Novel Ergot Alkaloids Production from Penicillium citrinum Employing Response Surface Methodology Technique
Abstract
:1. Introduction
2. Results
2.1. Strain Improvement
2.1.1. Impact of Physical and Chemical Mutagen on Penicillium citrinum
Impact of UV Irradiations
Impact of Ethyl Methane Sulfonate (EMS)
UV and EMS Mutated Strains for Ergot Alkaloids Synthesis
2.2. Response Surface Methodology
2.2.1. Screening Step Using PBD
ANOVA for PBD Model
2.2.2. Identification of Significant Factors Using Box–Behnken design (BBD)
ANOVA for BBD Model
Regression Analysis for Ergot Alkaloids Production and Comparison between the Observed and Predicted Response
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Microorganism and Its Maintenance
5.2. Strain Improvement
5.2.1. Impact of Physical and Chemical Mutagens on Penicillium citrinum
Impact of UV Irradiations
Impact of Ethyl Methane Sulfonate (EMS)
5.2.2. Calculation of Survival Percentage of Colonies
5.3. Maintenance of Mutant Strain
5.3.1. Selection of Best UV Mutant for the Production of Ergot Alkaloids
5.3.2. Selection of Best EMS Mutant for the Production of Ergot Alkaloids
5.3.3. Response Surface Methodology for Ergot Alkaloids Synthesis
5.4. Preparation of Inoculum
5.5. Response Surface Methodology
5.5.1. Plackett–Burman design (PBD) for Screening of Fermentation Factors
5.5.2. Identification of Significant Factors Using Box–Behnken Design (BBD)
5.5.3. Statistical Analyses of RSM
5.6. Ergot Alkaloids Determination
5.6.1. Ergot Alkaloids in Fermented Broth Extract
5.6.2. Ergot Alkaloids in Extract of Mycelia
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pichersky, E.; Gang, D.R. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000, 5, 439–445. [Google Scholar] [CrossRef]
- Shahid, M.G.; Nadeem, M.; Baig, S.; Cheema, T.A.; Atta, S.; Ghafoor, G. Screening and optimization of some inorganic salts for the production of ergot alkaloids from Penicillium species using surface culture fermentation process. Pak. J. Pharma. Sci. 2016, 29, 407–414. [Google Scholar]
- Shahid, M.G.; Baig, S.; Nadeem, M.; Cheema, T.A.; Nelofar, R.; Saleem, M. Biosynthesis of ergot alkaloids from Penicillium commune using response surface methodology (RSM). Pak. J. Bot. 2017, 49, 1569–1578. [Google Scholar]
- Zafar, A.M.; Waseemuddin, S.A.; Azhar, I.; Sualeh, M.; Baig, M.T.; Zoha, S.M.S. Bioactive alkaloids produced by fungi: Updates on alkaloids from the species of the genera Boletus, Fusarium and Psilocybe (Review). Pak. J. Pharma. Sci. 2010, 23, 349–357. [Google Scholar]
