Indoxyl Sulfate Elevated Lnc-SLC15A1-1 Upregulating CXCL10/CXCL8 Expression in High-Glucose Endothelial Cells by Sponging MicroRNAs
Abstract
:1. Introduction
2. Results
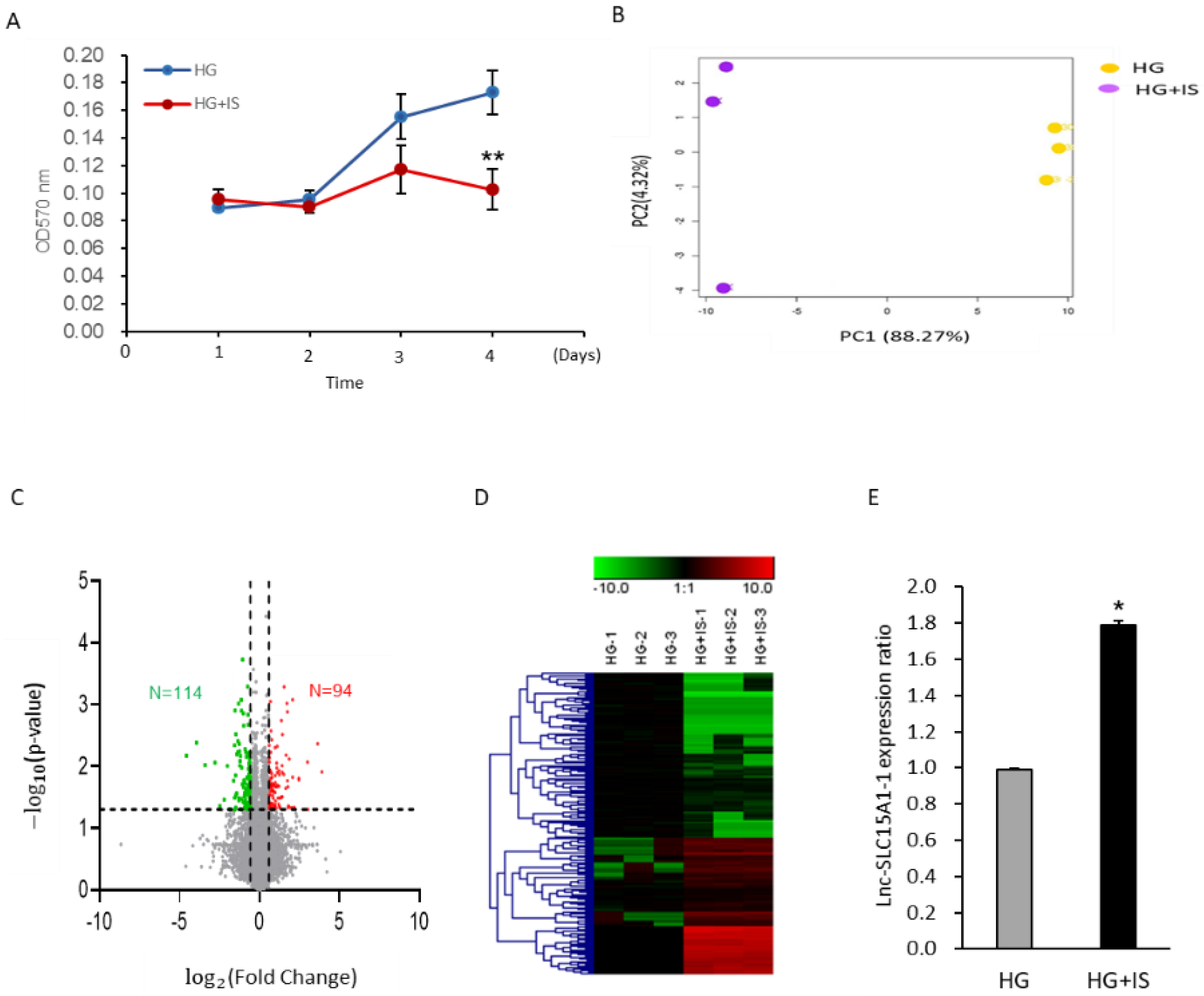
2.1. Upregulation of Lnc-SLC15A1-1 in High-Glucose-Cultured HUVECs after IS Treatment
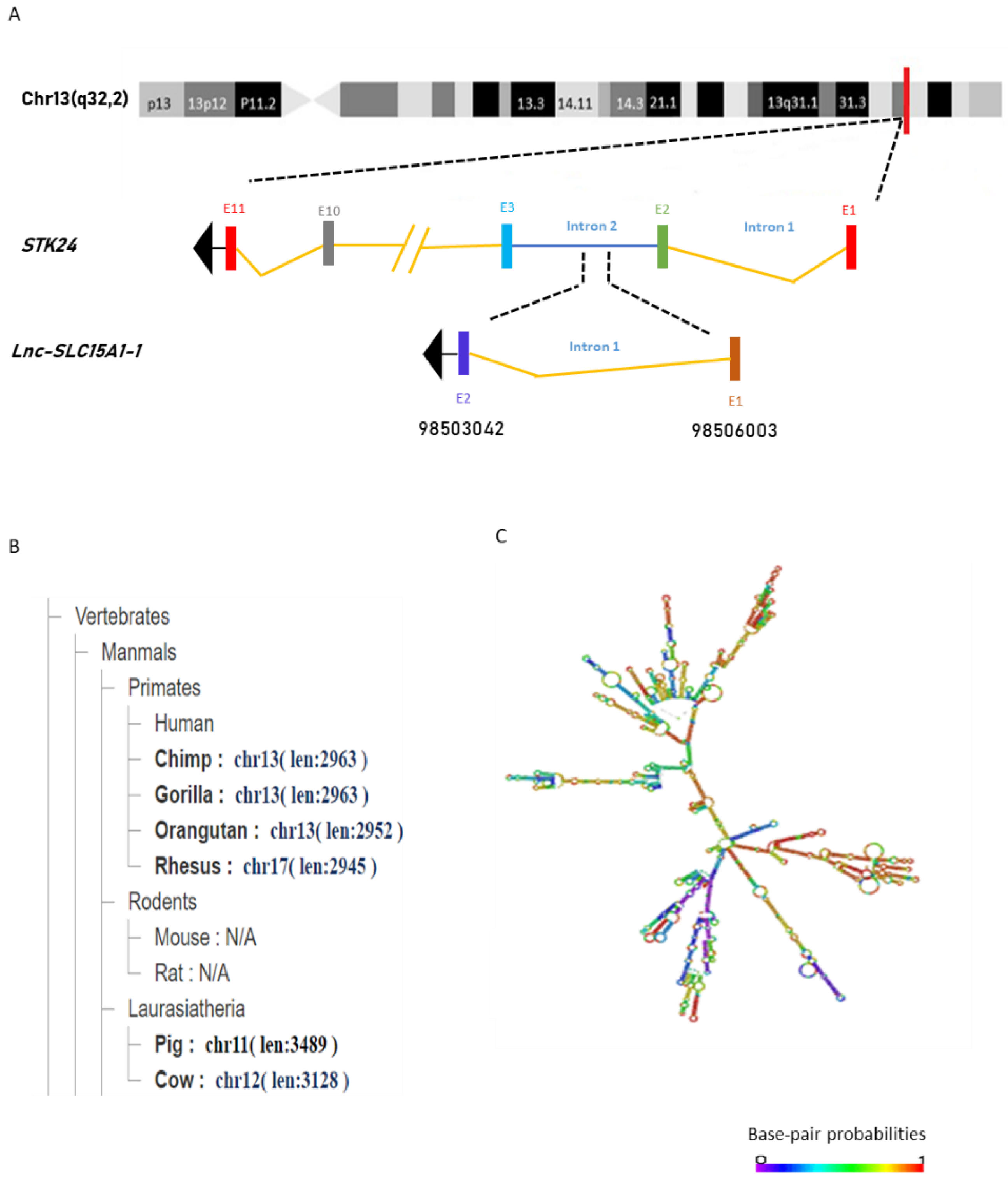
2.2. General Characterization of lnc-SLC15A1-1
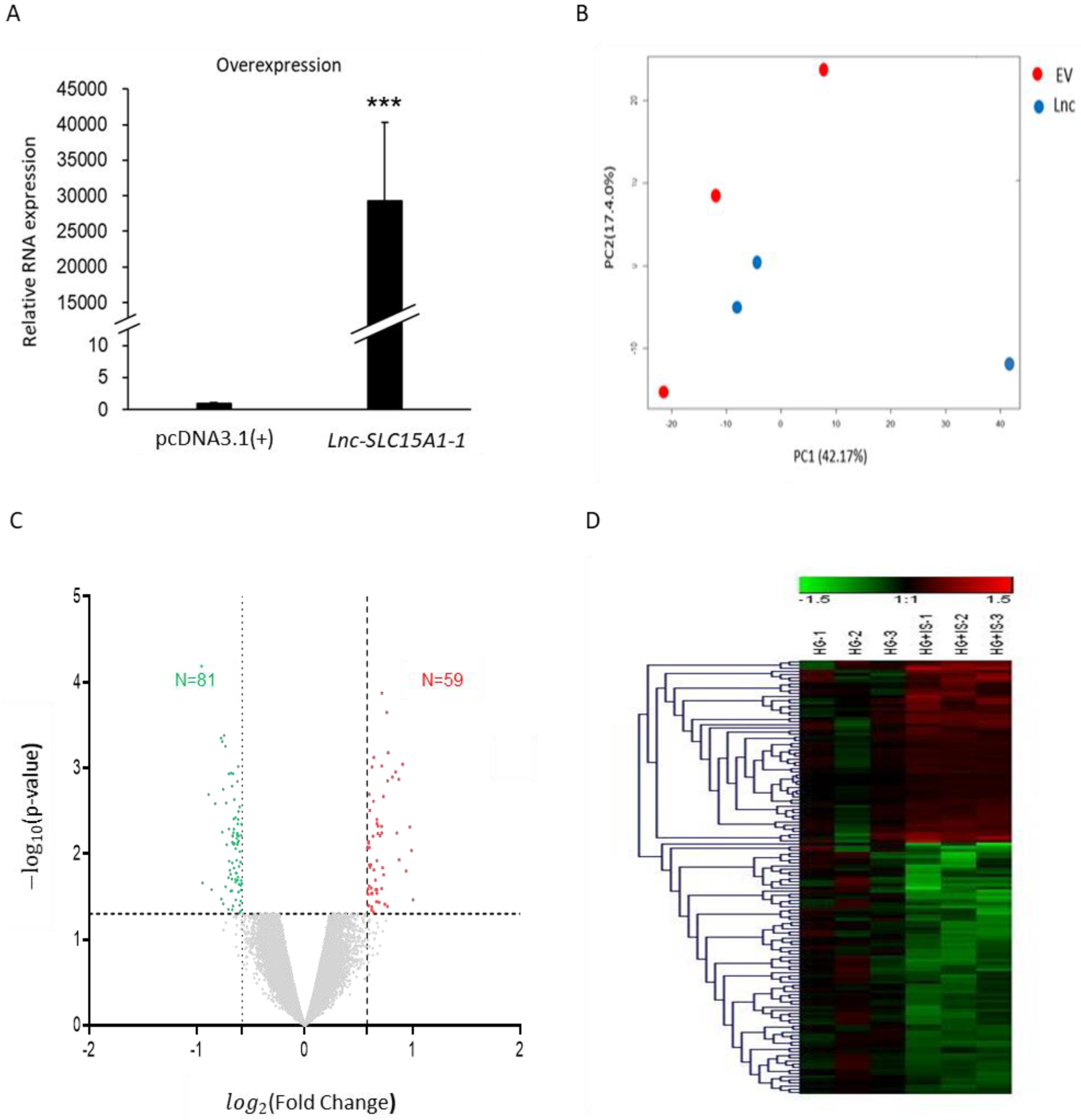
2.3. Identification of Genes Regulated by Lnc-SLC15A1-1 in HUVECs by Microarray Analysis
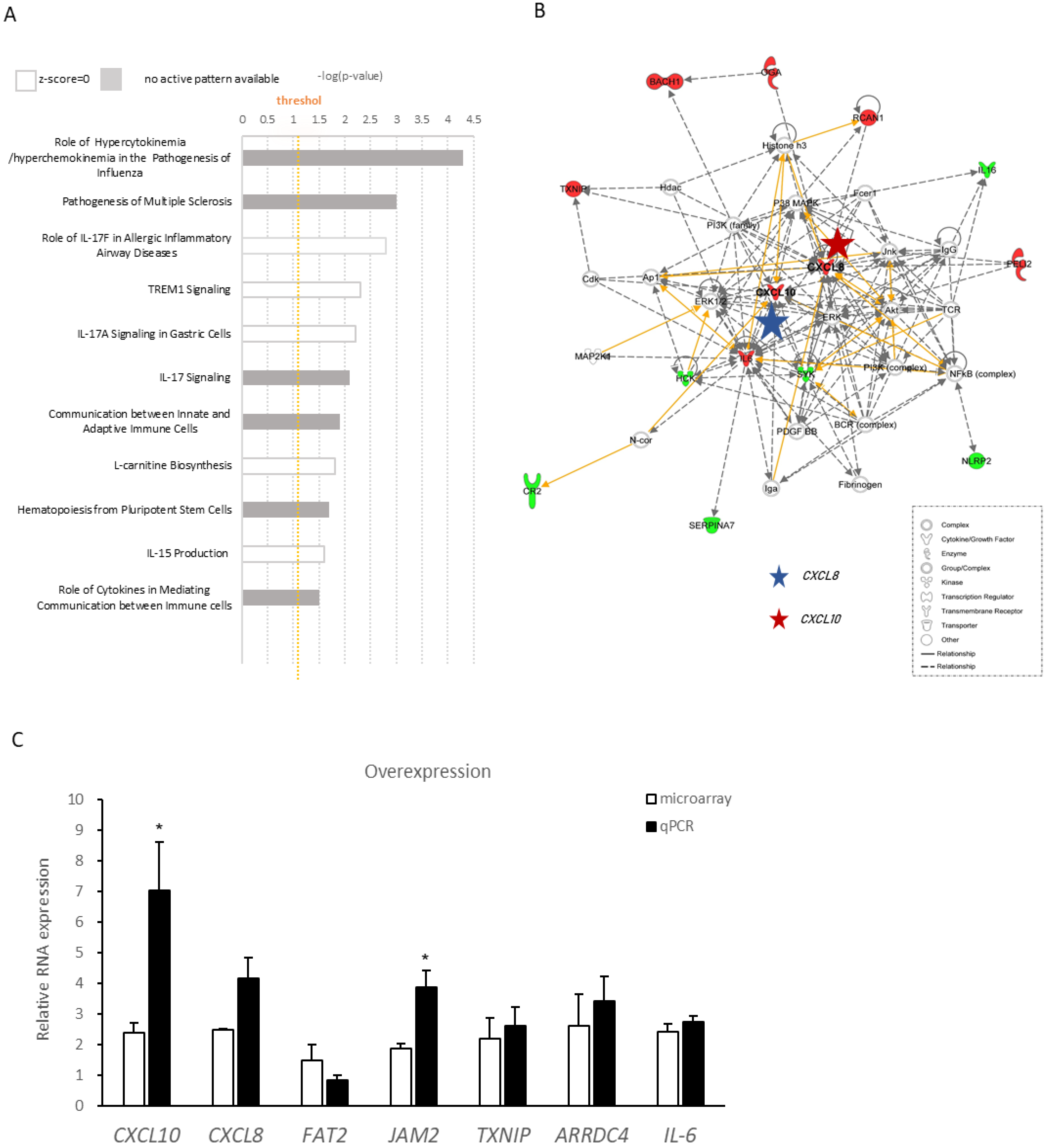
2.4. Ingenuity Pathway Analysis of Differentially Expressed Genes Regulated by lnc-SLC15A1-1
2.5. Upregulation and Secretion of CXC10 and CXCL8 in HUVECs by lnc-SLC15A1-1
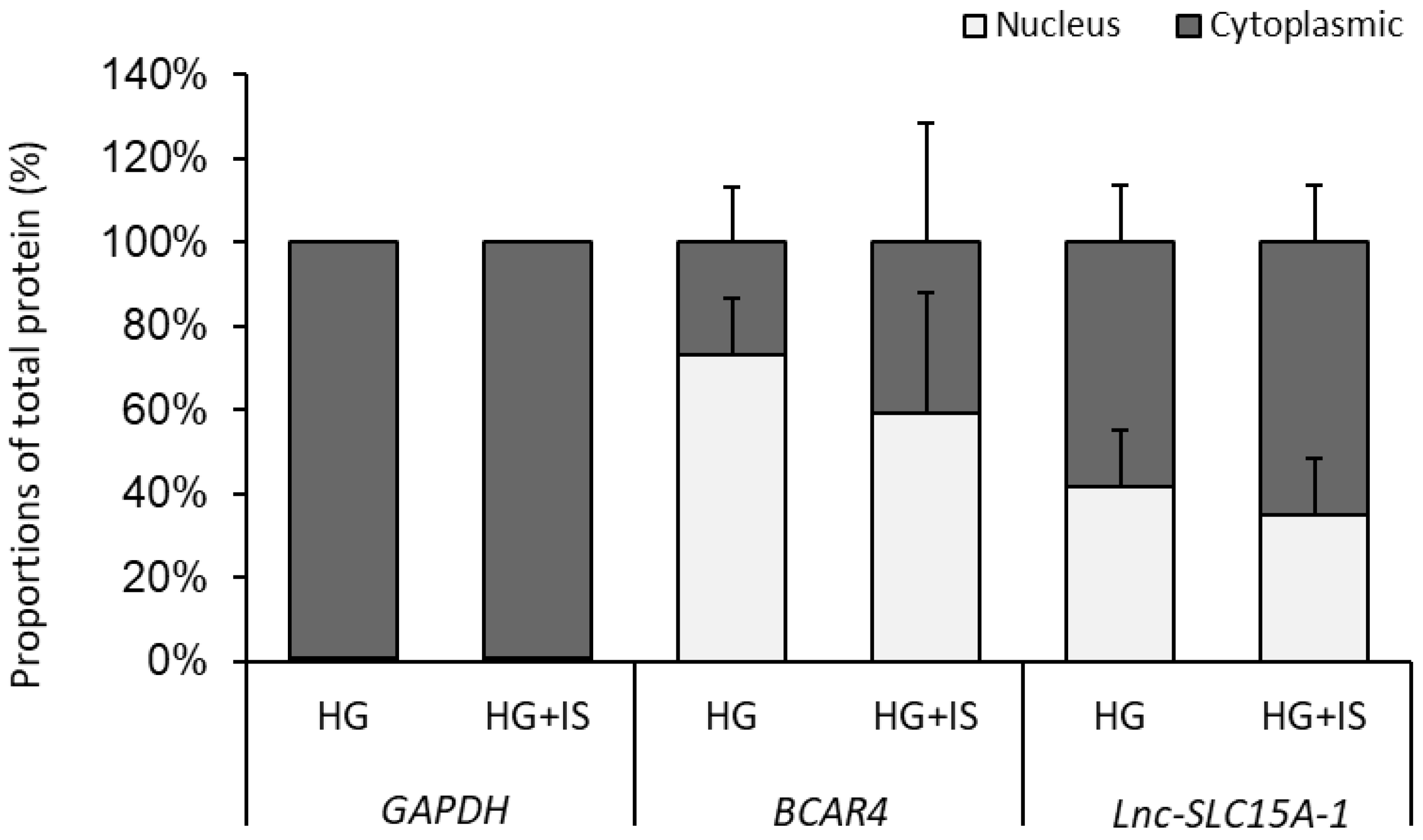
2.6. Lnc-SLC15A-1 Distribution in HUVECs
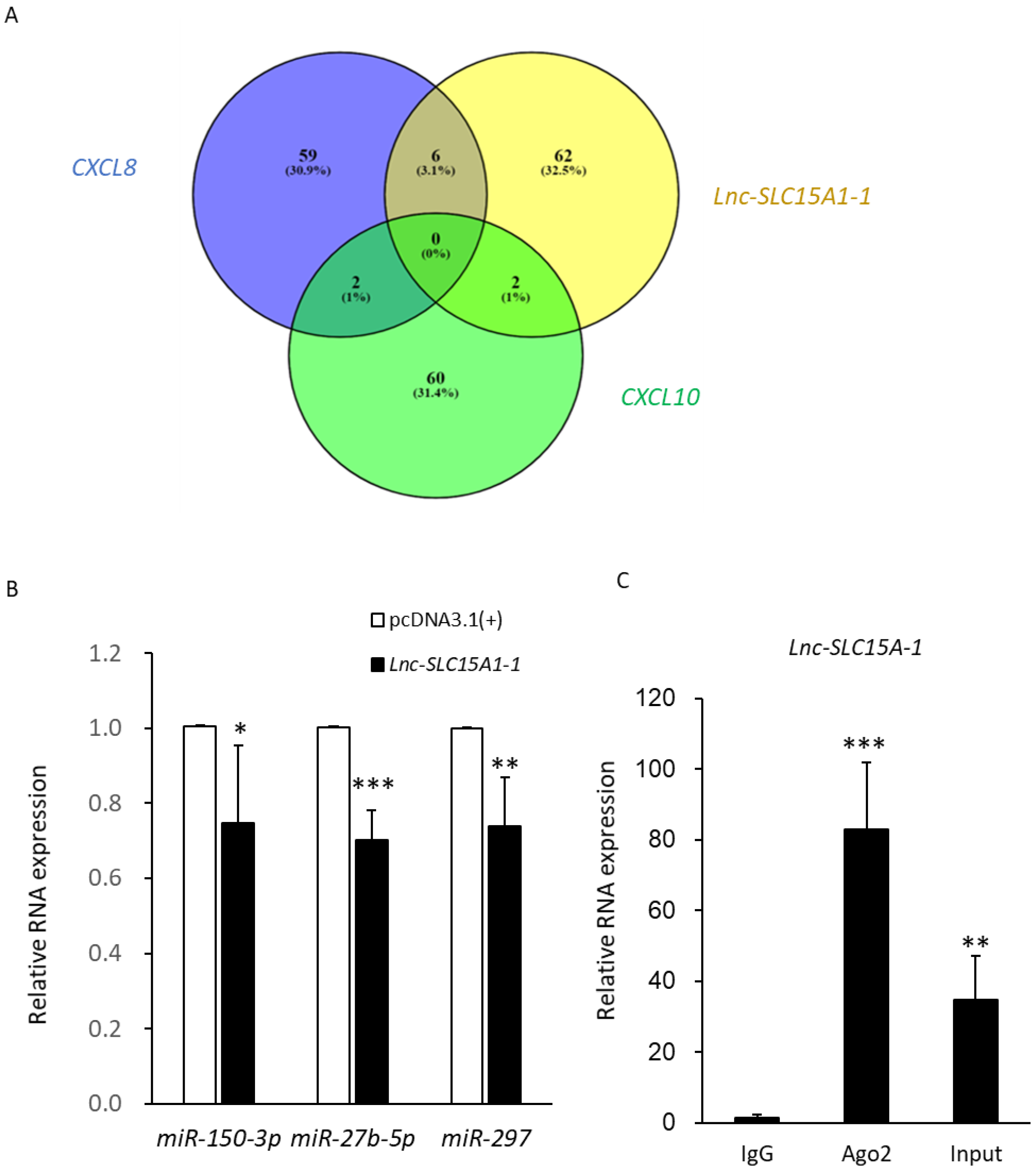
2.7. MiRNA Sponge Function of lnc-SLC15A1-1
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Culture and Treatments
5.2. Cell Proliferation Assay
5.3. RNA Extraction, Reverse Transcription, and Quantitative RT-PCR
5.4. RNA Sequencing Library Preparation
5.5. Screening and Cluster Analysis of Differentially Expressed lncRNAs
5.6. Plasmid DNA Construction and Transfection
5.7. Isolation and Amplification of Total RNA for Gene Expression Profiling by Microarrays
5.8. Analysis of Functional Enrichment and the lncRNA-Gene Network
5.9. Western Blotting
5.10. ELISA
5.11. Nuclear Cytoplasmic RNA Fractionation
5.12. MicroRNA Extraction, Reverse Transcription, and Quantitative RT-PCR
5.13. RNA Immunoprecipitation (RIP)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kondapally, R.; Seshasai, S.; Kaptoge, S.; Thompson, A.; Di Angelantonio, E.; Gao, P.; Sarwar, N.; Whincup, P.H.; Mukamal, K.J.; Gillum, R.F.; et al. Diabetes mellitus, fasting glucose, and risk of cause-specific death. N. Engl. J. Med. 2011, 364, 829–841. [Google Scholar] [CrossRef] [Green Version]
- Ikonomidis, I.; Michalakeas, C.A.; Parissis, J.; Paraskevaidis, I.; Ntai, K.; Papadakis, I.; Anastasiou-Nana, M.; Lekakis, J. Inflammatory markers in coronary artery disease. BioFactors 2012, 38, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Naguib, G.; Al-Mashat, H.; Desta, T.; Graves, D.T. Diabetes prolongs the inflammatory response to a bacterial stimulus through cytokine dysregulation. J. Investig. Dermatol. 2004, 123, 87–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansson, G.K.; Libby, P. The immune response in atherosclerosis: A double-edged sword. Nat. Rev. Immunol. 2006, 6, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Graves, D.T.; Kayal, R.A. Diabetic complications and dysregulated innate immunity. Front. Biosci. Landmark 2008, 13, 1227–1239. [Google Scholar] [CrossRef] [Green Version]
