tsuneda S, Noda N. MazF Endoribonucleolytic Toxin Conserved in Nitrospira Specifically Cleaves the AACU, AACG, and AAUU Motifs. Toxins. 2020; 12(5):287. https://doi.org/10.3390/toxins12050287
Chicago/Turabian StyleAoi, Rie, Tatsuki Miyamoto, Akiko Yokota, Yuri Ota, Hirotsugu Fujitani, Satoshi Tsuneda, and Naohiro Noda. 2020. "MazF Endoribonucleolytic Toxin Conserved in Nitrospira Specifically Cleaves the AACU, AACG, and AAUU Motifs" Toxins 12, no. 5: 287. https://doi.org/10.3390/toxins12050287