- Gulliamon, J.M.; Sabate, J.; Barrio, E.; Cano, J.; Querol, A. Rapid identification of nine Yeast species based on RFLP analysis of ribosomal internal transcribed spacer (ITS) regions. Arch. Microbiol. 1998, 169, 387–392. [Google Scholar]
- Tiwari, K.L.; Jadhav, S.K.; Fatima, A. Culture conditions for the production of thermostable amylase by Penicillium rugulosum. Glob. J. Biotechnol. Biochem. 2007, 2, 21–24. [Google Scholar]
- Kozlovsky, A.G.; Zhelifonova, V.P.; Antipova, T.V. Biologically active metabolites of Penicillium fungi. Org. Biomol. Chem. 2013, 1, 11–21. [Google Scholar]
- Nina, G.; Neubauer, L.; Tudzynski, P.; Shu-Ming, L. Biosynthesis pathways of ergot alkaloids. Toxins 2014, 6, 3281–3295. [Google Scholar]
- Khurana, S.; Kapoor, M.; Gupta, S.; Kuhad, R.C. Statistical optimization of alkaline xylanase production from Streptomyces violaceoruber under submerged fermentation using response surface methodology. Ind. J. Microbiol. 2007, 47, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Trejo, H.M.R.; Lonsane, B.K.; Raimbault, M.; Roussost, S. Spectra of ergot alkaloids produced by Claviceps purpurea 1029c in solid-state fermentation system: Influence of the composition of liquid medium used for impregnating sugar-cane pith bagasse. Proc. Biochem. 1993, 28, 23–27. [Google Scholar] [CrossRef]
- Plackett, R.L.; Burman, J.P. The design of optimum multifactorial experiments. Biometrika 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Box, G.E.P.; Behnken, D.W. Some new three level designs for the study of quantitative variables. Technometrics 1960, 2, 455–475. [Google Scholar] [CrossRef]
- Naveena, B.J.; Altaf, M.D.; Bhadriah, K. Selection of medium components by Plackett–Burman design for production of L (+) lactic acid by Lactobacillus amylophilus GV6 in SSF using wheatbran. Bioresour. Technol. 2005, 96, 485–490. [Google Scholar] [CrossRef]
- Venil, C.K.; Lakshmanaperumalsamy, P. Application of statistical design to the optimization of culture medium for prodigiosin production by Serratia marcescens SWML08. Malays. J. Microbiol. 2009, 5, 55–61. [Google Scholar]
- Shahid, M.G.; Baig, S.; Saleem, M.; Arif, R.; Ghafoor, G.; Liaqat, A. Qualitative and quantitative analysis of ergot alkaloids produced by Aspergillus niger through surface culture fermentation process. Pak. J. Bot. 2018, 50, 2423–2428. [Google Scholar]
