The hokW-sokW Locus Encodes a Type I Toxin–Antitoxin System That Facilitates the Release of Lysogenic Sp5 Phage in Enterohemorrhagic Escherichia coli O157
Abstract
:1. Introduction
2. Results
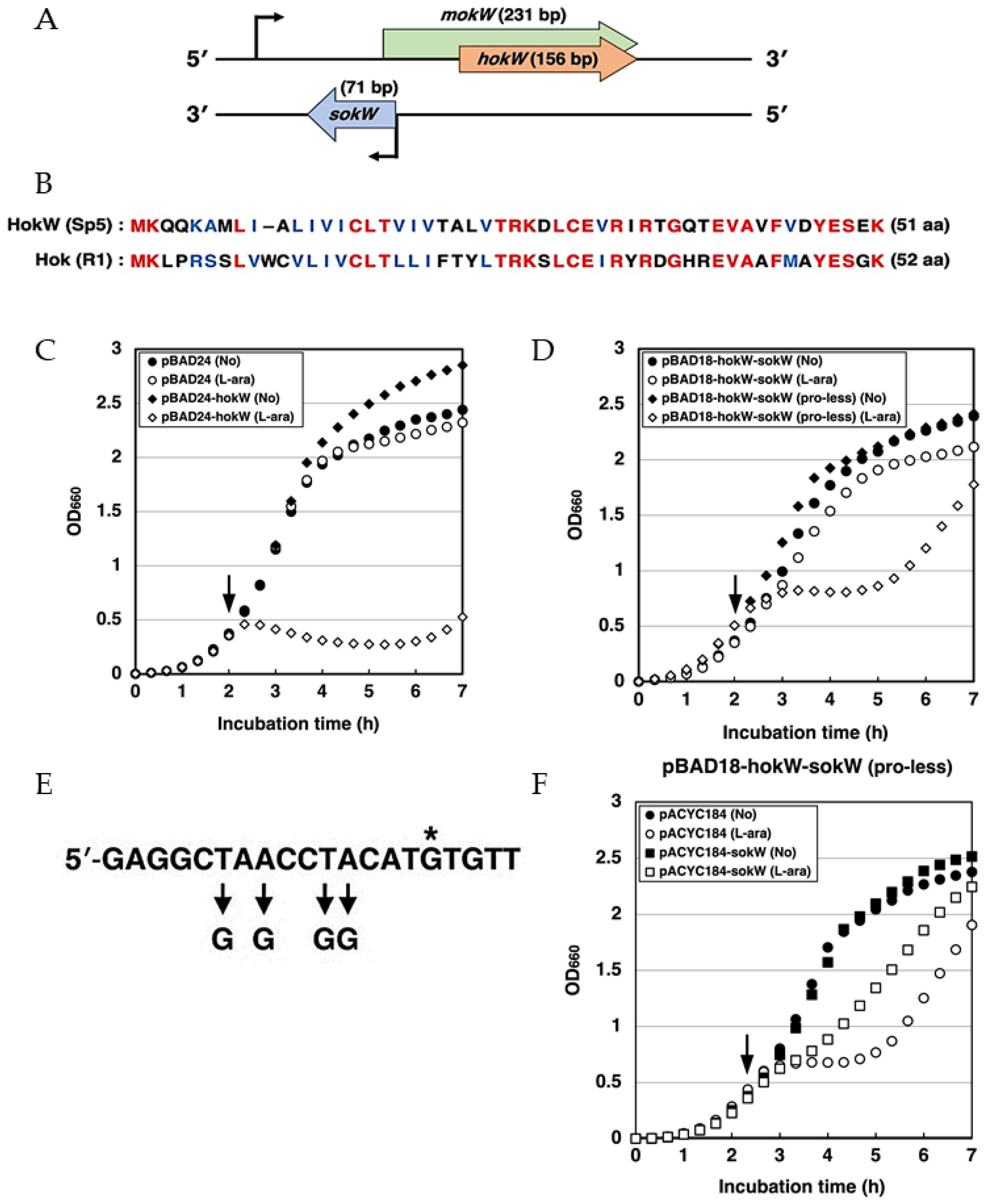
2.1. hokW-sokW Functions as a TA System
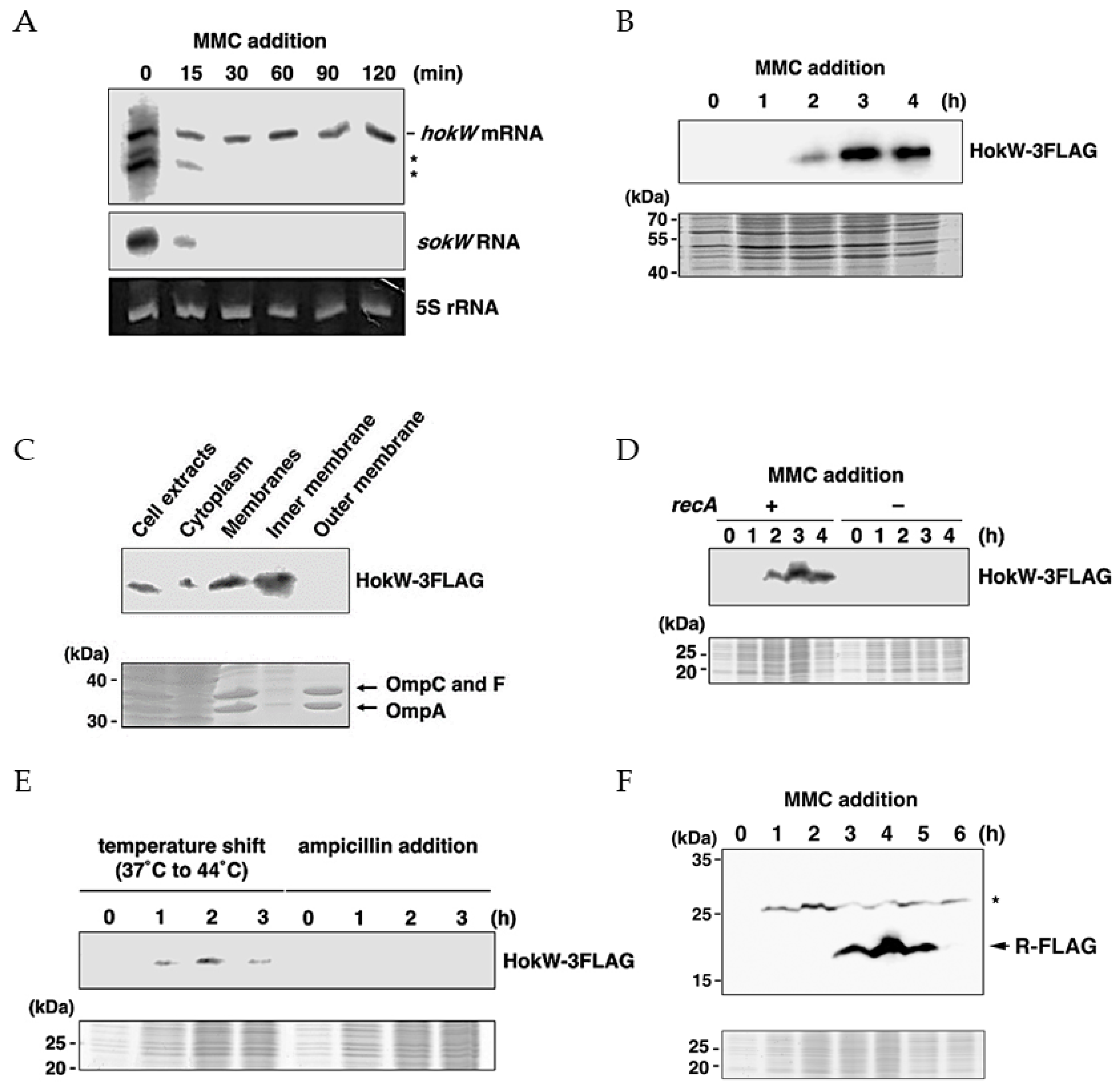
2.2. hokW-sokW Is a RNase III and Hfq Dependent Type I TA System
2.3. hokW-sokW Assists Growth Retardation and Reduces Cell Viability after Mitomycin C Treatment
2.4. MMC Triggers HokW Toxin Production in a RecA Dependent Manner
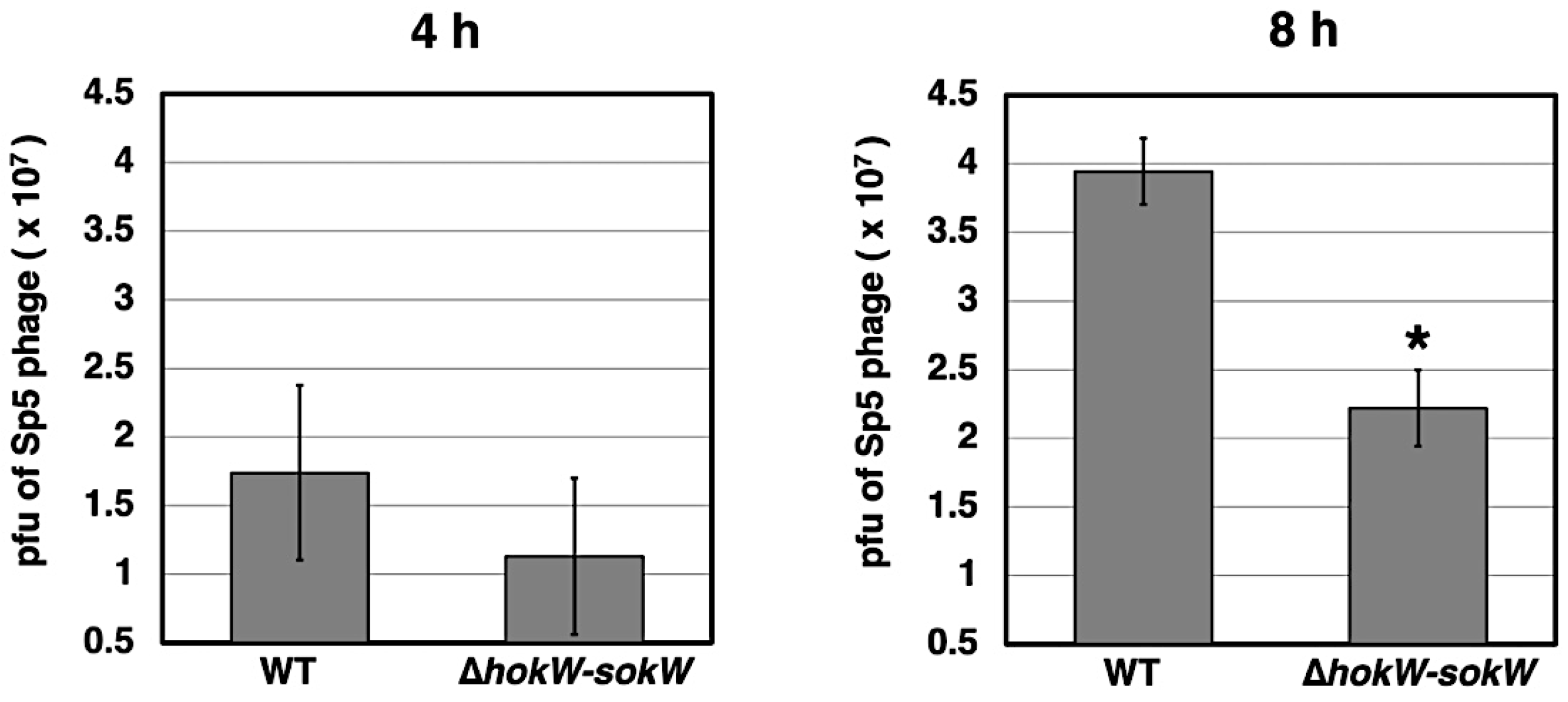
2.5. hokW-sokW Facilitates the Release of Sp5 Phage Progeny
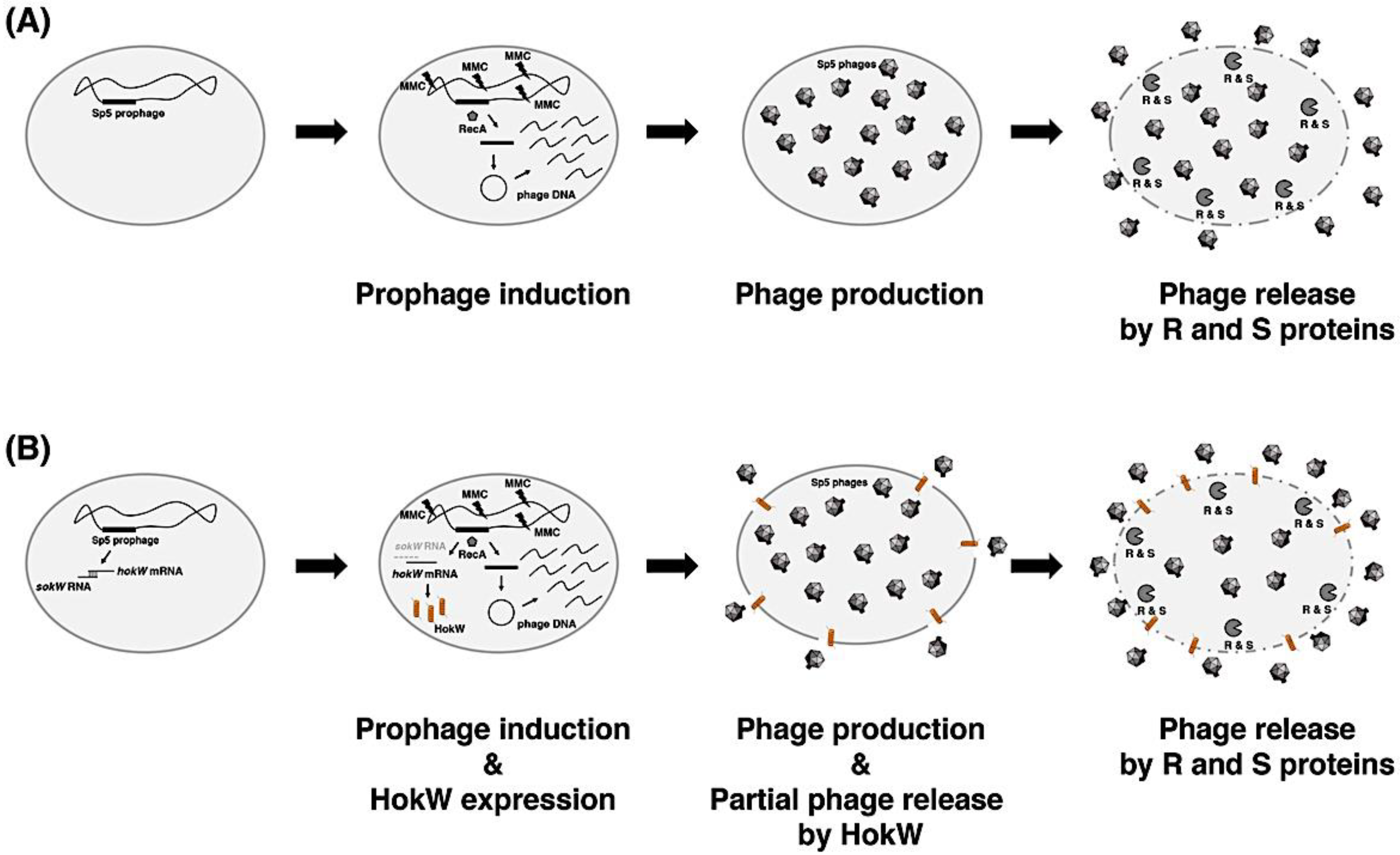
3. Discussion
4. Materials and Methods
4.1. E. coli Strains
4.2. Plasmid Construction
4.3. E. coli Growth and CFU Assay
4.4. Northern Blotting Analysis
4.5. Fractionation of Cell Extracts and Western Blotting Analysis
4.6. Plaque Formation Assay of Sp5 Phages
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandey, D.P. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef] [Green Version]
- Thisted, T.; Gerdes, K. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J. Mol. Biol. 1992, 223, 41–54. [Google Scholar] [CrossRef]
- Germain, E.; Castro-Roa, D.; Zenkin, N.; Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 2013, 52, 248–254. [Google Scholar] [CrossRef]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef]
