Diet Diversity in Carnivorous Terebrid Snails Is Tied to the Presence and Absence of a Venom Gland
Abstract
:1. Introduction
2. Results
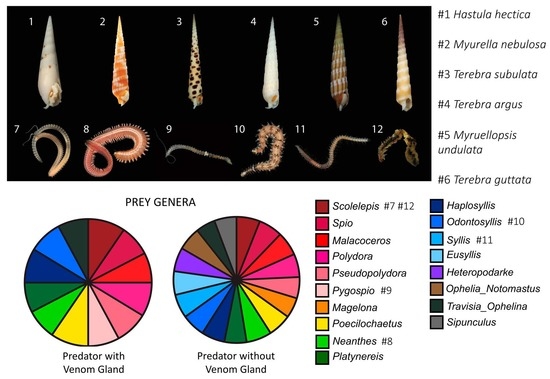
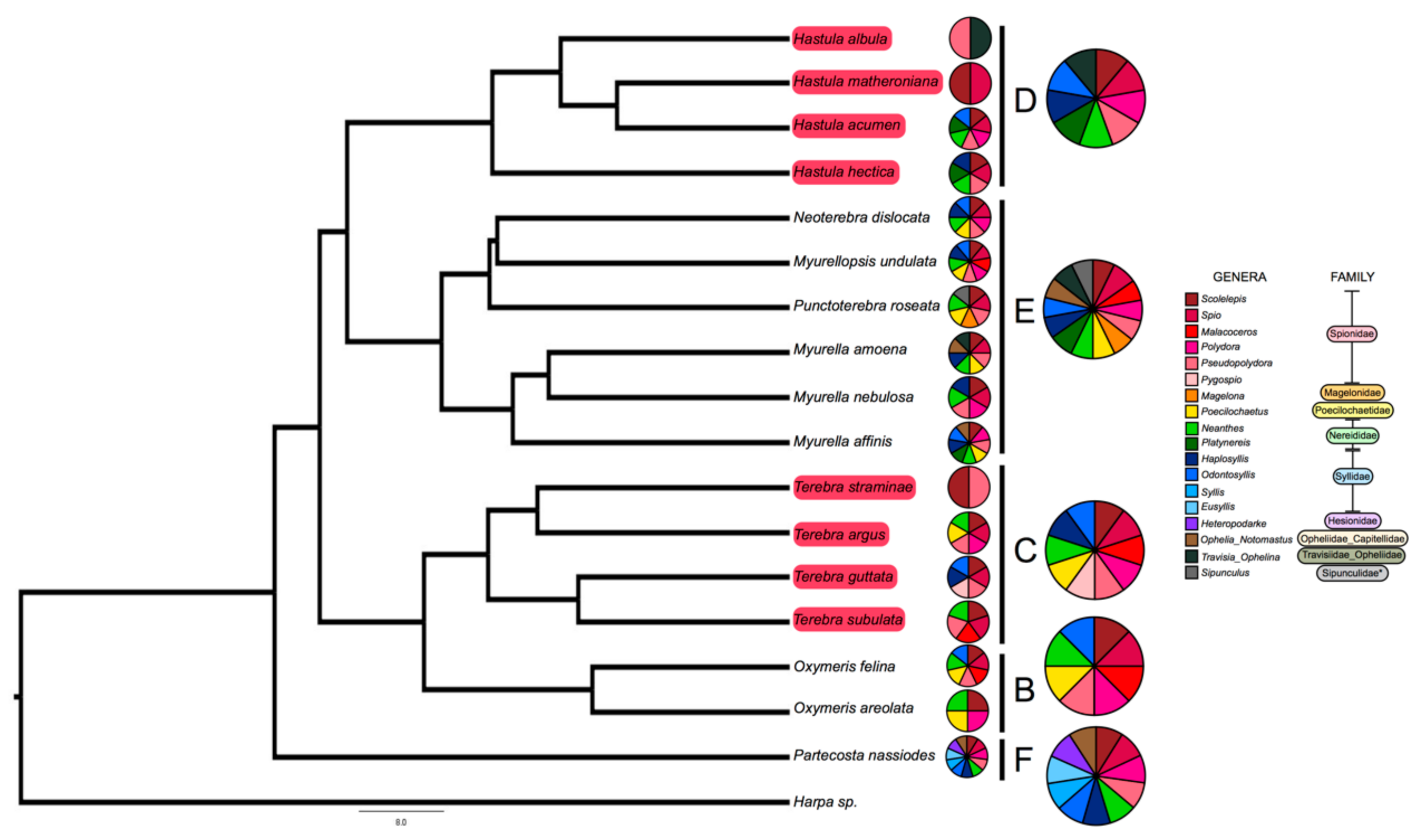
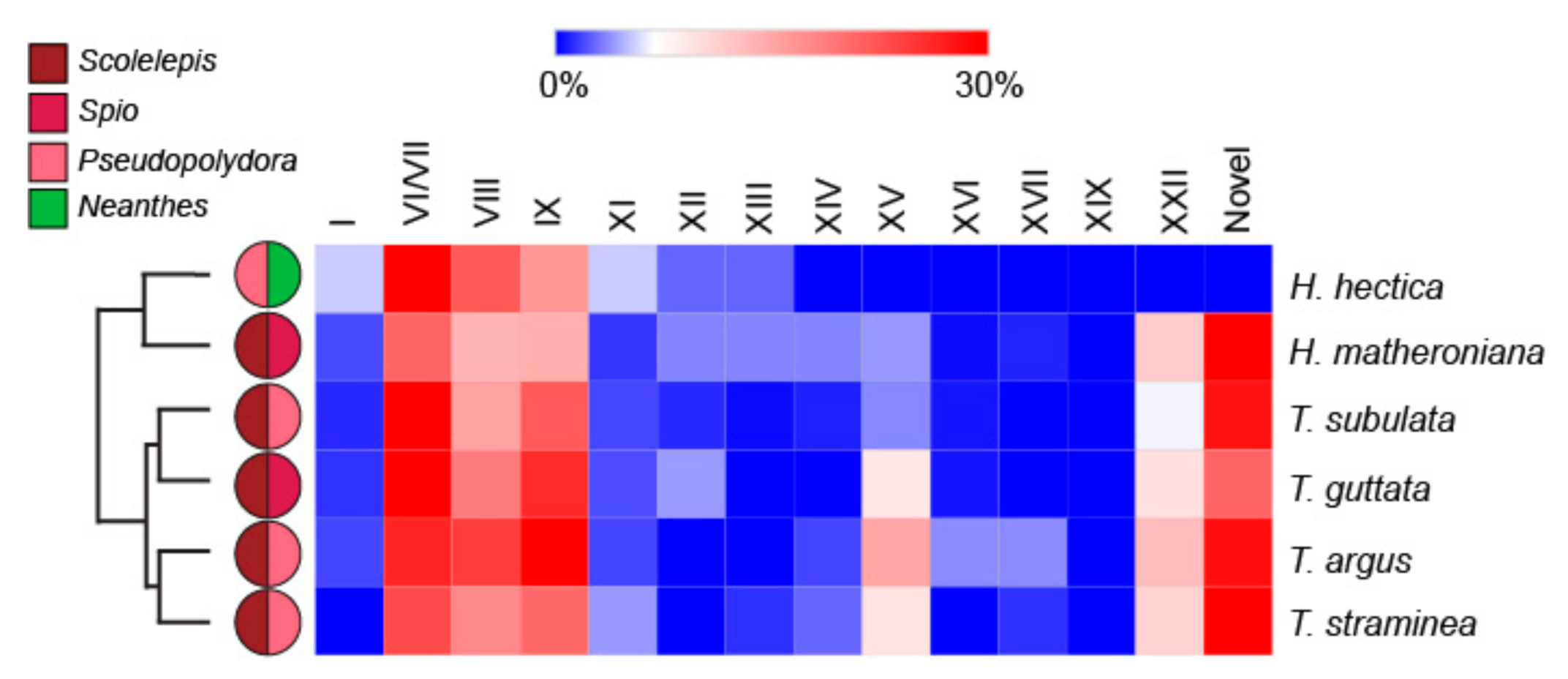
2.1. Terebrid Gut Content Reveals Diverse Annelid Prey
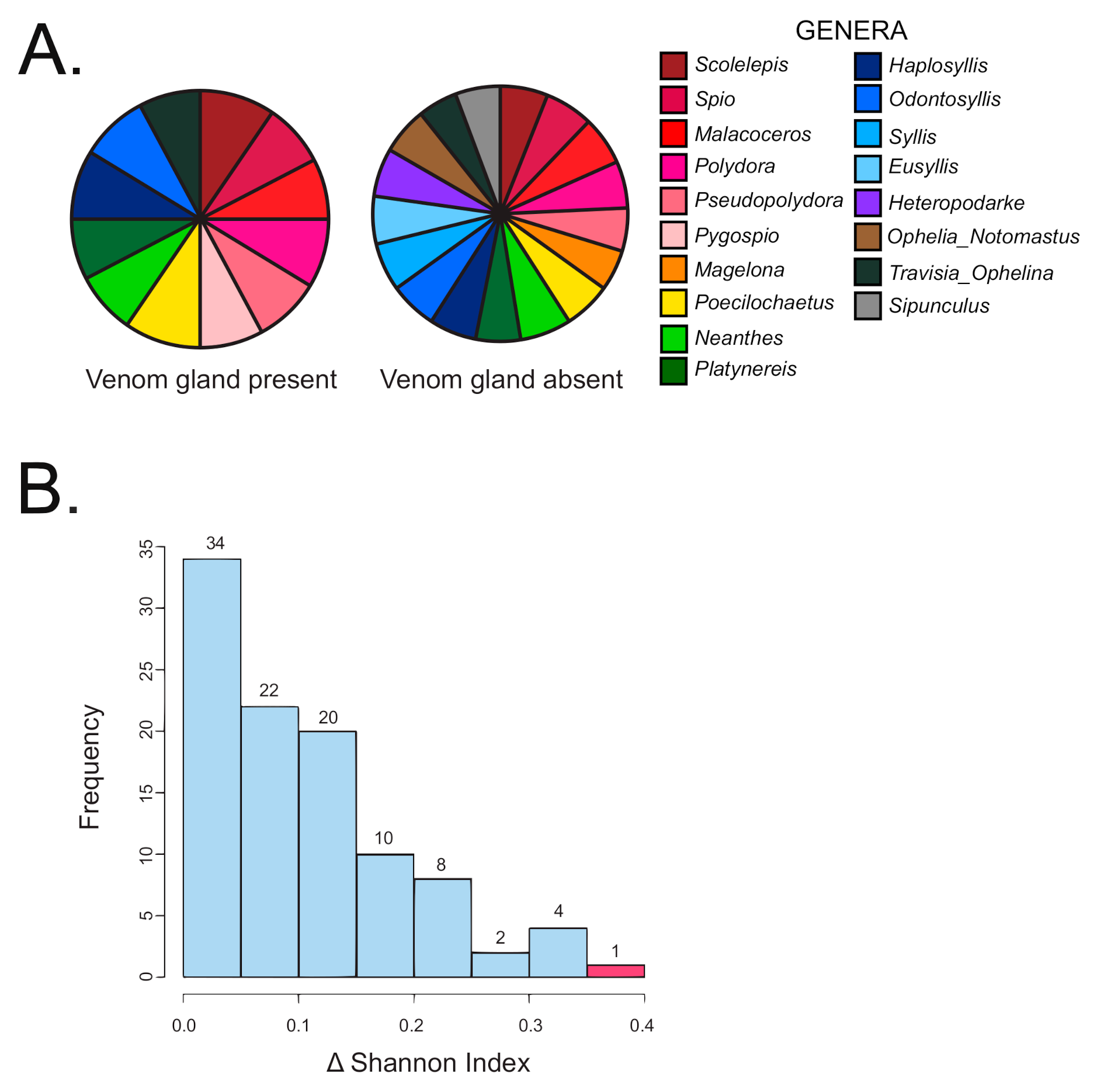
2.2. Strong Correlation between Presence and Absence of a Venom Apparatus and Terebrid Diet Diversity
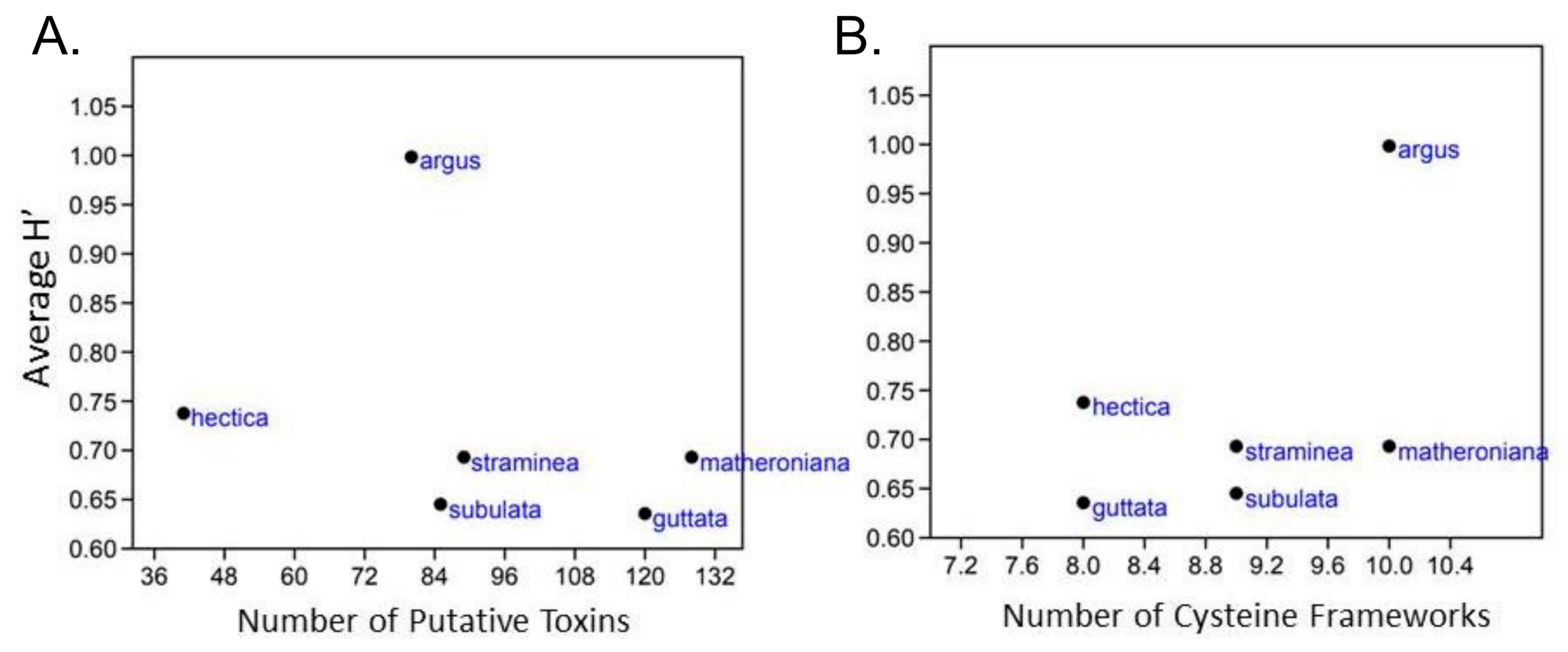
2.3. No Correlation between Terebrid Venom Complexity and Diet Diversity
2.4. No Correlation between Shell Length and Diet Diversity
3. Discussion
4. Materials and Methods
4.1. DNA Extraction and Amplification for Next Generation Sequencing
4.2. PCR Primers and Amplification
4.3. Library Preparation and Sequencing
4.4. Bioinformatics Pipeline
4.5. Reads Identification
4.6. Diet Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A. The Diversity of Venom: The Importance of Behavior and Venom System Morphology in Understanding Its Ecology and Evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pekár, S.; Toft, S.; Hrusková, M.; Mayntz, D. Dietary and prey-capture adaptations by which Zodarion germanicum, an ant-eating spider (Araneae: Zodariidae), specialises on the Formicinae. Naturwissenschaften 2008, 95, 233–239. [Google Scholar] [PubMed]
- Pucca, M.B.; Amorim, F.G.; Cerni, F.A.; Bordon, K.D.C.F.; Cardoso, I.A.; Anjolette, F.A.P.; Arantes, E.C. Influence of post-starvation extraction time and prey-specific diet in Tityus serrulatus scorpion venom composition and hyaluronidase activity. Toxicon 2014, 90, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Daltry, J.C.; Wüster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Remigio, E.A.; Duda, T.F., Jr. Evolution of ecological specialization and venom of a predatory marine gastropod. Mol. Ecol. 2008, 17, 1156–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyons, K.; Dugon, M.M.; Healy, K. Diet Breadth Mediates the Prey Specificity of Venom Potency in Snakes. Toxins 2020, 12, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phuong, M.A.; Mahardika, G.N.; Alfaro, M.E. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genom. 2016, 17, 401. [Google Scholar] [CrossRef] [Green Version]
- Vonk, F.J.; Jackson, K.; Doley, R.; Madaras, F.; Mirtschin, P.J.; Vidal, N. Snake venom: From fieldwork to the clinic: Recent insights into snake biology, together with new technology allowing high-throughput screening of venom, bring new hope for drug discovery. Bioessays 2011, 33, 269–279. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Arbuckle, K. From molecules to macroevolution: Venom as a model system for evolutionary biology across levels of life. Toxicon X 2020, 6, 100034. [Google Scholar] [CrossRef]
- Li, M.; Fry, B.G.; Kini, R.M. Eggs-only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii). J. Mol. Evol. 2005, 60, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Pahari, S.; Bickford, D.; Fry, B.G.; Kini, R.M. Expression pattern of three-finger toxin and phospholipase A2 genes in the venom glands of two sea snakes, Lapemis curtus and Acalyptophis peronii: Comparison of evolution of these toxins in land snakes, sea kraits and sea snakes. BMC Evol. Biol. 2007, 7, 175. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, N.J.; Aird, S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2001, 128, 425–456. [Google Scholar] [CrossRef]