- Devi, N.N.; Prabakaran, J.J. Bioactive metabolites from an endophytic fungus Penicillium sp. isolated from Centella asiatica. Curr. Res. Environ. Appl. Mycol. 2014, 4, 34–43. [Google Scholar] [CrossRef]
- Roberts, M.R.; Wink, M. Alkaloids: Biochemistry, Ecology and Medical Applications; Plenum: New York, NY, USA, 1998; pp. 135–146. [Google Scholar]
- Onyegeme-Okerenta, B.M.; Okochi, V.I.; Chinedu, S.N. Penicillin production by Penicillium chrysogenum PCL 501: Effect of UV induced mutation. Int. J. Microbiol. 2013, 12, 1–10. [Google Scholar]
- Veerapagu, M.; Jeya, K.R.; Ponmurugan, K. Mutational Effect of Penicillium chrysogenum on Antibiotic Production; Advanced Biotech: Totowa, NJ, USA, 2008; pp. 16–19. [Google Scholar]
- Moussa, L.A.A. Effect of some factor including irradiation on the ergot alkaloids production by members of Penicillium. J. Biol. Res. 2003, 3, 65–81. [Google Scholar]
- El-Bondkly, A.M.; Abeer, A.K. UV- and EMS- induced mutations affecting synthesis of alkaloids and lipase in Penicillium roquefortii. Arab. J. Biotechnol. 2007, 10, 241–248. [Google Scholar]
- Hamad, A.; Haq, I.; Qadeer, M.A.; Javed, I. Screening of Bacillus licheniformis mutants for improved production of alpha-amylase. Pak. J. Bot. 2001, 33, 517–525. [Google Scholar]
- Nadeem, M. Biotechnological Production of Alkaline Protease for Industrial Use. Ph.D. Thesis, Department of Zoology, University of the Punjab, New Campus, Lahore, Pakistan, 2009. [Google Scholar]
- Rao, J.M.; Kim, C.; Rhee, S. Statistical optimization of medium for the production of recombinant hirudin from Sacchromyces cerevisiae using response surface methodology. Proc. Biochem. 2000, 35, 639–647. [Google Scholar] [CrossRef]
- Venil, C.K.; Lakshmanaperumalsamy, P. Applications of response surface methodology in medium optimization for protease production by the new strain of Serratia marcescens SWML08. Pol. J. Microbiol. 2009, 58, 117–124. [Google Scholar]
- Mao, X.B.; Eksriwong, T.; Chauvatcharin, S.; Zhong, J.J. Optimization of carbon source and carbon/nitrogen ratio for cordycepin production by submerged cultivation of medicinal mushroom Cordyceps militaris. Proc. Biochem. 2005, 40, 1667–1672. [Google Scholar] [CrossRef]
- Wu, Q.; Yong-Chun, S.; Xu, H.; Guo, Y.; Li, J.; Ren-Xiang, T. Medium optimization for enhanced co-production of two bioactive metabolites in the same fermentation by a statistical approach. J. Asian Nat. Prod. Res. 2011, 13, 1110–1121. [Google Scholar]
