Lipid-Binding Aegerolysin from Biocontrol Fungus Beauveria bassiana
Abstract
:1. Introduction
2. Results
2.1. Genomes of Entomopathogenic Fungi Encode Aegerolysins
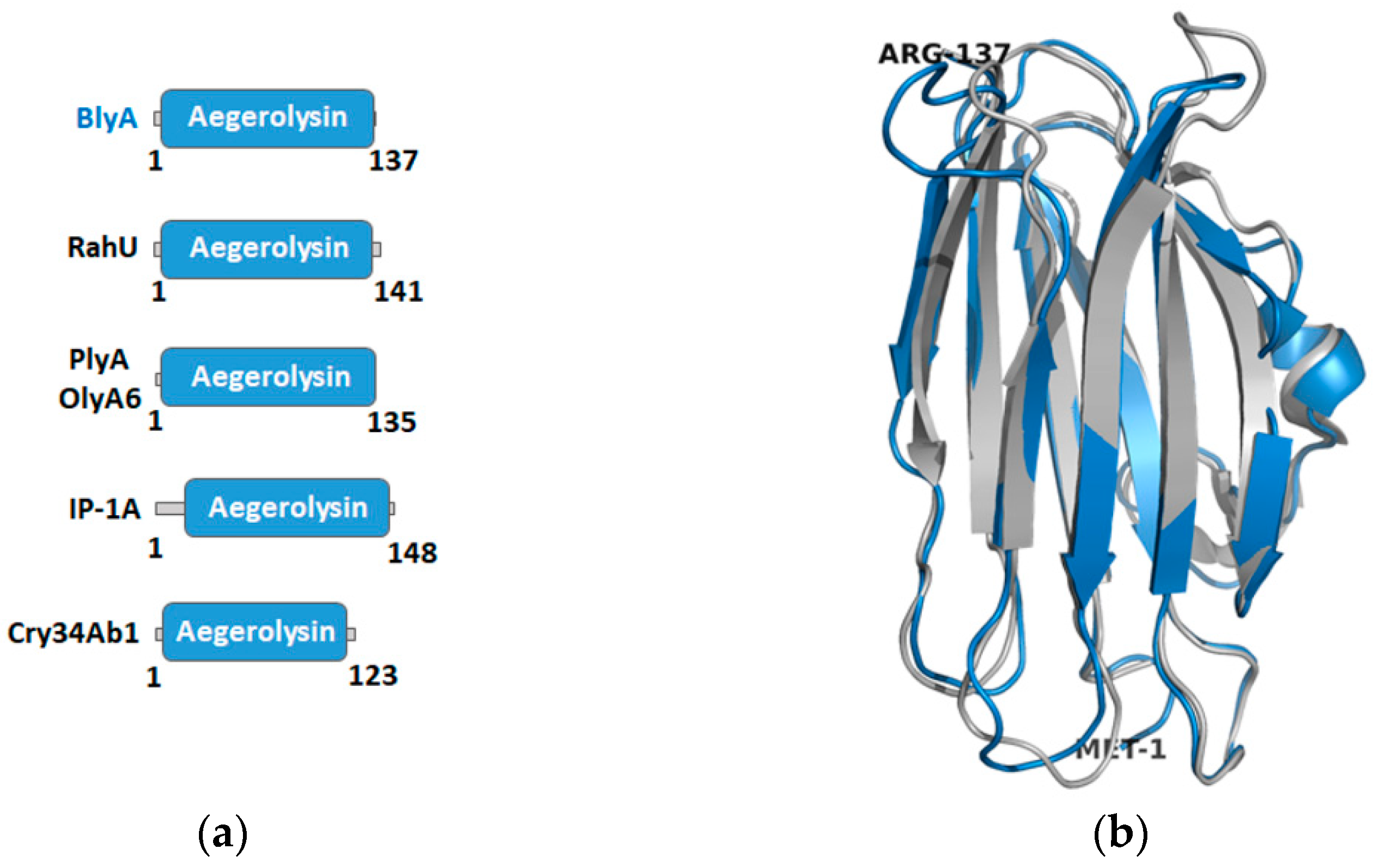
2.2. Building a Protein Model of Beauveriolysin A
2.3. Isolation and Characterisation of Beauveriolysin A
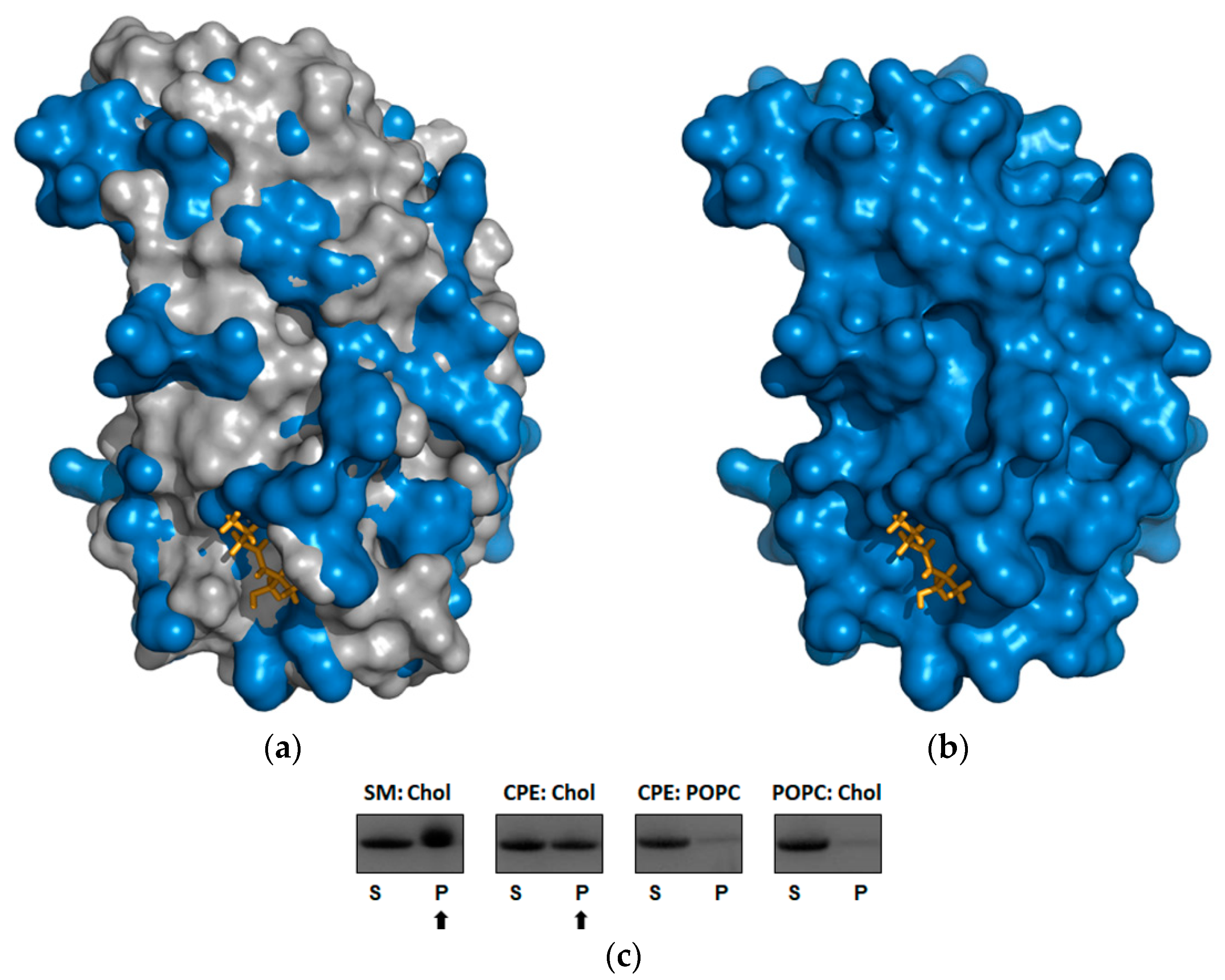
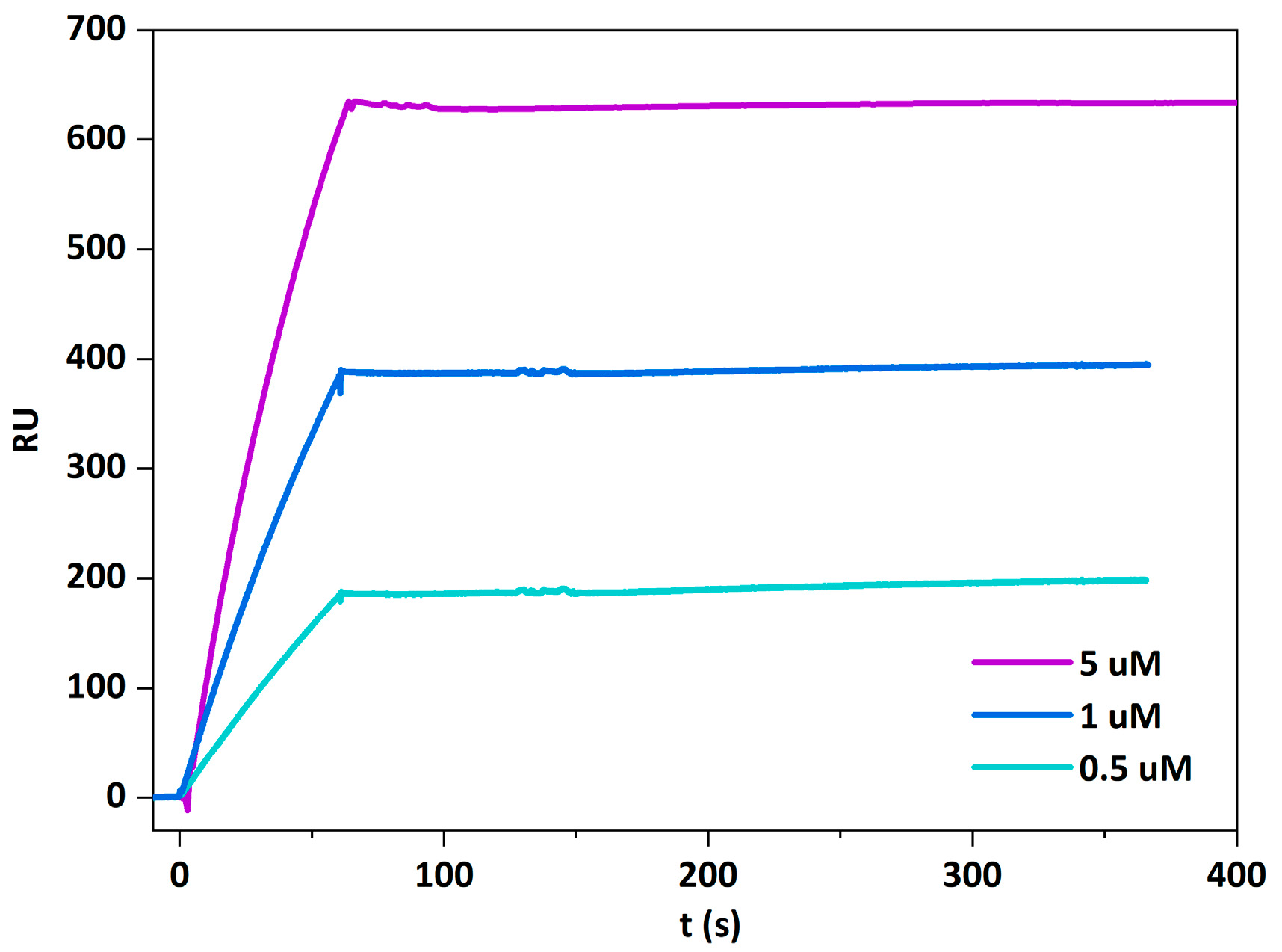
2.4. Beauveriolysin A Binds to Ceramide-Phosphoethanolamine or to Sphingomyelin in Combination with Cholesterol
2.5. Prediction of Secretion, Localization and Effector Function of Beauveriolysin A
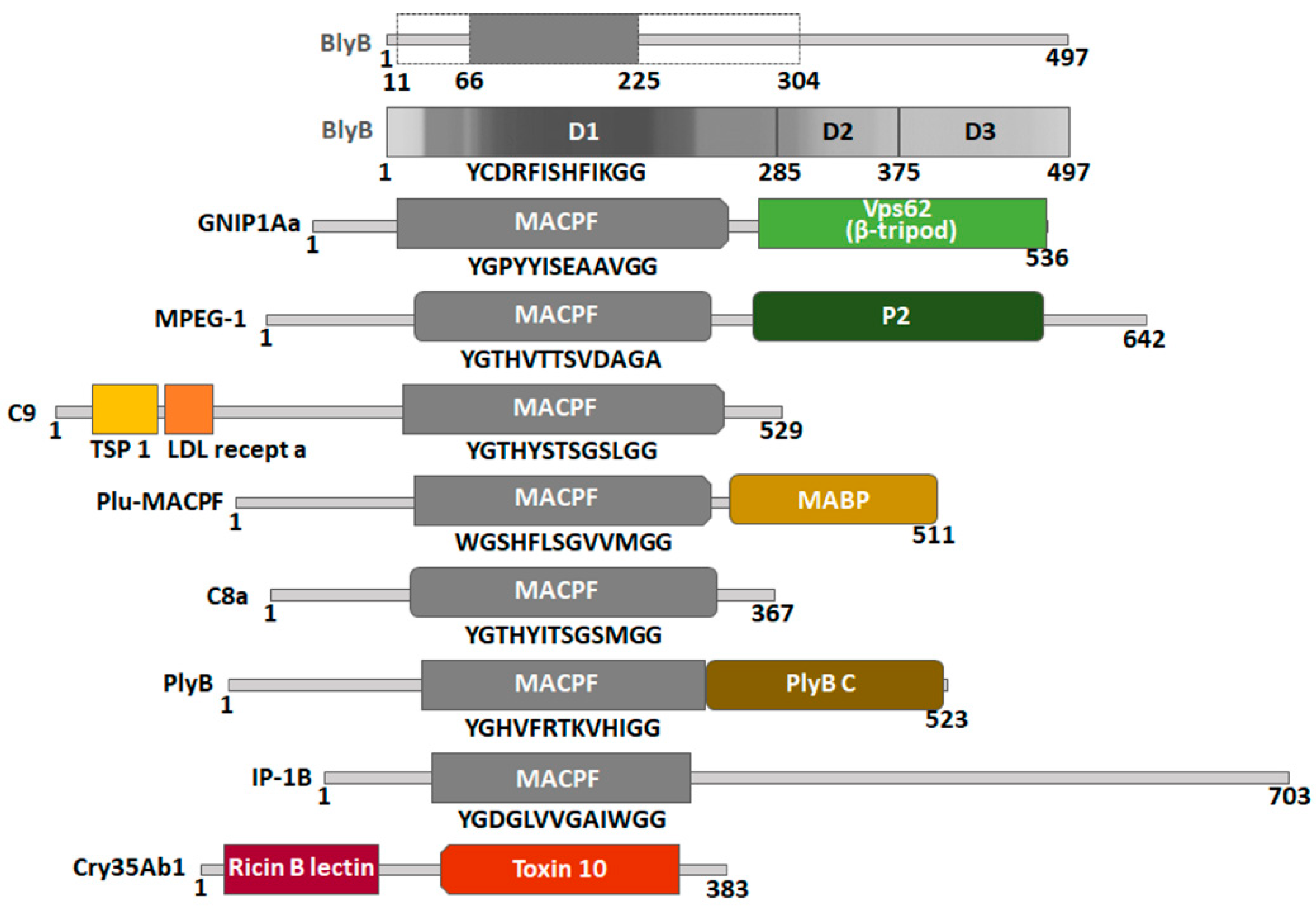
2.6. Construction of a Protein Model of Beauveriolysin B
3. Discussion
3.1. The Genomes of Entomopathogenic Fungi Encode Binary Toxins
3.2. Beauveriolysin A
