Prioritization of Mycotoxins Based on Their Genotoxic Potential with an In Silico-In Vitro Strategy
Abstract
:1. Introduction
2. Results
2.1. Structural Analysis
2.1.1. Group 1. Furo[2 ,3-b]benzofuran derivatives, AFB1 and STER
2.1.2. Group 2. 1,5-Dimethylspiro[8-oxatricyclo[7.2.1.02,7]dodec-5-ene-12,2’-oxirane] derivatives: NIV, DON, 3ADON, 15ADON, F-X, T-2 and HT-2 (trichothecenes)
2.1.3. Group 3. Structurally Unrelated Toxins, OTA, FB1 and ZEA
2.2. In Silico Toxicology (Phase 1)
2.2.1. Group 1 (AFB1 and STER)
2.2.2. Group 2 (Trichothecenes: NIV, DON, 3ADON, 15ADON, F-X, T-2 and HT-2 Toxins)
2.2.3. Group 3 (OTA, FB1 and ZEA)
2.3. In Vitro Genotoxicity Screening (Phase 2): SOS/umu Test
2.3.1. Exposure Concentrations and Solubility
2.3.2. SOS/Umu Test
2.3.3. Group 1 (AFB1 and STER)

2.3.4. Group 2 (NIV, DON, 3ADON, 15ADON, F-X, T-2 and HT-2 Toxins)


2.3.5. Group 3 (OTA, FB1 and ZEA)

3. Discussion
4. Conclusions
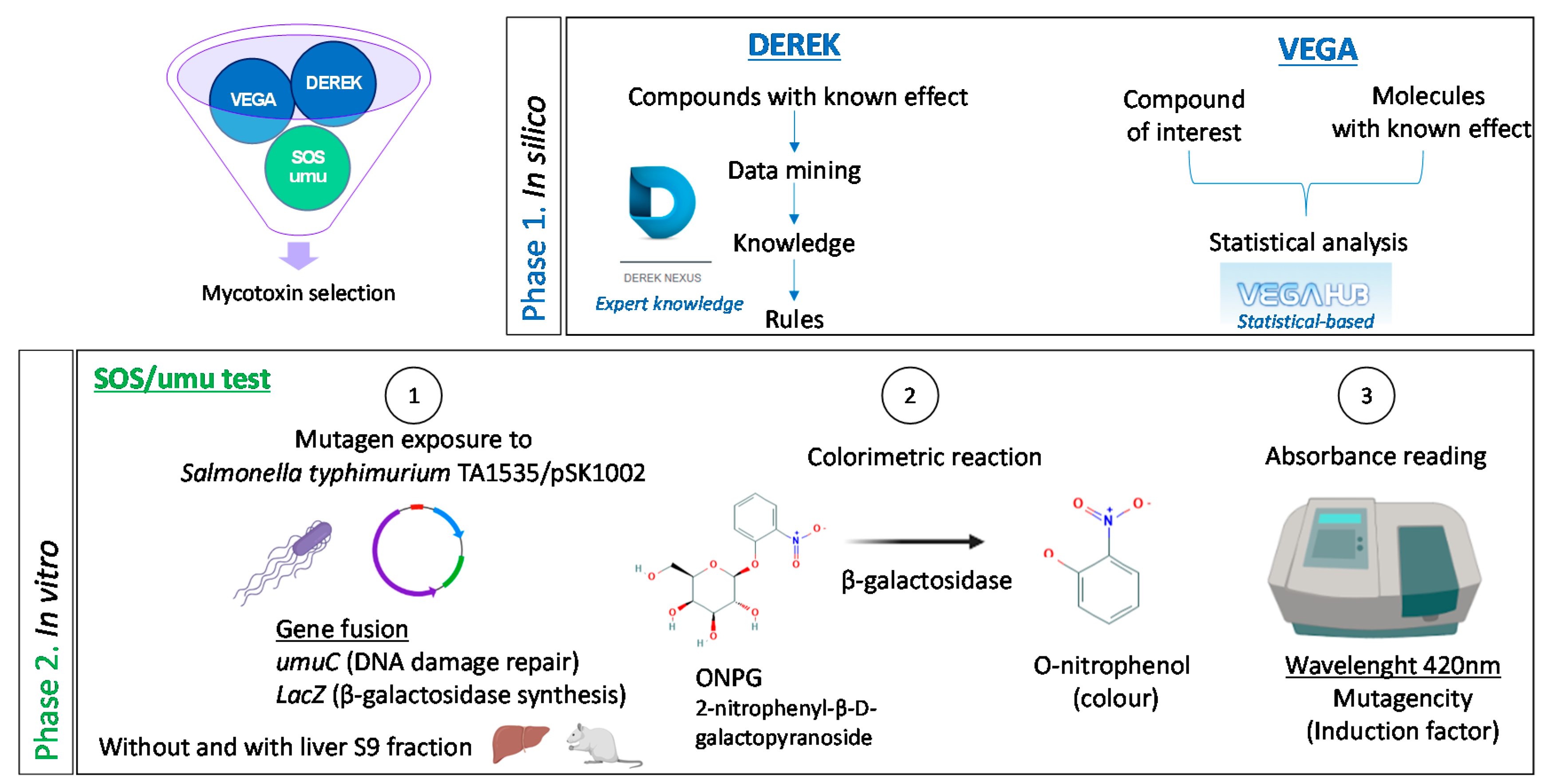
5. Materials and Methods
5.1. Chemicals and Reagents
5.2. In Silico Toxicology (Phase 1)
5.2.1. DEREK Nexus®
5.2.2. VEGA QSAR©
5.3. In Vitro Genotoxicity Screening (Phase 2)
5.3.1. Exposure Concentrations and Solubility Test
5.3.2. SOS/Umu Test
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Commission (EC). Commission Regulation (EC) No 1881 of 19 of December 2006 Setting Maximum Levels for Certain Contaminants in Foodstuffs (Text with EEA Relevance); European Commission: Brussels, Belgium, 2006; pp. 1–20. [Google Scholar]
- Logrieco, A.F.; Miller, J.D.; Eskola, M.; Krska, R.; Ayalew, A.; Bandyopadhyay, R.; Battilani, P.; Bhatnagar, D.; Chulze, S.; De Saeger, S.; et al. The Mycotox Charter: Increasing Awareness of, and Concerted Action for, Minimizing Mycotoxin Exposure Worldwide. Toxins 2018, 10, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited “FAO estimate” of 25%. Crit. Rev. Food. Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef] [PubMed]
- Maggiore, A.; Afonso, A.; Barrucci, F.; Sanctis, G.D.; EFSA (European Food Safety Authority). Climate change as a driver of emerging risks for food and feed safety, plant, animal health and nutritional quality. EFSA Support. Publ. 2020, 17, 146. [Google Scholar] [CrossRef]
- Battilani, P.; Toscano, P.; Van Der Fels-Klerx, H.J.; Moretti, A.; Leggieri, M.C.; Brera, C.; Rortais, A.; Goumperis, T.; Robinson, T. Aflatoxin B1 contamination in maize in Europe increases due to climate change. Sci. Rep. 2016, 6, 24328. [Google Scholar] [CrossRef] [Green Version]
- Penagos-Tabares, F.; Khiaosa-Ard, R.; Nagl, V.; Faas, J.; Jenkins, T.; Sulyok, M.; Zebeli, Q. Mycotoxins, Phytoestrogens and Other Secondary Metabolites in Austrian Pastures: Occurrences, Contamination Levels and Implications of Geo-Climatic Factors. Toxins 2021, 13, 460. [Google Scholar] [CrossRef]
- IARC. Chemical agents and related occupations. Monogr. Eval. Carcinog. Risks Hum. 2012, 100 F, 225–248. [Google Scholar]
- EFSA (European Food Safety Authority). Scientific Opinion on the risks for public health related to the presence of zearalenone in food. EFSA J. 2011, 9, 2197. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Knutsen, H.K.; Alexander, J.; Barregård, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Grasl-Kraupp, B.; et al. Risks to human and animal health related to the presence of deoxynivalenol and its acetylated and modified forms in food and feed. EFSA J. 2017, 15, 4718. [Google Scholar] [CrossRef]
- Arce-López, B.; Lizarraga, E.; Vettorazzi, A.; González-Peñas, E. Human Biomonitoring of Mycotoxins in Blood, Plasma and Serum in Recent Years: A Review. Toxins 2020, 12, 147. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.-C.; Madec, S.; Coton, E.; Hymery, N. Natural Co-Occurrence of Mycotoxins in Foods and Feeds and Their in vitro Combined Toxicological Effects. Toxins 2016, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Jáuregui, M.; López de Cerain, A.; González-Peñas, E.; Vettorazzi, A. Chapter 1: Combined Toxicity of Aflatoxins and Ochratoxin A. In Aflatoxins: Biochemistry, Toxicology, Public Health, Policies and Modern Methods of Analysis; Nova Science Publishers: New York, NY, USA, 2019; pp. 3–28. [Google Scholar]