- Rubina, N.; Ramnan, R.N.; Rahman, R.N.Z.R.; Basri, M.; Ariff, A.B. Sequential optimization of production of a thermostable and organic solvent tolerant lipase by recombinant Escherichia coli. Ann. Microbiol. 2011, 61, 535–544. [Google Scholar]
- Liu, Y.; Wang, F.; Tan, T. Cyclic resolution of racemic ibuprofen via coupled efficient lipase and acid base catalysis. Chirality 2009, 21, 349–353. [Google Scholar] [CrossRef]
- Pan, H.; Xie, Z.; Bao, W.; Zhang, J. Optimization of culture conditions to enhance cis-epoxysuccinate hydrolase production in Escherichia coli by response surface methodology. Biochem. Eng. J. 2008, 42, 133–138. [Google Scholar] [CrossRef]
- Lee, D.W.; Koh, Y.S.; Kim, B.C.; Choi, H.J.; Kim, D.S.; Suhartono, M.T.; Pyun, Y.R. Isolation and characterization of thermophilic lipase from Bcillus thermoleovorans ID-1. FEMS Microbiol. Lett. 1999, 179, 393–400. [Google Scholar] [CrossRef]
- Wang, X.L.; Liu, G.Q. Preliminary select and optimization of submerged fermentation media of Ganoderma sinense. Food Sci. Technol. 2009, 34, 14–16. [Google Scholar]
- Krishnaa, D.; Krishnaa, K.S.; Padma, S.R. Response surface modeling and optimization of chromium (Vi) removal from aqueous solution using Borasus flabellifer coir powder. Int. J. Appl. Sci. Eng. 2013, 11, 213–226. [Google Scholar]
- Amara, A.A.F. Optimizing PHB and protease production by Box-Behnken design. J. IILUM Eng. 2013, 14, 15–28. [Google Scholar] [CrossRef]
- Yasin, S.; Curti, M.; Behary, N.; Perwuelz, A.; Giraud, S.; Rovero, G.; Guan, J.; Chen, G. Process optimization of eco-friendly flame retardant finish for cotton fabric: A response surface methodology approach. Surf. Rev. Lett. 2017, 24, 1750114. [Google Scholar] [CrossRef]
- Cheng, S.W.; Wang, Y.F.; Hong, B. Statistical optimization of mediumcompositions for chitosanaseproduction by a newly isolated Streptomyces albus. Braz. J. Chem. Eng. 2012, 29, 691–698. [Google Scholar] [CrossRef] [Green Version]
- Sreedevi, K.; Venkateswara, R.J.; Lakshmi, N.; Fareedullah, M. Strain improvement of Aspergillus terreus for the enhanced production of lovastatin, a HMG-COA reductase inhibitor. J. Microbiol. Biotechnol. Res. 2011, 1, 96–100. [Google Scholar]