3.3. Templates for the Beauveriolysin A and B Protein Models and Their Protein Partners Function as Insecticidal or Immunological Protein Complexes
3.4. Function of Additional Domains in Templates or Partner Proteins
3.5. Localization of Aegerolysins in Fungi
3.6. Future Research Directions to Improve the Properties of Biopesticides
3.7. Future Research Directions to Improve Antagonism against Plant Diseases
4. Conclusions
5. Materials and Methods
5.1. Comparative Genomics, Prediction and Phylogenetic Methods
5.2. Cloning of Beauveriolysin A
5.3. Expression and Isolation of Recombinant Beauveriolysin A
5.4. SDS-PAGE-Analysis and Blue Native Electrophoresis
5.5. Circular Dichroism Spectroscopy
5.6. Protein Sedimentation Assay with Multilamellar Lipid Vesicles
5.7. Surface Plasmon Resonance
5.8. Hemolysis
5.9. Sf9 Viability Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Butt, T.; Coates, C.; Dubovskiy, I.; Ratcliffe, N. Entomopathogenic Fungi. Adv. Genet. 2016, 94, 307–364. [Google Scholar] [CrossRef]
- Ladurner, E.; Benuzzi, M.; Fiorentini, F.; Franceschini, S. Beauveria bassiana strain ATCC 74040 (Naturalis®), a valuable tool for the control of the cherry fruit fly (Rhagoletis cerasi). In Ecofruit-13th International Conference on Cultivation Technique and Phytopathological Problems in Organic Fruit-Growing: Proceedings to the Conference from 18th February to 20th February 2008 at Weinsberg Germany; Boos, M., Ed.; Organic Eprints; 2008; pp. 93–97. Available online: https://orgprints.org/id/eprint/13651/1/093-097.pdf (accessed on 5 September 2021).