- Hazan, R.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genom. 2004, 272, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Fineran, P.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P.C. The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koga, M.; Otsuka, Y.; Lemire, S.; Yonesaki, T. Escherichia coli rnlA and rnlB Compose a Novel Toxin–Antitoxin System. Genetics 2011, 187, 123–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guegler, C.K.; Laub, M.T. Shutoff of host transcription triggers a toxin-antitoxin system to cleave phage RNA and abort infection. Mol. Cell 2021, 81, 2361–2373.e9. [Google Scholar] [CrossRef]
- Karoui, H.; Bex, F.; Drèze, P.; Couturier, M. Ham22, a mini-F mutation which is lethal to host cell and promotes recA-dependent induction of lambdoid prophage. EMBO J. 1983, 2, 1863–1868. [Google Scholar] [CrossRef]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [Green Version]
- Fozo, E.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of type I toxin–antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef] [PubMed]
- Lehnherr, H.; Yarmolinsky, M.B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 3274–3277. [Google Scholar] [CrossRef] [Green Version]
- Goeders, N.; Chai, R.; Chen, B.; Day, A.; Salmond, G.P. Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins 2016, 8, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wood, T.K. Cryptic prophages as targets for drug development. Drug Resist. Updat. 2016, 27, 30–38. [Google Scholar] [CrossRef] [Green Version]
- DeShazer, D. Genomic diversity of Burkholderia pseudomallei clinical isolates: Subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 2004, 186, 3938–3950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decewicz, P.; Dziewit, L.; Golec, P.; Kozlowska, P.; Bartosik, D.; Radlinska, M. Characterization of the virome of Paracoccus spp. (Alphaproteobacteria) by combined in silico and in vivo approaches. Sci. Rep. 2019, 9, 7899. [Google Scholar] [CrossRef]
- Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. type II toxin-antitoxin systems: Evolution and revolutions. J. Bacteriol. 2020, 202, 00763–19. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yao, J.; Sun, Y.-C.; Wood, T.K. Type VII toxin/antitoxin classification system for antitoxins that enzymatically neutralize toxins. Trends Microbiol. 2021, 29, 388–393. [Google Scholar] [CrossRef]
- Gerdes, K.; Wagner, E.G.H. RNA antitoxins. Curr. Opin. Microbiol. 2007, 10, 117–124. [Google Scholar] [CrossRef]
- Fozo, E.M.; Hemm, M.R.; Storz, G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008, 72, 579–589. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, K.; Bech, F.; Jørgensen, S.; Løbner-Olesen, A.; Rasmussen, P.; Atlung, T.; Boe, L.; Karlstrom, O.; Molin, S.; von Meyenburg, K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986, 5, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Osapay, G.; Selsted, M.E.; Wood, T.K. Antimicrobial properties of the Escherichia coli R1 plasmid host killing peptide. J. Biotechnol. 2003, 100, 1–12. [Google Scholar] [CrossRef]
- Gerdes, K.; Nielsen, A.; Thorsted, P.; Wagner, E.H. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable Hok, SrnB and PndA effector messenger RNAs. J. Mol. Biol. 1992, 226, 637–649. [Google Scholar] [CrossRef]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecota, D.C.; Wood, T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1996, 178, 2044–2050. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, M.; Kurokawa, K.; Hayashi, T. Diversification of Escherichia coli genomes: Are bacteriophages the major contributors? Trends Microbiol. 2001, 9, 481–485. [Google Scholar] [CrossRef]