- Ryan, K.L.; Christopher, T.M.; Panaccione, D.G. Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins 2013, 5, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Naude, T.W.; Botha, C.J.; Vorster, J.H.; Roux, C.; Van der linde, E.J.; Van der walt, S.L.; Rottinghaus, G.E.; Van jaarsveld, I.; Lawrence, A.N. Claviceps cyperi, a new cause of severe ergotism in dairy cattle consuming maize silage and teff hay contaminated with ergotised Cyperus esculentus (nut sedge) on the Highveld of South Africa. Onderstepoort J. Vet. Res. 2005, 72, 23–37. [Google Scholar] [CrossRef] [Green Version]
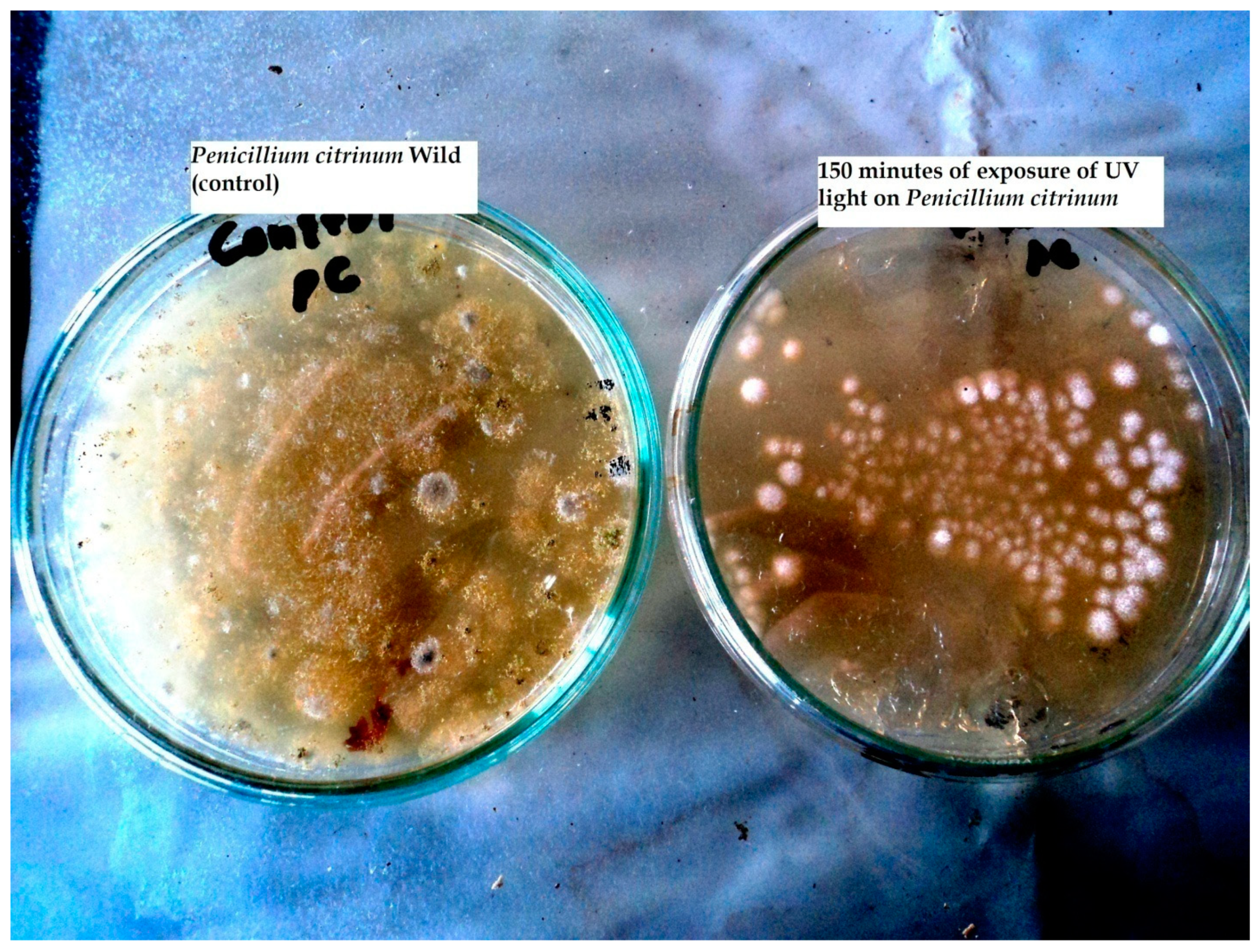







| UV Exposure Time (min) | Penicillium citrinum | |
|---|---|---|
| No. of Colonies | Survival Rate (%) | |
| 0 | 49 | 100 |
| 15 | 44 | 89.7 |
| 30 | 41 | 83.6 |
| 45 | 37 | 75.5 |
| 60 | 31 | 63.2 |
| 75 | 28 | 57.1 |
| 90 | 20 | 40.8 |
| 105 | 14 | 28.5 |
| 120 | 7 | 14.2 |
| 135 | 3 | 6.12 |
| 150 | 1 | 2.04 |
| EMS Exposure Time (min) | Penicillium citrinum | |
|---|---|---|
| No. of Colonies | Survival Rate (%) | |
| 0 | 31 | 100 |
| 10 | 25 | 80.6 |
| 15 | 14 | 45.1 |
| 20 | 7 | 22.5 |
| 25 | 1 | 3.2 |
| 30 | 0 | 0 |
| UV treated Strains of Penicillium citrinum | Extracellular Extract (mg/mL) | Intracellular Extract (mg/mL) | EMS treated Strains of Penicillium citrinum | Extracellular Extract (mg/mL) | Intracellular Extract (mg/mL) |
|---|---|---|---|---|---|
| PCUV-1 | 1.49 ± 0.01 | 1.05 ± 0.02 | PCEMS-1 | 1.84 ± 0.02 | 1.58 ± 0.01 |
| PCUV-2 | 1.68 ± 0.02 | 1.65 ± 0.03 | PCEMS-2 | 2.5 ±0.01 | 2.10 ± 0.03 |
| PCUV-3 | 2.56 ± 0.05 | 1.65 ± 0.01 | PCEMS-3 | 2.99 ± 0.005 * | 2.78 ± 0.04 * |
| PCUV-4 | 4.56 ± 0.01 * | 1.89 ± 0.03 * | Wild | 2.27 ± 0.02 | 2.18 ± 0.02 |
| PCUV-5 | 3.86 ± 0.02 | 1.34 ± 0.01 | |||
| Wild | 2.45 ± 0.03 | 1.66 ± 0.01 |
| Run. | Yield of Ergot Alkaloids (mg/mL) |
|---|---|
| 1 | 11.84 ± 0.1 |
| 2 | 14.76 ± 0.01 * |
| 3 | 0.36 ± 0.03 |
| 4 | 6.53 ± 0.01 |
| 5 | 10.96 ± 0.01 |
| 6 | 11.95 ± 0.02 |
| 7 | 0.74 ± 0.04 |
| 8 | 5.38 ± 0.1 |
| 9 | 11.79 ± 0.03 |
| 10 | 7.76 ± 0.05 |
| 11 | 0.24 ± 0.02 |
| 12 | 13.02 ± 0.03 |
| Source | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Intercept | 0.46 | 1 | 0.46 | 8.26 | 0.21 |
| Sucrose | 147.78 | 1 | 147.78 | 2670.95 | 0.012 |
| yeast extract | 22.54 | 1 | 22.54 | 4.7.44 | 0.032 |
| Succinic acid | 1.30 | 1 | 1.30 | 23.64 | 0.13 |
| MgSO4 | 1.16 | 1 | 1.16 | 20.96 | 0.14 |
| KH2PO4 | 0.16 | 1 | 0.16 | 2.86 | 0.34 |
| FeSO4 | 12.87 | 1 | 12.87 | 232.57 | 0.042 |
| ZnSO4 | 0.50 | 1 | 0.50 | 9.06 | 0.20 |
| Asparagine | 0.96 | 1 | 0.96 | 17.41 | 0.15 |
| Tryptophan | 0.36 | 1 | 0.36 | 6.47 | 0.24 |
| pH | 3.06 | 1 | 3.06 | 55.33 | 0.085 |
| Error | 0.06 | 1 | 0.06 |