- Xiang, F.; Cao, X.; Shen, B.; Chen, X.; Guo, M.; Ding, X.; Zou, J. Transcriptome Profiling Reveals Indoxyl Sulfate Should Be Culpable of Impaired T Cell Function in Chronic Kidney Disease. Front. Med. 2020, 7, 178. [Google Scholar] [CrossRef]
- Fernandez, E.J.; Lolis, E. Structure, function, and inhibition of chemokines. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 469–499. [Google Scholar] [CrossRef]
- Zernecke, A.; Shagdarsuren, E.; Weber, C. Chemokines in atherosclerosis: An update. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1897–1908. [Google Scholar] [CrossRef] [Green Version]
- Simonini, A.; Moscucci, M.; Muller, D.W.; Bates, E.R.; Pagani, F.D.; Burdick, M.D.; Strieter, R.M. IL-8 is an angiogenic factor in human coronary atherectomy tissue. Circulation 2000, 101, 1519–1526. [Google Scholar] [CrossRef]
- Barlic, J.; Murphy, P.M. Chemokine regulation of atherosclerosis. J. Leukoc. Biol. 2007, 82, 226–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Lee, K.; Ni, Z.; He, J.C. Diabetic Kidney Disease: Challenges, Advances, and Opportunities. Kidney Dis. 2020, 6, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Maciel, R.A.P.; Cunha, R.S.; Busato, V.; Franco, C.R.C.; Gregorio, P.C.; Dolenga, C.J.R.; Nakao, L.S.; Massy, Z.A.; Boullier, A.; Pecoits-Filho, R.; et al. Uremia Impacts VE-Cadherin and ZO-1 Expression in Human Endothelial Cell-to-Cell Junctions. Toxins 2018, 10, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leong, S.C.; Sirich, T.L. Indoxyl Sulfate—Review of Toxicity and Therapeutic Strategies. Toxins 2016, 8, 358. [Google Scholar] [CrossRef]
- Tan, X.; Cao, X.; Zou, J.; Shen, B.; Zhang, X.; Liu, Z.; Lv, W.; Teng, J.; Ding, X. Indoxyl sulfate, a valuable biomarker in chronic kidney disease and dialysis. Hemodial. Int. 2017, 21, 161–167. [Google Scholar] [CrossRef]
- Cohen, G.; Glorieux, G.; Thornalley, P.; Schepers, E.; Meert, N.; Jankowski, J.; Jankowski, V.; Argiles, A.; Anderstam, B.; Brunet, P.; et al. Review on uraemic toxins III: Recommendations for handling uraemic retention solutes in vitro—Towards a standardized approach for research on uraemia. Nephrol. Dial. Transplant. 2007, 22, 3381–3390. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Campbell, K.L.; Johnson, D.W.; Stanton, T.; Vesey, D.A.; Coombes, J.S.; Weston, K.S.; Hawley, C.M.; McWhinney, B.C.; Ungerer, J.P.J.; et al. Protein-bound Uremic Toxins, Inflammation and Oxidative Stress: A Cross-sectional Study in Stage 3–4 Chronic Kidney Disease. Arch. Med. Res. 2014, 45, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Atoh, K.; Itoh, H.; Haneda, M. Serum indoxyl sulfate levels in patients with diabetic nephropathy: Relation to renal function. Diabetes Res. Clin. Pract. 