- National Toxicology Program (NTP). Toxicology and Carcinogenesis Studies of Ochratoxin A (CAS No. 303-47-9) in F344/N Rats. Natl. Toxicol. Program. Tech. Rep. Ser. 1989, 358, 1–142. [Google Scholar]
- Ruan, H.; Lu, Q.; Wu, J.; Qin, J.; Sui, M.; Sun, X.; Shi, Y.; Luo, J.; Yang, M. Hepatotoxicity of food-borne mycotoxins: Molecular mechanism, anti-hepatotoxic medicines and target prediction. Crit. Rev. Food Sci. Nutr. 2021, 1–28. [Google Scholar] [CrossRef]
- Li, L.; Zhang, T.; Ren, X.; Li, B.; Wang, S. Male reproductive toxicity of zearalenone—meta-analysis with mechanism review. Ecotoxicol. Environ. Saf. 2021, 221, 112457. [Google Scholar] [CrossRef]
- Chen, J.; Wei, Z.; Wang, Y.; Long, M.; Wu, W.; Kuca, K. Fumonisin B1: Mechanisms of toxicity and biological detoxification progress in animals. Food Chem. Toxicol. 2021, 149, 111977. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Xue, D.; Zhang, C.; Rajput, S.A.; Qi, D. Mechanism of deoxynivalenol mediated gastrointestinal toxicity: Insights from mitochondrial dysfunction. Food Chem. Toxicol. 2021, 153, 112214. [Google Scholar] [CrossRef]
- Pickova, D.; Ostry, V.; Toman, J.; Malir, F. Aflatoxins: History, Significant Milestones, Recent Data on Their Toxicity and Ways to Mitigation. Toxins 2021, 13, 399. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Durston, W.E.; Yamasaki, E.; Lee, F.D. Carcinogens are Mutagens: A Simple Test System Combining Liver Homogenates for Activation and Bacteria for Detection. Proc. Natl. Acad. Sci. USA 1973, 70, 2281–2285. [Google Scholar] [CrossRef] [Green Version]
- McCann, J.; Spingarn, N.E.; Kobori, J.; Ames, B.N. Detection of carcinogens as mutagens: Bacterial tester strains with R factor plasmids. Proc. Natl. Acad. Sci. USA 1975, 72, 979–983. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.J.; Hsieh, D.P. Mutagenicity of aflatoxins related to their metabolism and carcinogenic potential. Proc. Natl. Acad. Sci. USA 1976, 73, 2241–2244. [Google Scholar] [CrossRef] [Green Version]
- Tang, T.; Friedman, M.A. Carcinogen activation by human liver enzymes in the ames mutagenicity test. Mutat. Res. Mutagen. Relat. Subj. 1977, 46, 387–394. [Google Scholar] [CrossRef]
- Kuczuk, M.H.; Benson, P.M.; Heath, H.; Hayes, A.W. Evaluation of the mutagenic potential of mycotoxins using Salmonella typhimurium and Saccharomyces cerevisiae. Mutat. Res. Mutagen. Relat. Subj. 1978, 53, 11–20. [Google Scholar] [CrossRef]
- Wehner, F.C.; Marasas, W.F.O.; Thiel, P.G. Lack of mutagenicity to Salmonella typhimurium of some Fusarium mycotoxins. Appl. Environ. Microbiol. 1978, 35, 659–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehner, F.; Thiel, P.; Van Rensburg, S.; Demasius, I.P. Mutagenicity to Salmonella typhimurium of some Aspergillus and Penicillium mycotoxins. Mutat. Res. Toxicol. 1978, 58, 193–203. [Google Scholar] [CrossRef]
- Lee, L.S.; Dunn, J.J.; Delucca, A.J.; Ciegler, A. Role of lactone ring of aflatoxin B1 in toxicity and mutagenicity. Cell. Mol. Life Sci. 1981, 37, 16–17. [Google Scholar] [CrossRef]
- Wheeler, L. A comparison of aflatoxin B1-induced cytotoxicity, mutagenicity and prophage induction in Salmonella typhimurium mutagen tester strains TA1535, TA1538, TA98 and TA100. Mutat. Res. Mol. Mech. Mutagen. 1981, 83, 39–48. [Google Scholar] [CrossRef]
- Beaune, P.; Lemestré-Cornet, R.; Kremers, P.; Albert, A.; Gielen, J. The Salmonella/mammalian microsome mutagenicity test: Comparison of human and rat livers as activating systems. Mutat. Res. Toxicol. 1985, 156, 139–146. [Google Scholar] [CrossRef]