- Vega, F.E.; Goettel, M.S.; Blackwell, M.; Chandler, D.; Jackson, M.A.; Keller, S.; Koike, M.; Maniania, N.K.; Monzón, A.; Ownley, B.; et al. Fungal entomopathogens: New insights on their ecology. Fungal Ecol. 2009, 2, 149–159. [Google Scholar] [CrossRef] [Green Version]
- Rasool, S.; Vidkjær, N.H.; Hooshmand, K.; Jensen, B.; Fomsgaard, I.S.; Meyling, N.V. Seed inoculations with entomopathogenic fungi affect aphid populations coinciding with modulation of plant secondary metabolite profiles across plant families. New Phytol. 2020, 229, 1715–1727. [Google Scholar] [CrossRef]
- Sánchez-Pérez, L.D.C.; Barranco-Florido, J.E.; Rodríguez-Navarro, S.; Cervantes-Mayagoitia, J.F.; Ramos-López, M.Á. Enzymes of Entomopathogenic Fungi, Advances and Insights. Adv. Enzym. Res. 2014, 02, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Amobonye, A.; Bhagwat, P.; Pandey, A.; Singh, S.; Pillai, S. Biotechnological potential of Beauveria bassiana as a source of novel biocatalysts and metabolites. Crit. Rev. Biotechnol. 2020, 40, 1019–1034. [Google Scholar] [CrossRef]
- Quesada-Moraga, E.; Vey, A. Bassiacridin, a protein toxic for locusts secreted by the entomopathogenic fungus Beauveria bassiana. Mycol. Res. 2004, 108, 441–452. [Google Scholar] [CrossRef] [Green Version]
- Mascarin, G.M.; Jaronski, S.T. The production and uses of Beauveria bassiana as a microbial insecticide. World J. Microbiol. Biotechnol. 2016, 32, 177. [Google Scholar] [CrossRef] [PubMed]
- List of Authorised Plant Protection Products. Available online: https://agromet.mko.gov.si/FFS/REGSR/EN/FFS_Descr.asp?CODE=865 (accessed on 5 September 2021).
- Conclusion on the peer review of the pesticide risk assessment of the active substances Beauveria bassiana strains ATCC-74040 and GHA. EFSA J. 2013, 11, 3031. [CrossRef]
- Gao, Q.; Jin, K.; Ying, S.-H.; Zhang, Y.; Xiao, G.; Shang, Y.; Duan, Z.; Hu, X.; Xie, X.-Q.; Zhou, G.; et al. Genome Sequencing and Comparative Transcriptomics of the Model Entomopathogenic Fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011, 7, e1001264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Xiao, G.; Zheng, P.; Shang, Y.; Su, Y.; Zhang, X.; Liu, X.; Zhan, S.; Leger, R.J.S.; Wang, C. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, 16796–16801. [Google Scholar] [CrossRef] [Green Version]
- Xiao, G.; Ying, S.-H.; Zheng, P.; Wang, Z.-L.; Zhang, S.; Xie, X.-Q.; Shang, Y.; Leger, R.J.S.; Zhao, G.-P.; Wang, C.; et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 483. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef] [Green Version]
- Shu, R.; Zhang, J.; Meng, Q.; Zhang, H.; Zhou, G.; Li, M.; Wu, P.; Zhao, Y.; Chen, C.; Qin, Q. A New High-Quality Draft Genome Assembly of the Chinese Cordyceps Ophiocordyceps sinensis. Genome Biol. Evol. 2020, 12, 1074–1079. [Google Scholar] [CrossRef] [PubMed]
- Berne, S.; Lah, L.; Sepčić, K. Aegerolysins: Structure, function, and putative biological role. Protein Sci. 2009, 18, 694–706. [Google Scholar] [CrossRef] [Green Version]
- Novak, M.; Kraševec, N.; Skočaj, M.; Maček, P.; Anderluh, G.; Sepčić, K. Fungal aegerolysin-like proteins: Distribution, activities, and applications. Appl. Microbiol. Biotechnol. 2014, 99, 601–610. [Google Scholar] [CrossRef]
- Butala, M.; Novak, M.; Kraševec, N.; Skočaj, M.; Veranič, P.; Maček, P.; Sepčić, K. Aegerolysins: Lipid-binding proteins with versatile functions. Semin. Cell Dev. Biol. 2017, 72, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Sepčić, K.; Berne, S.; Rebolj, K.; Batista, U.; Plemenitaš, A.; Šentjurc, M.; Maček, P. Ostreolysin, a pore-forming protein from the oyster mushroom, interacts specifically with membrane cholesterol-rich lipid domains. FEBS Lett. 2004, 575, 81–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ota, K.; Leonardi, A.; Mikelj, M.; Skočaj, M.; Wohlschlager, T.; Künzler, M.; Aebi, M.; Narat, M.; Križaj, I.; Anderluh, G.; et al. Membrane cholesterol and sphingomyelin, and ostreolysin A are obligatory for pore-formation by a MACPF/CDC-like pore-forming protein, pleurotolysin B. Biochimie 2013, 95, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Skočaj, M.; Resnik, N.; Grundner, M.; Ota, K.; Rojko, N.; Hodnik, V.; Anderluh, G.; Sobota, A.; Maček, P.; Veranič, P.; et al. Tracking Cholesterol/Sphingomyelin-Rich Membrane Domains with the Ostreolysin A-mCherry Protein. PLoS ONE 2014, 9, e92783. [Google Scholar] [CrossRef] [Green Version]
- Bhat, H.B.; Ishitsuka, R.; Inaba, T.; Murate, M.; Abe, M.; Makino, A.; Kohyama-Koganeya, A.; Nagao, K.; Kurahashi, A.; Kishimoto, T.; et al. Evaluation of aegerolysins as novel tools to detect and visualize ceramide phosphoethanolamine, a major sphingolipid in invertebrates. FASEB J. 2015, 29, 3920–3934. [Google Scholar] [CrossRef] [Green Version]
- Lukoyanova, N.; Kondos, S.C.; Farabella, I.; Law, R.; Reboul, C.; Caradoc-Davies, T.T.; Spicer, B.A.; Kleifeld, O.; Traore, D.; Ekkel, S.M.; et al. Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin. PLoS Biol. 2015, 13, e1002049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundner, M.; Panevska, A.; Sepčić, K.; Skočaj, M. What Can Mushroom Proteins Teach Us about Lipid Rafts? Membranes 2021, 11, 264. [Google Scholar] [CrossRef] [PubMed]
- Panevska, A.; Skočaj, M.; Križaj, I.; Maček, P.; Sepčić, K. Ceramide phosphoethanolamine, an enigmatic cellular membrane sphingolipid. Biochim. Biophys. Acta (BBA)-Biomembr. 2019, 1861, 1284–1292. [Google Scholar] [CrossRef]
- Endapally, S.; Frias, D.; Grzemska, M.; Gay, A.; Tomchick, D.; Radhakrishnan, A. Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes. Cell 2019, 176, 1040–1053. [Google Scholar] [CrossRef] [Green Version]
- Novak, M.; Krpan, T.; Panevska, A.; Shewell, L.K.; Day, C.J.; Jennings, M.P.; Guella, G.; Sepčić, K. Binding specificity of ostreolysin A6 towards Sf9 insect cell lipids. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183307. [Google Scholar] [CrossRef] [PubMed]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-forming protein complexes from Pleurotus mushrooms kill western corn rootworm and Colorado potato beetle through targeting membrane ceramide phosphoethanolamine. Sci. Rep. 2019, 9, 5073. [Google Scholar] [CrossRef] [Green Version]
- Cho, E.-M.; Boucias, D.; Keyhani, N.O. EST analysis of cDNA libraries from the entomopathogenic fungus Beauveria (Cordyceps) bassiana. II. Fungal cells sporulating on chitin and producing oosporein. Microbiology 2006, 152, 2855–2864. [Google Scholar] [CrossRef] [Green Version]
- MycoCosm-The Fungal Genomics Resouce (DOE Joint Genome Institute). Available online: https://mycocosm.jgi.doe.gov/mycocosm/home (accessed on 24 June 2021).