- Ogura, Y.; Mondal, S.I.; Islam, M.S.; Mako, T.; Arisawa, K.; Katsura, K.; Ooka, T.; Gotoh, Y.; Murase, K.; Ohnishi, M.; et al. The Shiga toxin 2 production level in enterohemorrhagic Escherichia coli O157: H7 is correlated with the subtypes of toxin-encoding phage. Sci. Rep. 2015, 5, 16663. [Google Scholar] [CrossRef] [Green Version]
- Kavita, K.; De Mets, F.; Gottesman, S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018, 42, 53–61. [Google Scholar] [CrossRef]
- Fuchs, S.; Mühldorfer, I.; Donohue-Rolfe, A.; Kerényi, M.; Emődy, L.; Alexiev, R.; Nenkov, P.; Hacker, J. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 1999, 27, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Ennis, D.G.; Fisher, B.; Edmiston, S.; Mount, D.W. Dual role for Escherichia coli RecA protein in SOS mutagenesis. Proc. Natl. Acad. Sci. USA 1985, 82, 3325–3329. [Google Scholar] [CrossRef] [Green Version]
- Bell, J.; Kowalczykowski, S.C. RecA: Regulation and mechanism of a molecular search engine. Trends Biochem. Sci. 2016, 41, 491–507. [Google Scholar] [CrossRef] [Green Version]
- Alawneh, A.M.; Qi, D.; Yonesaki, T.; Otsuka, Y. An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol. Microbiol. 2015, 99, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Franch, T.; Gerdes, K. Programmed cell death in bacteria: Translational repression by mRNA end-pairing. Mol. Microbiol. 1996, 21, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Brielle, R.; Pinel-Marie, M.