- Auffray, Y.; Boutibonnes, P. Evaluation of the genotoxic activity of some mycotoxins using Escherichia coli in the SOS spot test. Mutat. Res. Toxicol. 1986, 171, 79–82. [Google Scholar] [CrossRef]
- Auffray, Y.; Boutibonnes, P. Induction of SOS function inEscherichia coli by some mycotoxins. Environ. Toxicol. Water Qual. 1988, 3, 371–378. [Google Scholar] [CrossRef]
- Reis, J. Detection of genotoxic properties of mycotoxins with the SOS chromotest. Naturwissenschaften 1986, 73, 677–678. [Google Scholar] [CrossRef]
- Krivobok, S.; Olivier, P.; Marzin, D.; Seigle-Murandi, F.; Steiman, R. Study of the genotoxic potential of 17 mycotoxins with the SOS Chromotest. Mutagenesis 1987, 2, 433–439. [Google Scholar] [CrossRef]
- Nakamura, S.; Oda, Y.; Shimada, T.; Oki, I.; Sugimoto, K. SOS-inducing activity of chemical carcinogens and mutagens in Salmonella typhimurium TA1535/pSK1002: Examination with 151 chemicals. Mutat. Res. Lett. 1987, 192, 239–246. [Google Scholar] [CrossRef]
- Foster, P.L.; Groopman, J.D.; Eisenstadt, E. Induction of base substitution mutations by aflatoxin B1 is mucAB dependent in Escherichia coli. J. Bacteriol. 1988, 170, 3415–3420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambamurti, K.; Callahan, J.; Luo, X.; Perkins, C.P.; Jacobsen, J.S.; Humayun, M.Z. Mechanisms of mutagenesis by a bulky DNA lesion at the guanine N7 position. Genetics 1988, 120, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Baertschi, S.W.; Raney, K.D.; Shimada, T.; Harris, T.M.; Guengerich, F.P. Comparison of rates of enzymic oxidation of aflatoxin B1, aflatoxin G1, and sterigmatocystin and activities of the epoxides in forming guanyl-N7 adducts and inducing different genetic responses. Chem. Res. Toxicol. 1989, 2, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Sakai, M.; Abe, K.-I.; Okumura, H.; Kawamura, O.; Sugiura, Y.; Horie, Y.; Ueno, Y. Genotoxicity of fungi evaluated by SOS microplate assay. Nat. Toxins 1992, 1, 27–34. [Google Scholar] [CrossRef]
- Bailey, E.A.; Iyer, R.S.; Stone, M.; Harris, T.M.; Essigmann, J.M. Mutational properties of the primary aflatoxin B1-DNA adduct. Proc. Natl. Acad. Sci. USA 1996, 93, 1535–1539. [Google Scholar] [CrossRef] [Green Version]
- Shimada, T.; Hayes, C.L.; Yamazaki, H.; Amin, S.; Hecht, S.S.; Guengerich, F.P.; Sutter, T.R. Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res. 1996, 56, 2979–2984. [Google Scholar]
- IARC. Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Man: Some Naturally Occurring Substances; IARC: Lyon, France, 1976; Volume 10, pp. 1–353. [Google Scholar]
- Ueno, Y. Mode of action of trichothecenes. Pergamon 1978, 49, 1737–1745. [Google Scholar] [CrossRef]
- Shimada, T.; Yamazaki, H.; Mimura, M.; Guengerich, F. Rat pulmonary microsomal cytochrome P-450 enzymes involved in the activation of procarcinogens. Mutat. Res. Mol. Mech. Mutagen. 1992, 284, 233–241. [Google Scholar] [CrossRef]
- IARC. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. Monogr. Eval. Carcinog. Risks Hum. 1993, 56, 397–444. [Google Scholar]
- Knasmüller, S.; Bresgen, N.; Kassie, F.; Mersch-Sundermann, V.; Gelderblom, W.; Zöhrer, E.; Eckl, P. Genotoxic effects of three Fusarium mycotoxins, fumonisin B1, moniliformin and vomitoxin in bacteria and in primary cultures of rat hepatocytes. Mutat. Res. Toxicol. Environ. Mutagen. 1997, 391, 39–48. [Google Scholar] [CrossRef]
- Takakura, N.; Nesslany, F.; Fessard, V.; Le Hegarat, L. Absence of in vitro genotoxicity potential of the mycotoxin deoxynivalenol in bacteria and in human TK6 and HepaRG cell lines. Food Chem. Toxicol. 2014, 66, 113–121. [Google Scholar] [CrossRef]
- Bendele, A.; Neal, S.; Oberly, T.; Thompson, C.; Bewsey, B.; Hill, L.; Rexroat, M.; Carlton, W.; Probst, G. Evaluation of ochratoxin A for mutagenicity in a battery of bacterial and mammalian cell assays. Food Chem. Toxicol. 1985, 23, 911–918. [Google Scholar] [CrossRef]