2009, 83, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.A.; Lu, L.F.; Yu, T.H.; Hung, W.C.; Chung, F.M.; Tsai, I.T.; Yang, C.Y.; Hsu, C.C.; Lu, Y.C.; Wang, C.P.; et al. Increased levels of total P-Cresylsulphate and indoxyl sulphate are associated with coronary artery disease in patients with diabetic nephropathy. Rev. Diabet. Stud. 2010, 7, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Katakami, N.; Omori, K.; Taya, N.; Arakawa, S.; Takahara, M.; Matsuoka, T.-A.; Tsugawa, H.; Furuno, M.; Bamba, T.; Fukusaki, E.; et al. Plasma metabolites associated with arterial stiffness in patients with type 2 diabetes. Cardiovasc. Diabetol. 2020, 19, 75. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Groop, L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009, 58, 2718–2725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keating, S.T.; El-Osta, A. Epigenetics and metabolism. Circ. Res. 2015, 116, 715–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siasos, G.; Bletsa, E.; Stampouloglou, P.K.; Oikonomou, E.; Tsigkou, V.; Paschou, S.A.; Vlasis, K.; Marinos, G.; Vavuranakis, M.; Stefanadis, C.; et al. MicroRNAs in cardiovascular disease. Hell. J. Cardiol. 2020, 61, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zheng, D.; Li, Y.; Liu, G.; Zhou, H.; Liu, Y. Long noncoding RNA MANTIS relieved the protein-bound uremic toxin-induced injury on human umbilical vein endothelial cells in chronic kidney disease and end-stage renal disease. Int. J. Clin. Exp. Pathol. 2018, 11, 3236–3246. [Google Scholar] [PubMed]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zornig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef]
- Bell, G.I.; Polonsky, K.S. Diabetes mellitus and genetically programmed defects in beta-cell function. Nature 2001, 414, 788–791. [Google Scholar] [CrossRef]
- Allen, H.L.; Flanagan, S.E.; Shaw-Smith, C.; De Franco, E.; Akerman, I.; Caswell, R.; Ferrer, J.; Hattersley, A.T.; Ellard, S.; The International Pancreatic Agenesis Consortium. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat. Genet. 2012, 44, 20–22. [Google Scholar] [CrossRef]
- Moran, I.; Akerman, I.; van de Bunt, M.; Xie, R.; Benazra, M.; Nammo, T.; Arnes, L.; Nakic, N.; Garcia-Hurtado, J.; Rodriguez-Segui, S.; et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012, 16, 435–448. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef] [Green Version]