- Pel, H.J.; de Winde, H.; Archer, D.B.; Dyer, P.; Hofmann, G.; Schaap, P.; Turner, G.; de Vries, R.; Albang, R.; Albermann, K.; et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513. Nat. Biotechnol. 2007, 25, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Novak, M.; Čepin, U.; Hodnik, V.; Narat, M.; Jamnik, M.; Kraševec, N.; Sepčić, K.; Anderluh, G. Functional studies of aegerolysin and MACPF-like proteins in Aspergillus niger. Mol. Microbiol. 2019, 112, 1253–1269. [Google Scholar] [CrossRef]
- Alfaro, M.; Castanera, R.; Lavín, J.L.; Grigoriev, I.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative and transcriptional analysis of the predicted secretome in the lignocellulose-degrading basidiomycete fungus Pleurotus ostreatus. Environ. Microbiol. 2016, 18, 4710–4726. [Google Scholar] [CrossRef]
- Lakkireddy, K.K.R.; Navarro-González, M.; Velagapudi, R.; Kües, U. Proteins expressed during hyphal aggregation for fruting body formation in basidiomycetes. In Proceedings of the 7th International Conference on Mushroom Biology and Mushroom Products, Arcachon, France, 4–7 October 2011; Savoie, J.-M., Foulongne-Oriol, M., Largeteau, M., Barroso, G., Eds.; pp. 82–94. [Google Scholar]
- Roberts, R.G.; Bischoff, J.F.; Reymond, S.T. Differential gene expression in Alternaria gaisen exposed to dark and light. Mycol. Prog. 2011, 11, 373–382. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Kočar, E.; Lenarčič, T.; Hodnik, V.; Panevska, A.; Huang, Y.; Bajc, G.; Kostanjšek, R.; Naren, A.P.; Maček, P.; Anderluh, G.; et al. Crystal structure of RahU, an aegerolysin protein from the human pathogen Pseudomonas aeruginosa, and its interaction with membrane ceramide phosphorylethanolamine. Sci. Rep. 2021, 11, 6572. [Google Scholar] [CrossRef] [PubMed]
- Yalpani, N.; Altier, D.; Barry, J.; Kassa, A.; Nowatzki, T.M.; Sethi, A.; Zhao, J.; Diehn, S.; Crane, V.; Sandahl, G.; et al. An Alcaligenes strain emulates Bacillus thuringiensis producing a binary protein that kills corn rootworm through a mechanism similar to Cry34Ab1/Cry35Ab1. Sci. Rep. 2017, 7, 3063. [Google Scholar] [CrossRef] [PubMed]
- Kelker, M.S.; Berry, C.; Evans, S.L.; Pai, R.; McCaskill, D.; Wang, N.X.; Russell, J.C.; Baker, M.D.; Yang, C.; Pflugrath, J.W.; et al. Structural and Biophysical Characterization of Bacillus thuringiensis Insecticidal Proteins Cry34Ab1 and Cry35Ab1. PLoS ONE 2014, 9, e112555. [Google Scholar] [CrossRef]
- EMBL-EBI Pfam. Available online: http://pfam.xfam.org/ (accessed on 26 August 2021).
- The Molecular Graphics System PyMOL (Schrödinger, LLC). Available online: https://pymol.org/2/ (accessed on 4 October 2021).
- Humphreys, M.J.; Berry, C. Variants of the Bacillus sphaericus Binary Toxins: Implications for Differential Toxicity of Strains. J. Invertebr. Pathol. 1998, 71, 184–185. [Google Scholar] [CrossRef]
- Narva, K.E.; Wang, N.X.; Herman, R. Safety considerations derived from Cry34Ab1/Cry35Ab1 structure and function. J. Invertebr. Pathol. 2017, 142, 27–33. [Google Scholar] [CrossRef]
- Kraševec, N.; Novak, M.; Barat, S.; Skočaj, M.; Sepčić, K.; Anderluh, G. Unconventional Secretion of Nigerolysins A from Aspergillus Species. Microorganisms 2020, 8, 1973. [Google Scholar] [CrossRef]
- TMHMM Server v. 2.0 Prediction of Transmembrane Helices in Proteins. Available online: http://www.cbs.dtu.dk/services/TMHMM-2.0/ (accessed on 13 August 2021).
- PredαTM and PredβTM: Transmembrane Region Predictors. Available online: https://www.ki.si/odseki/d01-teoreticni-odsek/laboratorij-za-kemijsko-informatiko/transmembrane-region-predictors/ (accessed on 24 June 2021).
- TOPCONS. Available online: https://topcons.net/ (accessed on 30 August 2021).
- PrediSi PREDIction of SIgnal Peptides. Available online: http://www.predisi.de/ (accessed on 13 August 2021).
- The DeepLoc-1.0 Server. Available online: http://www.cbs.dtu.dk/services/DeepLoc/ (accessed on 24 June 2021).
- SignalP-5.0 Server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 13 August 2021).
- SecretomeP 2.0 Server Prediction of Non-Classical Protein Secretion. Available online: http://www.cbs.dtu.dk/services/SecretomeP (accessed on 13 August 2021).