- Zepnik, H. Ochratoxin A-Induced Tumor Formation: Is There a Role of Reactive Ochratoxin A Metabolites? Toxicol. Sci. 2001, 59, 59–67. [Google Scholar] [CrossRef]
- Föllmann, W.; Lucas, S. Effects of the mycotoxin ochratoxin A in a bacterial and a mammalian in vitro mutagenicity test system. Arch. Toxicol. 2003, 19, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Obrecht-Pflumio, S.; Chassat, T.; Dirheimer, G.; Marzin, D. Genotoxicity of ochratoxin A by Salmonella mutagenicity test after bioactivation by mouse kidney microsomes. Mutat. Res. Toxicol. Environ. Mutagen. 1999, 446, 95–102. [Google Scholar] [CrossRef]
- Malaveille, C.; Brun, G.; Bartsch, H. Structure-activity studies in E. coli strains on ochratoxin A (OTA) and its analogues implicate a genotoxic free radical and a cytotoxic thiol derivative as reactive metabolites☆. Mutat. Res. Mol. Mech. Mutagen. 1994, 307, 141–147. [Google Scholar] [CrossRef]
- Bartholomew, R.M.; Ryan, D.S. Lack of mutagenicity of some phytoestrogens in the Salmonella/mammalian microsome assay. Mutat. Res. Toxicol. 1980, 78, 317–321. [Google Scholar] [CrossRef]
- Ghédira-Chékir, L.; Maaroufi, K.; Zakhama, A.; Ellouz, F.; Dhouib, S.; Creppy, E.; Bacha, H. Induction of a SOS repair system in lysogenic bacteria by zearalenone and its prevention by vitamin E. Chem. Biol. Interact. 1998, 113, 15–25. [Google Scholar] [CrossRef]
- IARC. Monographs on the Evaluation of Carcinogenic Risks To Humans. IARC Monogr. Eval. Carcinog. Risks Hum. 2002, 96, 1–390. [Google Scholar]
- Park, D.L.; Rua, S.M.; Mirocha, C.J.; Abd-Alla, E.-S.; Weng, C.Y. Mutagenic potentials of fumonisin contaminated corn following ammonia decontamination procedure. Mycopathologia 1992, 117, 105–108. [Google Scholar] [CrossRef]
- Aranda, M.; Pérez-Alzola, L.; Ellahueñe, M.F.; Sepúlveda, C. Assessment of in vitro mutagenicity in Salmonella and in vivo genotoxicity in mice of the mycotoxin fumonisin B(1). Mutagenesis 2000, 15, 469–471. [Google Scholar] [CrossRef] [Green Version]
- ICH. Harmonised Guideline Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals To Limit Potential Carcinogenic Risk M7(R1); ICH: Geneva, Switzerland, 2017; Volume 7. [Google Scholar]
- ICH. Guideline S2 (R1) on Genotoxicity Testing and Data Interpretation for Pharmaceuticals Intended for Human Use; ICH: Geneva, Switzerland, 2008; Volume 2, pp. 1–28. [Google Scholar]
- EFSA (European Food Safety Authority). Scientific opinion on genotoxicity testing strategies applicable to food and feed safety assessment. EFSA J. 2011, 9, 1–68. [Google Scholar] [CrossRef]
- OECD. Test. No. 471: Bacterial Reverse Mutation Test.; (OECD Guidelines for the Testing of Chemicals, Section 4); OECD: Paris, France, 2020; pp. 1–24. [Google Scholar] [CrossRef] [Green Version]
- OECD. Test No. 487: In Vitro Mammalian Cell Micronucleus Test; (OECD Guidelines for the Testing of Chemicals, Section 4); OECD: Paris, France, 2016; pp. 1–29. [Google Scholar] [CrossRef]
- Oda, Y.; Nakamura, S.; Oki, I.; Kato, T.; Shinagawa, H. Evaluation of the new system (umu-test) for the detection of environmental mutagens and carcinogens. Mutat. Res. Mutagen. Relat Subj. 1985, 147, 219–229. [Google Scholar] [CrossRef]
- Reifferscheid, G.; Heil, J.; Oda, Y.; Zahn, R. A microplate version of the SOS/umu-test for rapid detection of genotoxins and genotoxic potentials of environmental samples. Mutat. Res. Mutagen. Relat. Subj. 1991, 253, 215–222. [Google Scholar] [CrossRef]
- Reifferscheid, G.; Heil, J. Validation of the SOS/umu test using test results of 486 chemicals and comparison with the Ames test and carcinogenicity data. Mutat. Res. Toxicol. 1996, 369, 129–145. [Google Scholar] [CrossRef]
- Weaver, S.; Gleeson, M.P. The importance of the domain of applicability in QSAR modeling. J. Mol. Graph. Model. 2008, 26, 1315–1326. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Donohoe, T.; Garnett, K.; Lansink, A.O.; Afonso, A.; Noteborn, H. Emerging risks identification on food and feed—EFSA. EFSA J. 2018, 16, 5359. [Google Scholar] [CrossRef] [Green Version]