- Boulberdaa, M.; Scott, E.; Ballantyne, M.; Garcia, R.; Descamps, B.; Angelini, G.D.; Brittan, M.; Hunter, A.; McBride, M.; McClure, J.; et al. A Role for the Long Noncoding RNA SENCR in Commitment and Function of Endothelial Cells. Mol. Ther. 2016, 24, 978–990. [Google Scholar] [CrossRef] [Green Version]
- Leisegang, M.S.; Fork, C.; Josipovic, I.; Richter, F.M.; Preussner, J.; Hu, J.; Miller, M.J.; Epah, J.; Hofmann, P.; Gunther, S.; et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation 2017, 136, 65–79. [Google Scholar] [CrossRef]
- Hofmann, P.; Sommer, J.; Theodorou, K.; Kirchhof, L.; Fischer, A.; Li, Y.; Perisic, L.; Hedin, U.; Maegdefessel, L.; Dimmeler, S.; et al. Long non-coding RNA H19 regulates endothelial cell aging via inhibition of STAT3 signalling. Cardiovasc. Res. 2019, 115, 230–242. [Google Scholar] [CrossRef] [Green Version]
- Jae, N.; Dimmeler, S. Noncoding RNAs in Vascular Diseases. Circ. Res. 2020, 126, 1127–1145. [Google Scholar] [CrossRef]
- Yang, J.; Richmond, A. The angiostatic activity of interferon-inducible protein-10/CXCL10 in human melanoma depends on binding to CXCR3 but not to glycosaminoglycan. Mol. Ther. 2004, 9, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-Y.; Hu, C.-J.; Chang, S.-M.; Tai, T.-Y.; Leu, S.-J. Aspirin inhibits monocyte chemoattractant protein-1 and interleukin-8 expression in TNF-α stimulated human umbilical vein endothelial cells. Atherosclerosis 2004, 174, 207–213. [Google Scholar] [CrossRef]
- Gencer, S.; Evans, B.R.; van der Vorst, E.P.C.; Döring, Y.; Weber, C. Inflammatory Chemokines in Atherosclerosis. Cells 2021, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Six, I.; Maizel, J.; Barreto, F.C.; Rangrez, A.Y.; Dupont, S.; Slama, M.; Tribouilloy, C.; Choukroun, G.; Maziere, J.C.; Bode-Boeger, S.; et al. Effects of phosphate on vascular function under normal conditions and influence of the uraemic state. Cardiovasc. Res. 2012, 96, 130–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattanakit, K.; Cushman, M.; Stehman-Breen, C.; Heckbert, S.R.; Folsom, A.R. Chronic kidney disease increases risk for venous thromboembolism. J. Am. Soc. Nephrol. 2008, 19, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.K.; Gardiner, T.A.; Archer, D.B. A morphologic and autoradiographic study of cell death and regeneration in the retinal microvasculature of normal and diabetic rats. Am. J. Ophthalmol. 1985, 100, 51–60. [Google Scholar] [CrossRef]