- Phobius A Combined Transmembrane Topology and Signal Peptide Predictor. Available online: http://phobius.binf.ku.dk/index.html (accessed on 24 June 2021).
- PredGPI GPI-Anchor Predictor. Available online: http://gpcr2.biocomp.unibo.it/gpipe/pred.htm (accessed on 24 June 2021).
- TargetP-2.0 Server. Available online: http://www.cbs.dtu.dk/services/TargetP/ (accessed on 24 June 2021).
- WOLF PSORT Protein Subcellular Localization Prediction. Available online: https://wolfpsort.hgc.jp/ (accessed on 30 August 2021).
- EffectorP: Predicting Fungal and Oomycete Effector Proteins from Secretomes using Machine Learning. Available online: http://effectorp.csiro.au (accessed on 30 August 2021).
- Anderluh, G.; Kisovec, M.; Kraševec, N.; Gilbert, R.J.C. Distribution of MACPF/CDC Proteins. Subcell. Biochem. 2014, 80, 7–30. [Google Scholar] [CrossRef]
- Zaitseva, J.; Vaknin, D.; Krebs, C.; Doroghazi, J.; Milam, S.L.; Balasubramanian, D.; Duck, N.B.; Freigang, J. Structure–function characterization of an insecticidal protein GNIP1Aa, a member of an MACPF and β-tripod families. Proc. Natl. Acad. Sci. USA 2019, 116, 2897–2906. [Google Scholar] [CrossRef] [Green Version]
- Pang, S.S.; Bayly-Jones, C.; Radjainia, M.; Spicer, B.A.; Law, R.H.P.; Hodel, A.; Parsons, E.S.; Ekkel, S.M.; Conroy, P.J.; Ramm, G.; et al. The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1. Nat. Commun. 2019, 10, 4288. [Google Scholar] [CrossRef] [PubMed]
- Spicer, B.A.; Law, R.H.P.; Caradoc-Davies, T.T.; Ekkel, S.M.; Bayly-Jones, C.; Pang, S.-S.; Conroy, P.J.; Ramm, G.; Radjainia, M.; Venugopal, H.; et al. The first transmembrane region of complement component-9 acts as a brake on its self-assembly. Nat. Commun. 2018, 9, 3266. [Google Scholar] [CrossRef] [Green Version]
- Rosado, C.J.; Buckle, A.M.; Law, R.H.P.; Butcher, R.E.; Kan, W.-T.; Bird, C.H.; Ung, K.; Browne, K.A.; Baran, K.; Bashtannyk-Puhalovich, T.A.; et al. A Common Fold Mediates Vertebrate Defense and Bacterial Attack. Science 2007, 317, 1548–1551. [Google Scholar] [CrossRef] [Green Version]
- Lovelace, L.L.; Cooper, C.L.; Sodetz, J.M.; Lebioda, L. Structure of Human C8 Protein Provides Mechanistic Insight into Membrane Pore Formation by Complement. J. Biol. Chem. 2011, 286, 17585–17592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, T.; Jiao, F.; Yu, X.; Aden, S.; Ginger, L.; Williams, S.I.; Bai, F.; Pražák, V.; Karia, D.; Stansfeld, P.; et al. Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity. Sci. Adv. 2020, 6, eaax8286. [Google Scholar] [CrossRef] [Green Version]
- PfamScan. Available online: https://www.ebi.ac.uk/Tools/pfa/pfamscan/ (accessed on 25 August 2021).
- InterPro. Available online: https://www.ebi.ac.uk/interpro/search/sequence/ (accessed on 25 August 2021).
- FUpred: Detecting Protein Domains through Deep-Learning Based Contact Map Prediction. Available online: https://zhanggroup.org//FUpred/ (accessed on 24 June 2021).
- EMBL-EBI HmmerWeb. Available online: https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan (accessed on 23 June 2021).
- Yamaji-Hasegawa, A.; Hullin-Matsuda, F.; Greimel, P.; Kobayashi, T. Pore-forming toxins: Properties, diversity, and uses as tools to image sphingomyelin and ceramide phosphoethanolamine. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1858, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Jotić, M.M.; Panevska, A.; Iacovache, I.; Kostanjšek, R.; Mravinec, M.; Skočaj, M.; Zuber, B.; Pavšič, A.; Razinger, J.; Modic, Š.; et al. Dissecting Out the Molecular Mechanism of Insecticidal Activity of Ostreolysin A6/Pleurotolysin B Complexes on Western Corn Rootworm. Toxins 2021, 13, 455. [Google Scholar] [CrossRef]
- Panevska, A.; Glavan, G.; Kokalj, A.J.; Kukuljan, V.; Trobec, T.; Žužek, M.; Vrecl, M.; Drobne, D.; Frangež, R.; Sepčić, K. Effects of Bioinsecticidal Aegerolysin-Based Cytolytic Complexes on Non-Target Organisms. Toxins 2021, 13, 457. [Google Scholar] [CrossRef] [PubMed]
- Starkey, M.; Rahme, L.G. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat. Protoc. 2009, 4, 117–124. [Google Scholar] [CrossRef]
- Bano, N.; Musarrat, J. Characterization of a New Pseudomonas aeruginosa Strain NJ-15 as a Potential Biocontrol Agent. Curr. Microbiol. 2003, 46, 324–328. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- De Souza, R.F.; Aravind, L. UMA and MABP domains throw light on receptor endocytosis and selection of endosomal cargoes. Bioinformatics 2010, 26, 1477–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iruela-Arispe, M.L.; Lombardo, M.; Krutzsch, H.C.; Lawler, J.; Roberts, D.D. Inhibition of Angiogenesis by Thrombospondin-1 Is Mediated by 2 Independent Regions within the Type 1 Repeats. Circulation 1999, 100, 1423–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, N.; Krutzsch, H.C.; Inman, J.K.; Roberts, D.D. Thrombospondin 1 and type I repeat peptides of thrombospondin 1 specifically induce apoptosis of endothelial cells. Cancer Res. 1997, 57, 1735–1742. [Google Scholar] [PubMed]