- Van Bossuyt, M.; Van Hoeck, E.; Raitano, G.; Vanhaecke, T.; Benfenati, E.; Mertens, B.; Rogiers, V. Performance of In Silico Models for Mutagenicity Prediction of Food Contact Materials. Toxicol. Sci. 2018, 163, 632–638. [Google Scholar] [CrossRef]
- Ferrari, T.; Gini, G. An open source multistep model to predict mutagenicity from statistical analysis and relevant structural alerts. Chem. Cent. J. 2010, 4, S2. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Cheng, F.; Chen, L.; Du, Z.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. In silico Prediction of Chemical Ames Mutagenicity. J. Chem. Inf. Model. 2012, 52, 2840–2847. [Google Scholar] [CrossRef]
- Benigni, R.; Battistelli, C.L.; Bossa, C.; Tcheremenskaia, O.; Crettaz, P. New perspectives in toxicological information management, and the role of ISSTOX databases in assessing chemical mutagenicity and carcinogenicity. Mutagenesis 2013, 28, 401–409. [Google Scholar] [CrossRef] [Green Version]
- Vian, M.; Raitano, G.; Roncaglioni, A.; Benfenati, E. In silico model for mutagenicity (Ames test), taking into account metabolism. Mutagenesis 2019. [Google Scholar] [CrossRef]
- Hillebrecht, A.; Muster, W.; Brigo, A.; Kansy, M.; Weiser, T.; Singer, T. Comparative Evaluation of in SilicoSystems for Ames Test Mutagenicity Prediction: Scope and Limitations. Chem. Res. Toxicol. 2011, 24, 843–854. [Google Scholar] [CrossRef] [PubMed]
- McDaniels, A.E.; Reyes, A.L.; Wymer, L.J.; Rankin, C.C.; Stelma, G.N. Comparison of the Salmonella (Ames) test, Umu tests, and the sos chromotests for detecting genotoxins. Environ. Mol. Mutagen. 1990, 16, 204–215. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority); Schrenk, D.; Bignami, M.; Bodin, L.; Chipman, J.K.; del Mazo, J.; Grasl-Kraupp, B.; Hogstrand, C.; Hoogenboom, L.; Leblanc, J.-C.; et al. Risk assessment of aflatoxins in food. EFSA J. 2020, 18, 6040. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Opinion of the Sscientific Panel on Contaminants in the Food Chain on a request from the Commission related to the potential increase of consumer health risk by a possible increase of the existing maximum levels for aflatoxins in almonds, hazelnuts and pistachios and derived products. EFSA J. 2007, 446, 1–127. [Google Scholar] [CrossRef]
- Díaz Nieto, C.H.; Granero, A.M.; Zon, M.A.; Fernández, H. Sterigmatocystin: A mycotoxin to be seriously considered. Food Chem. Toxicol. 2018, 118, 460–470. [Google Scholar] [CrossRef]
- Theumer, M.; Henneb, Y.; Khoury, L.; Snini, S.; Tadrist, S.; Canlet, C.; Puel, O.; Oswald, I.; Audebert, M. Genotoxicity of aflatoxins and their precursors in human cells. Toxicol. Lett. 2018, 287, 100–107. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Scientific Opinion on the risk for public and animal health related to the presence of sterigmatocystin in food and feed. EFSA J. 2013, 11, 3254. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Schrenk, D.; Bodin, L.; Chipman, J.K.; del Mazo, J.; Grasl-Kraupp, B.; Hogstrand, C.; Hoogenboom, L.; Leblanc, J.-C.; Nebbia, C.S.; et al. Risk assessment of ochratoxin A in food. EFSA J. 2020, 18, 6113. [Google Scholar] [CrossRef]
- Mostrom, M.S.; Raisbeck, M.F. Trichothecenes. In Veterinary Toxicology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 951–976. [Google Scholar]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From Simple to Complex Mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority). Scientific Opinion on the risks for animal and public health related to the presence of T-2 and HT-2 toxin in food and feed. EFSA J. 2011, 19, 2481. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Le Hégarat, L.; Takakura, N.; Simar, S.; Nesslany, F.; Fessard, V. The in vivo genotoxicity studies on nivalenol and deoxynivalenol. EFSA Support. Publ. 2014, 11, 1–33. [Google Scholar] [CrossRef] [Green Version]