| Runs | Sucrose (g/100 mL) | Yeast Extract (g/100 mL) | FeSO4 (g/100 mL) | Alkaloids Yield (Observed) mg/ml | Alkaloids Yield (Predicted) mg/ml |
|---|---|---|---|---|---|
| 1. | 41 | 5 | 0.06 | 22.50 | 21.75 |
| 2. | 41 | 39 | 0.06 | 16.00 | 17.42 |
| 3. | 41 | 22 | 0.01 | 24.55 | 24.32 |
| 4. | 41 | 22 | 0.11 | 27.79 | 27.79 |
| 5. | 5 | 5 | 0.06 | 18.90 | 17.94 |
| 6. | 5 | 39 | 0.06 | 13.50 * | 14.16 * |
| 7. | 5 | 22 | 0.01 | 20.40 | 20.06 |
| 8. | 5 | 22 | 0.11 | 25.20 | 25.30 |
| 9. | 23 | 5 | 0.01 | 16.87 | 16.65 |
| 10. | 23 | 39 | 0.01 | 17.90 | 17.56 |
| 11. | 23 | 5 | 0.11 | 25.20 | 25.53 |
| 12. | 23 | 39 | 0.11 | 17.60 | 16.75 |
| 13. | 23 | 22 | 0.06 | 35.60 | 35.60 |
| Variable | Sum of Square | Degree of Freedom | Means Square | F-Value | p-Value | t-Value |
|---|---|---|---|---|---|---|
| Intercept | 175.21 | 1 | 175.21 | 167.58 | 0.001 | −12.55 |
| Sucrose | 121.31 | 1 | 121.31 | 11.33 | 0.002 | 10.07 |
| Sucrose2 | 116.22 | 1 | 116.22 | 119.58 | 0.002 | −10.24 |
| yeast Extract | 294.94 | 1 | 294.94 | 259.87 | 0.000 | 16.12 |
| yeast Extract2 | 308.41 | 1 | 308.41 | 285.67 | 0.000 | −16.63 |
| FeSO4 | 75.62 | 1 | 75.62 | 76.79 | 0.004 | 8.17 |
| FeSO4 2 | 49.91 | 1 | 49.91 | 39.89 | 0.006 | −6.91 |
| Sucrose, yeast Extract | 3.16 | 1 | 3.16 | 2.01 | 0.251 | −1.42 |
| Sucrose, FeSO4 | 0.03 | 1 | 0.03 | 0.04 | 0.862 | −0.18 |
| yeast Extract, FeSO4 | 24.61 | 1 | 24.61 | 21.01 | 0.019 | −4.58 |
| Ingredients. | g/100 mL |
| NH4Cl | 0.2 |
| Succinic Acid | 0.5 |
| Sucrose | 5 |
| KH2PO4 | 0.5 |
| Asparagine | 0.5 |
| Tryptophan | 0.5 |
| yeast Extract | 0.5 |
| MgSO4. 7H2O | 0.03 |
| FeSO4 | 0.01 |
| ZnSO4 | 0.002 |
| Fermentation Conditions | |
| Incubation Time (Days) | 21 |
| Inoculum Size (ml) | 5 |
| pH | 5 |
| Incubation Temperature (°C) | 25 |
| Range and Level | Fermentation Factor | |
|---|---|---|
| −1 | +1 | |
| 5 | 35 | Sucrose, X1 |
| 5 | 30 | yeast Extract, X2 |
| 0.1 | 1 | Succinic acid, X3 |
| 0.1 | 1 | Asparagine, X4 |
| 0.1 | 1 | Tryptophan, X5 |
| 0.1 | 1 | KH2PO4, X6 |
| 0.25 | 0.625 | MgSO4, X7 |
| 0.01 | 0.1 | FeSO4, X8 |
| 0.02 | 0.2 | ZnSO4, X9 |
| 3 | 5 | pH, X 10 |
| Runs | Variables (x) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | |
| Sucrose | Yeast Extract | Succinic Acid | Asparagine | Tryptophan | MgSO4 | KH2PO4 | ZnSO4 | FeSO4 | pH | |
| 1. | 35 | 5 | 0.1 | 0.1 | 1 | 0.625 | 0.1 | 0.2 | 0.1 | 5 |
| 2. | 35 | 30 | 0.1 | 0.1 | 0.1 | 0.625 | 1 | 0.02 | 0.1 | 5 |
| 3. | 5 | 5 | 0.1 | 0.1 | 0.1 | 0.25 | 0.1 | 0.02 | 0.01 | 3 |