- Nykjaer, A.; Willnow, T. The low-density lipoprotein receptor gene family: A cellular Swiss army knife? Trends Cell Biol. 2002, 12, 273–280. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Dutta, S.K.; Kasai, K.-I. Novel Galactose-binding Proteins in Annelida: Characterization of 29-kDa tandem repeat-type lectins from the earthworm Lumbricus terrestris. J. Biol. Chem. 1998, 273, 14450–14460. [Google Scholar] [CrossRef] [Green Version]
- Hazes, B.; Read, R.J. A mosquitocidal toxin with a ricin-like cell-binding domain. Nat. Struct. Mol. Biol. 1995, 2, 358–359. [Google Scholar] [CrossRef]
- Hazes, B. The (QxW) 3domain: A flexible lectin scaffold. Protein Sci. 1996, 5, 1490–1501. [Google Scholar] [CrossRef] [Green Version]
- Sabotič, J.; Ohm, R.A.; Künzler, M. Entomotoxic and nematotoxic lectins and protease inhibitors from fungal fruiting bodies. Appl. Microbiol. Biotechnol. 2015, 100, 91–111. [Google Scholar] [CrossRef] [Green Version]
- Rosana, A.R.R.; Pokorny, S.; Klutsch, J.G.; Ibarra-Romero, C.; Sanichar, R.; Engelhardt, D.; van Belkum, M.J.; Erbilgin, N.; Bohlmann, J.; Carroll, A.L.; et al. Selection of entomopathogenic fungus Beauveria bassiana (Deuteromycotina: Hyphomycetes) for the biocontrol of Dendroctonus ponderosae (Coleoptera: Curculionidae, Scolytinae) in Western Canada. Appl. Microbiol. Biotechnol. 2021, 105, 2541–2557. [Google Scholar] [CrossRef]
- Fang, W.; Azimzadeh, P.; Leger, R.J.S. Strain improvement of fungal insecticides for controlling insect pests and vector-borne diseases. Curr. Opin. Microbiol. 2012, 15, 232–238. [Google Scholar] [CrossRef]
- Lacey, L.; Grzywacz, D.; Shapiro-Ilan, D.; Frutos, R.; Brownbridge, M.; Goettel, M. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Lovett, B.; Fang, W. Genetically Engineering Entomopathogenic Fungi. In Genetics and Molecular Biology of Entomopathogenic Fungi; Lovett, B., Leger, R.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 137–163. [Google Scholar]
- Karabörklü, S.; Azizoglu, U.; Azizoglu, Z.B. Recombinant entomopathogenic agents: A review of biotechnological approaches to pest insect control. World J. Microbiol. Biotechnol. 2017, 34, 14. [Google Scholar] [CrossRef]
- Lovett, B.; Leger, R.J.S. Genetically engineering better fungal biopesticides. Pest Manag. Sci. 2017, 74, 781–789. [Google Scholar] [CrossRef]
- Deng, S.-Q.; Zou, W.-H.; Li, D.-L.; Chen, J.-T.; Huang, Q.; Zhou, L.-J.; Tian, X.-X.; Chen, Y.-J.; Peng, H.-J. Expression of Bacillus thuringiensis toxin Cyt2Ba in the entomopathogenic fungus Beauveria bassiana increases its virulence towards Aedes mosquitoes. PLoS Neglected Trop. Dis. 2019, 13, e0007590. [Google Scholar] [CrossRef] [Green Version]
- Achari, T.S.; Barik, T.K.; Acharya, U.R. Toxins of Bacillus thuringiensis: A Novel Microbial Insecticide for Mosquito Vector Control. In Molecular Identification of Mosquito Vectors and Their Management; Springer: Singapore, 2020; pp. 89–116. [Google Scholar]
- Cohen, S.; Dym, O.; Albeck, S.; Ben-Dov, E.; Cahan, R.; Firer, M.; Zaritsky, A. High-Resolution Crystal Structure of Activated Cyt2Ba Monomer from Bacillus thuringiensis subsp. israelensis. J. Mol. Biol. 2008, 380, 820–827. [Google Scholar] [CrossRef]
- Verma, M.; Brar, S.K.; Tyagi, R.; Surampalli, R.; Valéro, J. Antagonistic fungi, Trichoderma spp.: Panoply of biological control. Biochem. Eng. J. 2007, 37, 1–20. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Genet. 2004, 2, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Tombolini, R.; Woo, S.; Zeilinger, S.; Lorito, M.; Jansson, J.K. In Vivo Study of Trichoderma-Pathogen-Plant Interactions, Using Constitutive and Inducible Green Fluorescent Protein Reporter Systems. Appl. Environ. Microbiol. 2004, 70, 3073–3081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macías-Rodríguez, L.; Guzmán-Gómez, A.; García-Juárez, P.; Contreras-Cornejo, H.A. Trichoderma atroviride promotes tomato development and alters the root exudation of carbohydrates, which stimulates fungal growth and the biocontrol of the phytopathogen Phytophthora cinnamomi in a tripartite interaction system. FEMS Microbiol. Ecol. 2018, 94, fiy137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Spada, F.; Stracquadanio, C.; Riolo, M.; Pane, A.; Cacciola, S.O. Trichoderma Counteracts the Challenge of Phytophthora nicotianae Infections on Tomato by Modulating Plant Defense Mechanisms and the Expression of Crinkler, Necrosis-Inducing Phytophthora Protein 1, and Cellulose-Binding Elicitor Lectin Pathogenic Effectors. Front. Plant Sci. 2020, 11, 583539. [Google Scholar] [CrossRef] [PubMed]
- Dubey, M.; Jensen, D.F.; Karlsson, M. Functional characterization of the AGL1 aegerolysin in the mycoparasitic fungus Trichoderma atroviride reveals a role in conidiation and antagonism. Mol. Genet. Genom. 2020, 296, 131–140. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.A.; Otillar, R.; Riley, R.; Salamov, A.A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2013, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; López, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Zhou, X.; Wuyun, Q.; Pearce, R.; Li, Y.; Zhang, Y. FUpred: Detecting protein domains through deep-learning-based contact map prediction. Bioinformatics 2020, 36, 3749–3757. [Google Scholar] [CrossRef]
- ClustalW (Kyoto University Bioinformatics Centre). Available online: https://www.genome.jp/tools-bin/clustalw (accessed on 1 October 2021).