-L.; Felden, B. Linking bacterial type I toxins with their actions. Curr. Opin. Microbiol. 2016, 30, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Ishikawa, T.; Takahashi, C.; Masuda, M. A short peptide derived from the ZorO Toxin Functions as an effective antimicrobial. Toxins 2019, 11, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nonin-Lecomte, S.; Fermon, L.; Felden, B.; Pinel-Marie, M.-L. Bacterial type I toxins: Folding and membrane interactions. Toxins 2021, 13, 490. [Google Scholar] [CrossRef]
- Emond, E.; Dion, E.; Walker, S.A.; Vedamuthu, E.R.; Kondo, J.K.; Moineau, S. AbiQ, an abortive infection mechanism from Lactococcus lactis. Appl. Environ. Microbiol. 1998, 64, 4748–4756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsunaka, S.; Sudo, N.; Sekine, Y. Lysogenisation of Shiga toxin-encoding bacteriophage represses cell motility. J. Gen. Appl. Microbiol. 2018, 64, 34–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzzau, S.; Figueroa-Bossi, N.; Rubino, S.; Bossi, L. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 2001, 98, 15264–15269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, G.G.; Young, K.K.Y.; Edlin, G.J.; Konigsberg, W. High-frequency generalised transduction by bacteriophage T4. Nat. Cell Biol. 1979, 280, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kai, T.; Selick, H.E.; Yonesaki, T. Destabilization of bacteriophage T4 mRNAs by a mutation of gene 61.5. Genetics 1996, 144, 7–14. [Google Scholar] [CrossRef]
- Fontaine, F.; Fuchs, R.T.; Storz, G. Membrane localization of small proteins in Escherichia coli. J. Biol. Chem. 2011, 286, 32464–32474. [Google Scholar] [CrossRef] [Green Version]