- EFSA (European Food Safety Authority). Scientific Opinion on risks for animal and public health related to the presence of nivalenol in food and feed. EFSA J. 2013, 11, 3262. [Google Scholar] [CrossRef]
- Rogowska, A.; Pomastowski, P.; Sagandykova, G.; Buszewski, B. Zearalenone and its metabolites: Effect on human health, metabolism and neutralisation methods. Toxicon 2019, 162, 46–56. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Opinion of the Scientific Panel on contaminants in the food chain [CONTAM] related to fumonisins as undesirable substances in animal feed. EFSA J. 2005, 3, 235. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Knutsen, H.-K.; Barregård, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Edler, L.; Grasl-Kraupp, B.; et al. Appropriateness to set a group health-based guidance value for fumonisins and their modified forms. EFSA J. 2018, 16, 5172. [Google Scholar] [CrossRef]
- Hideo, S.; Takeshi, K.; Tsuneo, I.; Kozo, M.; Atsuo, N. Cloning and characterization of the umu operon responsible for inducible mutagenesis in Escherichia coli. Gene 1983, 23, 167–174. [Google Scholar] [CrossRef]
- Ductless Fume Hoods Classic Range|CRUMA. Available online: https://cruma.es/en/ductless-fume-hoods-classic-range/ (accessed on 3 June 2020).
- Lhasa Limited. ICH M7 Prediction. Available online: https://www.lhasalimited.org/ich-m7.htm (accessed on 13 November 2019).
- Marchant, C.A.; Briggs, K.A.; Long, A. In Silico Tools for Sharing Data and Knowledge on Toxicity and Metabolism: Derek for Windows, Meteor, and Vitic. Toxicol. Mech. Methods 2008, 18, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Worth, A.; Lapenna, S.; Lo Piparo, E.; Mostrag-Szlichtyng, A.; Serafimova, R. The Applicability of Software Tools for Genotoxicity and Carcinogenicity Prediction: Case Studies Relevant to the Assessment of Pesticides; JRC Scientific and Technical Reports; EC Joint Research Centre Institute for Health and Consumer Protection: Ispra, Italy, 2010; pp. 18–19. [Google Scholar] [CrossRef]
- Benfenati, E.; Manganaro, A.; Gini, G. VEGA-QSAR: AI inside a platform for predictive toxicology. In Proceedings of the Popularize Artificial Intelligence 2013, CEUR, Turing, Italy, 5 December 2013; Volume 1107, pp. 21–28. [Google Scholar]
- Kühne, R.; Ebert, R.-U.; Schüürmann, G. Chemical Domain of QSAR Models from Atom-Centered Fragments. J. Chem. Inf. Model. 2009, 49, 2660–2669. [Google Scholar] [CrossRef] [PubMed]



| Mycotoxin | IARC Classification (Year) | Ames Test | SOS Test |
|---|---|---|---|
| Aflatoxin B1 | Group 1 (2012) [8] | Positive [20,21,22,23,24,25,26,27,28,29] | Positive [30,31,32,33,34,35,36,37,38,39] Negative [40] |
| Sterigmatocystin | Group 2B (1976) [41] | Positive [21,23,24,25,26,42] | Positive [32,33,37,38,43] Negative [40] |
| Nivalenol | Group 3 (1993) [44] | No data | No data |
| Deoxynivalenol | Group 3 (1993) [44] | Negative [25,45,46] | Negative [45] |
| 3-Acetyldeoxynivalenol | No classification | Negative [25] | No data |
| 15-Acetyldeoxynivalenol | No classification | No data | No data |
| Fusarenon-X | No clasiffication | Negative [42] | No data |
| T-2 toxin | Group 3 (1993) [44] | Negative [24,25,42] | Negative [30,33] |
| HT-2 toxin | No classification | No data | No data |
| Ochratoxin A | Group 2B (1993) [44] | Negative [24,26,47,48,49] Positive [50] | Negative: [32,38] Weak positive [33] Positive [51] |
| Zearalenone | Group 3 (1993) [44] | Negative [24,25,52] | Negative [30,31,32,33,38] Positive [53] |
| Fumonisin B1 | Group 2B (2002) [54] | Negative [45,55,56] | Negative [45] |
| Ref | Mutagenicity In Vitro | Mutagenicity In Vivo | Chromosome Damage (Mammal) | Non-Specific Genotoxicity In Vitro d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacterium | Mammal | Mammal | In Vitro | In Vivo | ||||||||