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [Green Version]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [Green Version]
- Hiller, K.; Grote, A.; Scheer, M.; Münch, R.; Jahn, D. PrediSi: Prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004, 32, W375–W379. [Google Scholar] [CrossRef]
- Perdih, A.; Choudhury, A.R.; Župerl, Š.; Sikorska, E.; Zhukov, I.; Solmajer, T.; Novič, M. Structural Analysis of a Peptide Fragment of Transmembrane Transporter Protein Bilitranslocase. PLoS ONE 2012, 7, e38967. [Google Scholar] [CrossRef]
- Choudhury, A.R.; Novič, M. PredβTM: A Novel β-Transmembrane Region Prediction Algorithm. PLoS ONE 2015, 10, e0145564. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [Green Version]
- Tsirigos, K.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sepčić, K.; Berne, S.; Potrich, C.; Turk, T.; Maček, P.; Menestrina, G. Interaction of ostreolysin, a cytolytic protein from the edible mushroom Pleurotus ostreatus, with lipid membranes and modulation by lysophospholipids. JBIC J. Biol. Inorg. Chem. 2003, 270, 1199–1210. [Google Scholar] [CrossRef] [Green Version]
- Šakanović, A.; Hodnik, V.; Anderluh, G. Surface plasmon resonance for measuring interactions of proteins with lipids and lipid membranes. In Lipid-Protein Interactions: Methods and Protocols, Methods in Molecular Biology; Kleinschmidt, J.H., Ed.; Springer Science+Business Media, LLC, Part of Springer Nature, Humana: New York, NY, USA, 2019; pp. 53–70. [Google Scholar]

| Short Name | Protein Name Structure ID | Origin | Putative Function | % Identity Residues Coverage Confidence | Partner Protein | Ref. |
|---|---|---|---|---|---|---|
| RahU | RahU protein PDB ID: 6ZC1 | Pseudomonas aeruginosa Proteobacteria | Toxin | 32 2–135 97% 100 | No data | [37] |
| PlyA | Pleurotolysin A Membrane embedded pleurotolysin pore with 13-fold symmetry PDB ID: 4V2T | Pleurotus ostreatus Agaricomycotina | Transport protein | 34 2–137 98% 100 | Pleurotolysin B; PlyB PDB ID: 4OEJ | [20,23] |
| IP-1A | Two-component insecticidal protein 16 kDa unit PDB ID: 5V3S | Alcaligenes faecalis Proteobacteria | Toxin | 36 2–137 98% 100 | Two-component insecticidal protein 77 kDa unit; IP-1B | [38] |
| Cry34Ab1 | 13.6 kDa Insecticidal crystal protein PDB ID: 4JOX | Bacillus thuringiensis Firmicutes | Toxin | 25 4–134 94% 99.1 | 43.8 kDa insecticidal crystal protein; Cry35Ab1 PDB ID: 4JP0 | [39] |
| Short Name | Protein Name Structure ID | Origin | Putative Function | % Identity Residues Coverage Confidence | Partner Protein | Ref. |
|---|---|---|---|---|---|---|
| GNIP1Aa | Gram-negative insecticidal protein PDB ID: 6FBM | Chromobacterium piscinae Proteobacteria | Insecticidal protein, specifically toxic to Diabrotica virgifera virgifera larvae upon feeding | 13 36–225 38% 95.1 | No data | [58] |
| MPEG-1 (Perforin-2) | Macrophage-expressed gene 1 protein L425K (and w. t.) PDB ID: 6U2W (and PDB ID: 6U23) | Homo sapiens Chordata | Immune system | 16 11–226 43% 94.6 | No | [59] |
| C9 | Complement component C9 PDB ID: 6CXO | Mus musculus Chordata | Immune system | 14 29–225 39% 93.2 | Membrane attack complex | [60] |
| Plu-MACPF | Photorhabdus luminescens MACPF protein PDB ID: 2QP2 | Photorhabdus luminescens Proteobacteria | Photorhabdus luminescens is a lethal pathogen of insects | 11 66–304 47% 93.1 | No data | [61] |
| C8a | Complement component C8 alpha chain PDB ID: 2RD7 | Homo sapiens Chordata | Immune system | 14 36–225 38% 90.7 | Membrane attack complex | [62] |
| Phylum | Organism (CPE + Additional Major Lipid) [25] |
|---|---|
| Arthropoda, Insecta | Drosophila melanogaster, Musca domestica, Aedes aegypti (+SM), Culex quinquefasciatus (+SM), Manduca sexta (+SM), Apis mellifera (+SM), Centruroides sculpturatus (+SM) |
| Mollusca | Heterogen longispira (+CAEP), Sinotaia histrica (+CAEP), Semisulcospira bensoni (+CAEP), Batillaria multiformis (+CAEP) |
| Ciliophora | Entodinium caudatum, Paramecium tetraurelia |
| Apicomplexa | Toxoplasma gondii |
| Euglenozoa | Trypanosoma brucei |
| Metamonada | Trichomonas vaginalis |
| Oomycota | Pythium ultimum, Phytophthora infestans, P. capsici |
| Bacteroidetes | Flectobacillus major (+CPG), Bacteroides fragilis (+CPG) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kraševec, N.; Panevska, A.; Lemež, Š.; Razinger, J.; Sepčić, K.; Anderluh, G.; Podobnik, M. Lipid-Binding Aegerolysin from Biocontrol Fungus Beauveria bassiana. Toxins 2021, 13, 820. https://doi.org/10.3390/toxins13110820
Kraševec N, Panevska A, Lemež Š, Razinger J, Sepčić K, Anderluh G, Podobnik M. Lipid-Binding Aegerolysin from Biocontrol Fungus Beauveria bassiana. Toxins. 2021; 13(11):820. https://doi.org/10.3390/toxins13110820
Chicago/Turabian StyleKraševec, Nada, Anastasija Panevska, Špela Lemež, Jaka Razinger, Kristina Sepčić, Gregor Anderluh, and Marjetka Podobnik. 2021. "Lipid-Binding Aegerolysin from Biocontrol Fungus Beauveria bassiana" Toxins 13, no. 11: 820. https://doi.org/10.3390/toxins13110820
APA StyleKraševec, N., Panevska, A., Lemež, Š., Razinger, J., Sepčić, K., Anderluh, G., & Podobnik, M. (2021). Lipid-Binding Aegerolysin from Biocontrol Fungus Beauveria bassiana. Toxins, 13(11), 820. https://doi.org/10.3390/toxins13110820