| Strains | Genotype | Source/Reference |
|---|---|---|
| TY0807 | sup0 araD139 hsdR ∆lacX74 rpsL araD+ | [9] |
| TY0807 ∆hfq | TY0807 ∆hfq::kmr | This study |
| BW25113 | rrnB3 ∆lacZ4787 hsdR514 ∆(araBAD)567 ∆(rhaBAD)568 rph-1 | NBRP-E. coli at NIG |
| JW2669 | BW25113 ∆recA::kmr | NBRP-E. coli at NIG |
| JW4130 | BW25113 ∆hfq::kmr | NBRP-E. coli at NIG |
| ME5413 | metB1 his rnc-105 ranA2074 | NBRP-E. coli at NIG |
| MG1655 | λ- ilvG- rfb-50 rph-1 | [40] |
| MG1655-Sp5 | MG1655-Sp5(∆stx2AB) | [40] |
| MG1655-Sp5(kmr) | MG1655-Sp5(∆stx2AB::kmr) | [40] |
| MG1655-Sp5(∆hokW-sokW) | MG1655-Sp5(∆stx2AB ∆hokW-sokW::kmr) | This study |
| MG1655-Sp5(∆r-s) | MG1655-Sp5(∆stx2AB ∆r-s::cat) | This study |
| MG1655-Sp5(∆hokW-sokW ∆r-s) | MG1655-Sp5(∆stx2AB ∆hokW-sokW::kmr ∆r-s::cat) | This study |
| MG1655-Sp5(hokW-3FLAG-kmr) | MG1655-Sp5(∆stx2AB hokW-3FLAG-kmr) | This study |
| MG1655-Sp5(hokW-3FLAG) | MG1655-Sp5(∆stx2AB hokW-3FLAG) | This study |
| MG1655 ∆recA-Sp5(hokW-3FLAG) | MG1655 ∆recA::kmr-Sp5(∆stx2AB hokW-3FLAG) | This study |
| MG1655-Sp5(r-FLAG) | MG1655-Sp5(∆stx2AB r-FLAG-cat) | This study |
| Primer Name | Sequence (5′–3′) |
|---|---|
| YO-349 | CTTGAGGCTATCTGCCTCGGGCATG |
| YO-350 | GCGTTGAGGATGCCTGACACATCAG |
| YO-351 | GCGGGTGCTTGAGGCTATCTGCCTCGGGCATGAACACCAACGGCAGATAGCATATGAATATCCTCCTTAG |
| YO-352 | CACATCAGAGGTGGCGGGAGATTACTCCCCCGCTTGGTCTCTTACTTCTCGTGTAGGCTGGAGCTGCTTC |
| YO-357 | AGGAATTCACCATGAAGCAGCAAAAGGCGATG |
| YO-369 | TCCTGCAGTTACTTCTCAGATTCGTAGTC |
| YO-406 | TCAAGCTTTCATAGCCTGCTTCTCCTTGCC |
| YO-421 | CCGGCTCGCCTCTTACGTGCCGAAAG |
| YO-422 | CATGTGTTCAGCATGGATTGAGCCTC |
| YO-432 | AGGAATTCGAATCAATGACCTGGCCTGAAGC |