| Alert Ref a | Prediction | Alert Ref a | Prediction | Alert Ref a | Prediction | Alert Ref a | Prediction | Alert Ref a | Prediction | Alert Ref a | Prediction | |
| AFB1 | 201 | PLAUSIBLE | 306 | PLAUSIBLE | 201 | EQUIVOCAL | 306, 309 | PLAUSIBLE | - b | - b | 306 | PLAUSIBLE |
| STER | 201 | PLAUSIBLE | 306 | PLAUSIBLE | 201 | EQUIVOCAL | 306 | PLAUSIBLE | - b | - b | 306 | PLAUSIBLE |
| NIV | 019 | PLAUSIBLE | - b | - b | 019 | PLAUSIBLE | 019, 309 | PLAUSIBLE | 019 | PLAUSIBLE | - b | - b |
| DON | 019 | PLAUSIBLE | - b | - b | 019 | PLAUSIBLE | 019, 309 | PLAUSIBLE | 019 | PLAUSIBLE | - b | - b |
| 3ADON | 019 | PLAUSIBLE | - b | - b | 019 | PLAUSIBLE | 019, 309 | PLAUSIBLE | 019 | PLAUSIBLE | - b | - b |
| 15ADON | 019 | PLAUSIBLE | - b | - b | 019 | PLAUSIBLE | 019, 309 | PLAUSIBLE | 019 | PLAUSIBLE | - b | - b |
| F-X | 019 | PLAUSIBLE | - b | - b | 019 | PLAUSIBLE | 019, 309 | PLAUSIBLE | 019 | PLAUSIBLE | - b | - b |
| T-2 | 019 | PLAUSIBLE | - b | - b | 019 | PLAUSIBLE | 019 | PLAUSIBLE | 019 | PLAUSIBLE | - b | - b |
| HT-2 | 019 | PLAUSIBLE | - b | - b | 019 | PLAUSIBLE | 019 | PLAUSIBLE | 019 | PLAUSIBLE | - b | - b |
| OTA | (*) c | INACTIVE | - b | - b | - b | - b | - b | - b | - b | - b | - b | - b |
| ZEA | (*) c | INACTIVE | - b | - b | - b | - b | - b | - b | - b | - b | - b | - b |
| FB1 | (*) c | INACTIVE | - b | - b | - b | - b | - b | - b | - b | - b | - b | - b |
| Mycotoxin | VEGA Tool | Exp a | Pdtion b | SA/SM c |
|---|---|---|---|---|
| AFB1 | CAESAR | M | M | - |
| SarPy/IRFMN | M | M | SM: 35, 141, 154, 158, 177, 187, 196 | |
| ISS | M | M | SA:30 | |
| KNN/Racross | M | M | ||
| STER | CAESAR | M | M | - |
| SarPy/IRFMN | M | M | SM: 35, 59, 84 | |
| ISS | M | M | SA:59 | |
| KNN/Racross | - | M | ||
| NIV | CAESAR | NM | suspect M | SA:7 |
| SarPy/IRFMN | NM | M | SM: 92, 97, 113, 162, 163, 169, 177, 182, 186 | |
| ISS | - | M | SA: 7, 10 | |
| KNN/Racross | NM | NM | ||
| DON | CAESAR | NM | suspect M | SA: 7 |
| SarPy/IRFMN | NM | M | SM: 92, 97, 113, 162, 163, 169, 177, 182, 186 | |
| ISS | - | M | SA: 7, 10 | |
| KNN/Racross | NM | NM | ||
| 3ADON | CAESAR | - | suspect M | SA: 7 |
| SarPy/IRFMN | - | M | SM: 92, 97, 113, 162, 163, 169, 177, 182, 186, 188 | |
| ISS | - | M | SA: 7, 10 | |
| KNN/Racross | - | NM | ||
| 15ADON | CAESAR | - | suspect M | SA: 7 |
| SarPy/IRFMN | - | M | SM: 92, 97, 113, 123, 162, 163, 169, 177, 182, 186, 188 | |
| ISS | - | M | SA: 7, 10 | |
| KNN/Racross | - | NM | ||
| F-X | CAESAR | NM | suspect M | SA: 7 |
| SarPy/IRFMN | NM | M | SM: 92, 97, 113, 162, 163, 169, 177, 182, 186, 188 | |
| ISS | - | M | SA: 7, 10 | |
| KNN/Racross | - | NM | ||
| T-2 | CAESAR | NM | suspect M | SA: 7 |
| SarPy/IRFMN | NM | M | SM: 92, 97, 113, 162, 163, 169, 178, 182, 186, 188 | |
| ISS | - | M | SA:7 | |
| KNN/Racross | - | NM | ||
| HT-2 | CAESAR | - | suspect M | SA:7 |
| SarPy/IRFMN | - | M | SM: 92, 97, 113, 123, 162, 163, 169, 178, 182, 186, 188 | |
| ISS | - | M | SA:7 | |
| KNN/Racross | - | NM | ||
| OTA | CAESAR | - | NM | - |
| SarPy/IRFMN | - | NM | SM: 146, 170, 189, 190 | |
| ISS | NM | NM | - | |
| KNN/Racross | NM | NM | ||
| ZEA | CAESAR | NM | NM | - |
| SarPy/IRFMN | NM | NM | SM: 146, 163, 170, 177, 189 | |
| ISS | NM | NM | - | |
| KNN/Racross | NM | NM | ||
| FB1 | CAESAR | - | NM | - |
| SarPy/IRFMN | - | NM | SM: 121, 137, 157, 163, 169, 178, 182, 188 | |
| ISS | NM | NM | - | |
| KNN/Racross | NM | NM |
| Ref | VEGA Tool | Prediction Quality VEGA Models Outcomes a | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AD | ADI | Reliability | CONSENSUS | Sim | Acc | Cc | DRC | ACF | ||
| AFB1 | CAESAR | INTO | 1 | good | M | 1 | 1 | 1 | True | 1 |
| SarPy/IRFMN | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| ISS | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| KNN/Racross | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| STER | CAESAR | INTO | 1 | good | M | 1 | 1 | 1 | True | 1 |