- Yamamoto, S.; Fuller, D.S.; Komaba, H.; Nomura, T.; Massy, Z.A.; Bieber, B.; Robinson, B.; Pisoni, R.; Fukagawa, M. Serum total indoxyl sulfate and clinical outcomes in hemodialysis patients: Results from the Japan Dialysis Outcomes and Practice Patterns Study. Clin. Kidney J. 2020, 14, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.C.; Brownlee, M.; Susztak, K.; Sharma, K.; Jandeleit-Dahm, K.A.M.; Zoungas, S.; Rossing, P.; Groop, P.-H.; Cooper, M.E. Diabetic kidney disease. Nat. Rev. Dis. Primers 2015, 1, 15018. [Google Scholar] [CrossRef]
- Kolseth, I.B.; Reine, T.M.; Parker, K.; Sudworth, A.; Witczak, B.J.; Jenssen, T.G.; Kolset, S.O. Increased levels of inflammatory mediators and proinflammatory monocytes in patients with type I diabetes mellitus and nephropathy. J. Diabetes Complicat. 2017, 31, 245–252. [Google Scholar] [CrossRef]
- Espi, M.; Koppe, L.; Fouque, D.; Thaunat, O. Chronic Kidney Disease-Associated Immune Dysfunctions: Impact of Protein-Bound Uremic Retention Solutes on Immune Cells. Toxins 2020, 12, 300. [Google Scholar] [CrossRef] [PubMed]
- Kamiński, T.W.; Pawlak, K.; Karbowska, M.; Myśliwiec, M.; Pawlak, D. Indoxyl sulfate—The uremic toxin linking hemostatic system disturbances with the prevalence of cardiovascular disease in patients with chronic kidney disease. BMC Nephrol. 2017, 18, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, T.; Katsuki, S.; Chen, M.; Decano, J.L.; Halu, A.; Lee, L.H.; Pestana, D.V.S.; Kum, A.S.T.; Kuromoto, R.K.; Golden, W.S.; et al. Uremic Toxin Indoxyl Sulfate Promotes Proinflammatory Macrophage Activation Via the Interplay of OATP2B1 and Dll4-Notch Signaling. Circulation 2019, 139, 78–96. [Google Scholar] [CrossRef]
- Lano, G.; Burtey, S.; Sallée, M. Indoxyl Sulfate, a Uremic Endotheliotoxin. Toxins 2020, 12, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregório, P.C.; Favretto, G.; Sassaki, G.L.; Cunha, R.S.; Becker-Finco, A.; Pecoits-Filho, R.; Souza, W.M.; Barreto, F.C.; Stinghen, A.E.M. Sevelamer reduces endothelial inflammatory response to advanced glycation end products. Clin. Kidney J. 2018, 11, 89–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinuani, I.; Beberashvili, I.; Averbukh, Z.; Sandbank, J. Role of IL-10 in the progression of kidney disease. World J. Transplant. 2013, 3, 91–98. [Google Scholar] [CrossRef]
- Roebuck, K.A. Oxidant stress regulation of IL-8 and ICAM-1 gene expression: Differential activation and binding of the transcription factors AP-1 and NF-kappaB (Review). Int. J. Mol. Med. 1999, 4, 223–230. [Google Scholar] [CrossRef]