| YO-433 | AGAAGCTTGAAATAAGTGCTGCAATCAATAC |
| YO-541 | AGGGATCCGCCTCGGGCATGAACACCAAC |
| YO-560 | GAACCGGTCAGACGGAGGTCGCTGTCTTCGTAGACTACGAATCTGAGAAGGACTACAAAGACCATGACGG |
| YO-561 | TCAGAGATGAACATTCAAACAGCATTTTCAGTATGGTAAAGCGCGGGTGCCATATGAATATCCTCCTTAG |
| YO-592 | [Dig]-CATTAATCTGAGGCTCAATCCATGCTGAAC |
| YO-597 | [Dig]-CCTTGCCTTTCGGCACGTAAGAGGCTAACC |
| YO-635 | ACAGCTGCTGGCCTTTTTCATGTTGTGAGCTTCCGGATTGCGGGAGACGGGTGTAGGCTGGAGCTGCTTC |
| YO-636 | TTCGTGTTATCCGTCCATGTAAGCAAACCTCATTTT TCAGCAAAATATTCCATATGAATATCCTCCTTAG |
| YO-637 | CCCGAATCGGTCATGATGCTGTAAC |
| YO-638 | AACTGTTTTGACTTTATTCACTTAC |
| YO-644 | TTTTGACTTTATTCACTTACATTTTGCCAATTTGCAGGATTTCGTGTTATCATATGAATATCCTCCTTAG |
| YO-657 | GTATCCCGTCGTGACCAGGAGAGCGCGCTGGCGTGCTGGGGAATCGACAGAGACTACAAAGATGACGACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takada, K.; Hama, K.; Sasaki, T.; Otsuka, Y. The hokW-sokW Locus Encodes a Type I Toxin–Antitoxin System That Facilitates the Release of Lysogenic Sp5 Phage in Enterohemorrhagic Escherichia coli O157. Toxins 2021, 13, 796. https://doi.org/10.3390/toxins13110796
Takada K, Hama K, Sasaki T, Otsuka Y. The hokW-sokW Locus Encodes a Type I Toxin–Antitoxin System That Facilitates the Release of Lysogenic Sp5 Phage in Enterohemorrhagic Escherichia coli O157. Toxins. 2021; 13(11):796. https://doi.org/10.3390/toxins13110796
Chicago/Turabian StyleTakada, Kosuke, Kotone Hama, Takaomi Sasaki, and Yuichi Otsuka. 2021. "The hokW-sokW Locus Encodes a Type I Toxin–Antitoxin System That Facilitates the Release of Lysogenic Sp5 Phage in Enterohemorrhagic Escherichia coli O157" Toxins 13, no. 11: 796. https://doi.org/10.3390/toxins13110796