| SarPy/IRFMN | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| ISS | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| KNN/Racross | could out | 0.76 | moderate | 0.963 | 0.49 | 0.75 | 1 | |||
| NIV | CAESAR | OUT | 0 | low | NM | 1 | 0 | 0 | True | 1 |
| SarPy/IRFMN | OUT | 0 | low | 1 | 0 | 0 | 1 | |||
| ISS | OUT | 0 | low | 0.80 | 0.51 | 0.51 | 1 | |||
| KNN/Racross | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| DON | CAESAR | OUT | 0 | low | NM | 1 | 0 | 0 | True | 1 |
| SarPy/IRFMN | OUT | 0 | low | 1 | 0 | 0 | 1 | |||
| ISS | OUT | 0.64 | low | 0.80 | 0.51 | 0.51 | 1 | |||
| KNN/Racross | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| 3ADON | CAESAR | OUT | 0 | low | NM | 0.96 | 0 | 0 | True | 1 |
| SarPy/IRFMN | OUT | 0 | low | 0.96 | 0 | 0 | 1 | |||
| ISS | OUT | 0.65 | low | 0.82 | 0.51 | 0.51 | 1 | |||
| KNN/Racross | INTO | 0.97 | good | 0.94 | 1 | 1 | 1 | |||
| 15ADON | CAESAR | OUT | 0 | low | NM | 0.96 | 0 | 0 | True | 1 |
| SarPy/IRFMN | OUT | 0 | low | 0.96 | 0 | 0 | 1 | |||
| ISS | OUT | 0.64 | low | 0.82 | 0.51 | 0.51 | 1 | |||
| KNN/Racross | INTO | 0.97 | good | 0.94 | 1 | 1 | 1 | |||
| F-X | CAESAR | OUT | 0 | low | NM | 1 | 0 | 0 | True | 1 |
| SarPy/IRFMN | OUT | 0 | low | 1 | 0 | 0 | 1 | |||
| ISS | could out | 0.90 | moderate | 1 | 1 | 1 | 1 | |||
| KNN/Racross | INTO | 0.97 | good | 0.93 | 1 | 1 | 1 | |||
| T-2 | CAESAR | OUT | 0 | low | NM | 1 | 0 | 0 | True | 1 |
| SarPy/IRFMN | OUT | 0 | low | 1 | 0 | 0 | 1 | |||
| ISS | could out | 0.86 | moderate | 0.78 | 1 | 1 | 1 | |||
| KNN/Racross | could out | 0.78 | moderate | 0.854 | 1 | 0.52 | 1 | |||
| HT-2 | CAESAR | OUT | 0 | low | M | 0.935 | 0 | 0 | True | 1 |
| SarPy/IRFMN | OUT | 0 | low | 0.935 | 0 | 0 | 1 | |||
| ISS | could out | 0.89 | moderate | 0.8 | 1 | 1 | 1 | |||
| KNN/Racross | could out | 0.87 | moderate | 0.877 | 1 | 0.75 | 1 | |||
| OTA | CAESAR | could out | 0.72 | moderate | NM | 0.83 | 1 | 0.38 | True | 1 |
| SarPy/IRFMN | could out | 0.72 | moderate | 0.83 | 1 | 0.38 | 1 | |||
| ISS | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| KNN/Racross | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| ZEA | CAESAR | INTO | 1 | good | NM | 1 | 1 | 1 | True | 1 |
| SarPy/IRFMN | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| ISS | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| KNN/Racross | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| FB1 | CAESAR | could out | 0.88 | moderate | NM | 0.908 | 1 | 0.71 | True | 1 |
| SarPy/IRFMN | could out | 0.80 | moderate | 0.908 | 0.71 | 0.71 | 1 | |||
| ISS | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
| KNN/Racross | INTO | 1 | good | 1 | 1 | 1 | 1 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alonso-Jauregui, M.; Font, M.; González-Peñas, E.; López de Cerain, A.; Vettorazzi, A. Prioritization of Mycotoxins Based on Their Genotoxic Potential with an In Silico-In Vitro Strategy. Toxins 2021, 13, 734. https://doi.org/10.3390/toxins13100734
Alonso-Jauregui M, Font M, González-Peñas E, López de Cerain A, Vettorazzi A. Prioritization of Mycotoxins Based on Their Genotoxic Potential with an In Silico-In Vitro Strategy. Toxins. 2021; 13(10):734. https://doi.org/10.3390/toxins13100734
Chicago/Turabian StyleAlonso-Jauregui, Maria, María Font, Elena González-Peñas, Adela López de Cerain, and Ariane Vettorazzi. 2021. "Prioritization of Mycotoxins Based on Their Genotoxic Potential with an In Silico-In Vitro Strategy" Toxins 13, no. 10: 734. https://doi.org/10.3390/toxins13100734
APA StyleAlonso-Jauregui, M., Font, M., González-Peñas, E., López de Cerain, A., & Vettorazzi, A. (2021). Prioritization of Mycotoxins Based on Their Genotoxic Potential with an In Silico-In Vitro Strategy. Toxins, 13(10), 734. https://doi.org/10.3390/toxins13100734