- Dou, L.; Jourde-Chiche, N.; Faure, V.; Cerini, C.; Berland, Y.; Dignat-George, F.; Brunet, P. The uremic solute indoxyl sulfate induces oxidative stress in endothelial cells. J. Thromb. Haemost. 2007, 5, 1302–1308. [Google Scholar] [CrossRef]
- Zhan, J.F.; Huang, H.W.; Huang, C.; Hu, L.L.; Xu, W.W. Long Non-Coding RNA NEAT1 Regulates Pyroptosis in Diabetic Nephropathy via Mediating the miR-34c/NLRP3 Axis. Kidney Blood Press. Res. 2020, 45, 589–602. [Google Scholar] [CrossRef]
- Zha, F.; Qu, X.; Tang, B.; Li, J.; Wang, Y.; Zheng, P.; Ji, T.; Zhu, C.; Bai, S. Long non-coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR-181a/Egr-1/TLR4 axis. Aging 2019, 11, 3716–3730. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.Y.; Dong, B.; Fang, Z.F.; Hu, X.Q.; Tang, L.; Zhou, S.H. Knockdown of lncRNA AK139328 alleviates myocardial ischaemia/reperfusion injury in diabetic mice via modulating miR-204-3p and inhibiting autophagy. J. Cell. Mol. Med. 2018, 22, 4886–4898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadi, H.A.R.; Carr, C.S.; Al Suwaidi, J. Endothelial dysfunction: Cardiovascular risk factors, therapy, and outcome. Vasc. Health Risk Manag. 2005, 1, 183–198. [Google Scholar] [PubMed]
- Schiffrin, E.L.; Lipman, M.L.; Mann, J.F. Chronic kidney disease: Effects on the cardiovascular system. Circulation 2007, 116, 85–97. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, R.T.; Mamoni, R.L.; Souza, J.R.; Fernandes, J.L.; Rios, F.J.; Gidlund, M.; Coelho, O.R.; Blotta, M.H. Differential expression of cytokines, chemokines and chemokine receptors in patients with coronary artery disease. Int. J. Cardiol. 2009, 136, 17–26. [Google Scholar] [CrossRef]
- Mantovani, A. The chemokine system: Redundancy for robust outputs. Immunol. Today 1999, 20, 254–257. [Google Scholar] [CrossRef]
- Lo, B.K.; Yu, M.; Zloty, D.; Cowan, B.; Shapiro, J.; McElwee, K.J. CXCR3/ligands are significantly involved in the tumorigenesis of basal cell carcinomas. Am. J. Pathol. 2010, 176, 2435–2446. [Google Scholar] [CrossRef] [Green Version]
- Sajadi, S.M.; Khoramdelazad, H.; Hassanshahi, G.; Rafatpanah, H.; Hosseini, J.; Mahmoodi, M.; Arababadi, M.K.; Derakhshan, R.; Hasheminasabzavareh, R.; Hosseini-Zijoud, S.M.; et al. Plasma levels of CXCL1 (GRO-alpha) and CXCL10 (IP-10) are elevated in type 2 diabetic patients: Evidence for the involvement of inflammation and angiogenesis/angiostasis in this disease state. Clin. Lab. 2013, 59, 133–137. [Google Scholar] [CrossRef]
- Hatanaka, E.; Monteagudo, P.T.; Marrocos, M.S.M.; Campa, A. Neutrophils and monocytes as potentially important sources of proinflammatory cytokines in diabetes. Clin. Exp. Immunol. 2006, 146, 443–447. [Google Scholar] [CrossRef]