| 4. | 5 | 30 | 0.1 | 1 | 1 | 0.25 | 0.1 | 0.02 | 0.1 | 5 |
| 5. | 35 | 5 | 0.1 | 1 | 0.1 | 0.25 | 1 | 0.2 | 0.01 | 3 |
| 6. | 35 | 5 | 1 | 1 | 1 | 0.25 | 1 | 0.02 | 0.1 | 3 |
| 7. | 5 | 5 | 1 | 0.1 | 1 | 0.625 | 1 | 0.02 | 0.01 | 3 |
| 8. | 5 | 30 | 0.1 | 1 | 1 | 0.625 | 1 | 0.2 | 0.01 | 3 |
| 9. | 35 | 30 | 1 | 0.1 | 1 | 0.25 | 0.1 | 0.2 | 0.01 | 5 |
| 10. | 5 | 30 | 1 | 0.1 | 0.1 | 0.25 | 1 | 0.2 | 0.1 | 3 |
| 11. | 5 | 5 | 1 | 1 | 0.1 | 0.625 | 0.1 | 0.2 | 0.1 | 5 |
| 12. | 35 | 30 | 1 | 1 | 0.1 | 0.625 | 0.1 | 0.02 | 0.01 | 5 |
| Level and Range | Fermentation Factor | ||
|---|---|---|---|
| −1 | 0 | +1 | |
| 5 | 23 | 41 | Sucrose, X1 |
| 5 | 22 | 39 | yeast Extract, X2 |
| 0.01 | 0.06 | 0.11 | FeSO4, X3 |
| Runs | Variables | ||
|---|---|---|---|
| X1 | X2 | X3 | |
| Sucrose | Yeast Extract | FeSO4 | |
| 1 | 41 | 5 | 0.06 |
| 2 | 41 | 39 | 0.06 |
| 3 | 41 | 22 | 0.01 |
| 4 | 41 | 22 | 0.11 |
| 5 | 5 | 5 | 0.06 |
| 6 | 5 | 39 | 0.06 |
| 7 | 5 | 22 | 0.01 |
| 8 | 5 | 22 | 0.11 |
| 9 | 23 | 5 | 0.01 |
| 10 | 23 | 39 | 0.01 |
| 11 | 23 | 5 | 0.11 |
| 12 | 23 | 39 | 0.11 |
| 13 | 23 | 22 | 0.06 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahid, M.G.; Nadeem, M.; Gulzar, A.; Saleem, M.; Rehman, H.u.; Ghafoor, G.Z.; Hayyat, M.U.; Shahzad, L.; Arif, R.; Nelofer, R. Novel Ergot Alkaloids Production from Penicillium citrinum Employing Response Surface Methodology Technique. Toxins 2020, 12, 427. https://doi.org/10.3390/toxins12070427
Shahid MG, Nadeem M, Gulzar A, Saleem M, Rehman Hu, Ghafoor GZ, Hayyat MU, Shahzad L, Arif R, Nelofer R. Novel Ergot Alkaloids Production from Penicillium citrinum Employing Response Surface Methodology Technique. Toxins. 2020; 12(7):427. https://doi.org/10.3390/toxins12070427
Chicago/Turabian StyleShahid, Memuna Ghafoor, Muhammad Nadeem, Ahmed Gulzar, Muhammad Saleem, Hafeez ur Rehman, Gul Zareen Ghafoor, Muhammad Umar Hayyat, Laila Shahzad, Rabia Arif, and Rubina Nelofer. 2020. "Novel Ergot Alkaloids Production from Penicillium citrinum Employing Response Surface Methodology Technique" Toxins 12, no. 7: 427. https://doi.org/10.3390/toxins12070427
APA StyleShahid, M. G., Nadeem, M., Gulzar, A., Saleem, M., Rehman, H. u., Ghafoor, G. Z., Hayyat, M. U., Shahzad, L., Arif, R., & Nelofer, R. (2020). Novel Ergot Alkaloids Production from Penicillium citrinum Employing Response Surface Methodology Technique. Toxins, 12(7), 427. https://doi.org/10.3390/toxins12070427