- Sun, C.-Y.; Hsu, H.-H.; Wu, M.-S. p-Cresol sulfate and indoxyl sulfate induce similar cellular inflammatory gene expressions in cultured proximal renal tubular cells. Nephrol. Dial. Transplant. 2012, 28, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Galkina, E.; Ley, K. Vascular Adhesion Molecules in Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2292–2301. [Google Scholar] [CrossRef]
- Bermudez, E.A.; Rifai, N.; Buring, J.; Manson, J.E.; Ridker, P.M. Interrelationships among circulating interleukin-6, C-reactive protein, and traditional cardiovascular risk factors in women. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 1668–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashizume, M.; Mihara, M. Atherogenic effects of TNF-α and IL-6 via up-regulation of scavenger receptors. Cytokine 2012, 58, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Spagnoli, V.; Tardif, J.C.; L’Allier, P.L. Novel anti-inflammatory therapies for the treatment of atherosclerosis. Atherosclerosis 2015, 240, 497–509. [Google Scholar] [CrossRef]
- Alfaddagh, A.; Martin, S.S.; Leucker, T.M.; Michos, E.D.; Blaha, M.J.; Lowenstein, C.J.; Jones, S.R.; Toth, P.P. Inflammation and cardiovascular disease: From mechanisms to therapeutics. Am. J. Prev. Cardiol. 2020, 4, 100130. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhou, X.; Wan, M.; Zeng, X.; Luo, J.; Xu, Y.; Ji, L.; Zhang, J.-A.; Fan, P.; Zhong, J.; et al. FoxP3-miR-150-5p/3p suppresses ovarian tumorigenesis via an IGF1R/IRS1 pathway feedback loop. Cell Death Dis. 2021, 12, 275. [Google Scholar] [CrossRef]
- Kefas, B.; Floyd, D.; Comeau, L.; Frisbee, A.; Dominguez, C.; Dipierro, C.; Guessous, F.; Abounader, R.; Purow, B. A miR-297/hypoxia/DGK-axis regulating glioblastoma survival. Neuro Oncol. 2013, 15, 1652–1663. [Google Scholar] [CrossRef]
- Rong, X.; Ge, D.; Shen, D.; Chen, X.; Wang, X.; Zhang, L.; Jia, C.; Zeng, J.; He, Y.; Qiu, H.; et al. miR-27b Suppresses Endothelial Cell Proliferation and Migration by Targeting Smad7 in Kawasaki Disease. Cell. Physiol. Biochem. 2018, 48, 1804–1814. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Huang, C.; Senavirathna, L.; Wang, P.; Liu, L. miR-27b inhibits fibroblast activation via targeting TGFβ signaling pathway. BMC Cell Biol. 2017, 18, 9. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Si, W.; Shen, J.; Du, C.; Lou, W.; Bao, C.; Zheng, H.; Pan, J.; Zhong, G.; Xu, L.; et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Jerez-Morales, A.; Merino, J.S.; Díaz-Castillo, S.T.; Smith, C.T.; Fuentealba, J.; Bernasconi, H.; Echeverría, G.; García-Cancino, A. The Administration of the Synbiotic Lactobacillus bulgaricus 6c3 Strain, Inulin and Fructooligosaccharide Decreases the Concentrations of Indoxyl Sulfate and Kidney Damage in a Rat Model. Toxins 2021, 13, 192. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Johnson, D.W.; Morrison, M.; Pascoe, E.; Coombes, J.S.; Forbes, J.M.; McWhinney, B.C.; Ungerer, J.P.J.; Dimeski, G.; Campbell, K.L. SYNbiotics Easing Renal failure by improving Gut microbiologY (SYNERGY): A protocol of placebo-controlled randomised cross-over trial. BMC Nephrol. 2014, 15, 106. [Google Scholar] [CrossRef] [Green Version]
- Nakada, Y.; Onoue, K.; Nakano, T.; Ishihara, S.; Kumazawa, T.; Nakagawa, H.; Ueda, T.; Nishida, T.; Soeda, T.; Okayama, S.; et al. AST-120, an Oral Carbon Absorbent, Protects against the Progression of Atherosclerosis in a Mouse Chronic Renal Failure Model by Preserving sFlt-1 Expression Levels. Sci. Rep. 2019, 9, 15571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Bush, K.T.; Nigam, S.K. Key Role for the Organic Anion Transporters, OAT1 and OAT3, in the in vivo Handling of Uremic Toxins and Solutes. Sci. Rep. 2017, 7, 4939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| CXCL10/lnc-SLC15A1-1 | PCR Validation | CXCL8/lnc-SLC15A1-1 | PCR Validation | |
|---|---|---|---|---|
| MicroRNA | miR-27b-5p miR-297 | ○ ○ | miR-150-3p miR-513a-3p miR-3606-3p miR-513c-3p miR-10523-5p miR-567 | ○ X X X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-C.; Tsai, T.-C.; Chang, C.-H.; Chang, K.-T.; Ko, P.-H.; Lai, L.-C. Indoxyl Sulfate Elevated Lnc-SLC15A1-1 Upregulating CXCL10/CXCL8 Expression in High-Glucose Endothelial Cells by Sponging MicroRNAs. Toxins 2021, 13, 873. https://doi.org/10.3390/toxins13120873
Huang Y-C, Tsai T-C, Chang C-H, Chang K-T, Ko P-H, Lai L-C. Indoxyl Sulfate Elevated Lnc-SLC15A1-1 Upregulating CXCL10/CXCL8 Expression in High-Glucose Endothelial Cells by Sponging MicroRNAs. Toxins. 2021; 13(12):873. https://doi.org/10.3390/toxins13120873
Chicago/Turabian StyleHuang, Yu-Chin, Tzu-Chun Tsai, Chia-Hsin Chang, Kuo-Ting Chang, Pin-Hao Ko, and Liang-Chuan Lai. 2021. "Indoxyl Sulfate Elevated Lnc-SLC15A1-1 Upregulating CXCL10/CXCL8 Expression in High-Glucose Endothelial Cells by Sponging MicroRNAs" Toxins 13, no. 12: 873. https://doi.org/10.3390/toxins13120873
APA StyleHuang, Y.-C., Tsai, T.-C., Chang, C.-H., Chang, K.-T., Ko, P.-H., & Lai, L.-C. (2021). Indoxyl Sulfate Elevated Lnc-SLC15A1-1 Upregulating CXCL10/CXCL8 Expression in High-Glucose Endothelial Cells by Sponging MicroRNAs. Toxins, 13(12), 873. https://doi.org/10.3390/toxins13120873