- Richards, D.P.; Barlow, A.; Wüster, W. Venom lethality and diet: Differential responses of natural prey and model organisms to the venom of the saw-scaled vipers (Echis). Toxicon 2012, 59, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Vidal, N.; Norman, J.A.; Vonk, F.J.; Scheib, H.; Ramjan, S.R.; Kuruppu, S.; Fung, K.; Hedges, S.B.; Richardson, M.K.; et al. Early evolution of the venom system in lizards and snakes. Nature 2006, 439, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Casewell, N.R.; Wüster, W.; Vidal, N.; Young, B.; Jackson, T.N. The structural and functional diversification of the Toxicofera reptile venom system. Toxicon 2012, 60, 434–448. [Google Scholar] [CrossRef]
- Modica, M.V.; Gorson, J.; Fedosov, A.E.; Malcolm, G.; Terryn, Y.; Puillandre, N.; Holford, M. Macroevolutionary Analyses Suggest That Environmental Factors, Not Venom Apparatus, Play Key Role in Terebridae Marine Snail Diversification. Syst. Biol. 2020, 69, 413–430. [Google Scholar] [CrossRef] [Green Version]
- Holford, M.; Puillandre, N.; Terryn, Y.; Cruaud, C.; Olivera, B.; Bouchet, P. Evolution of the Toxoglossa venom apparatus as inferred by molecular phylogeny of the Terebridae. Mol. Biol. Evol. 2009, 26, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Castelin, M.; Puillandre, N.; Kantor, Y.I.; Modica, M.V.; Terryn, Y.; Cruaud, C.; Bouchet, P.; Holford, M. Macroevolution of venom apparatus innovations in auger snails (Gastropoda; Conoidea; Terebridae). Mol. Phylogenet. Evol. 2012, 64, 21–44. [Google Scholar] [CrossRef] [Green Version]
- Gorson, J.; Holford, M. Small Packages, Big Returns: Uncovering the Venom Diversity of Small Invertebrate Conoidean Snails. Integr. Comp. Biol. 2016, 56, 962–972. [Google Scholar]
- Gorson, J.; Ramrattan, G.; Verdes, A.; Wright, E.M.; Kantor, Y.; Rajaram Srinivasan, R. Molecular Diversity and Gene Evolution of the Venom Arsenal of Terebridae Predatory Marine Snails. Genome Biol. Evol. 2015, 7, 1761–1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imperial, J.S.; Kantor, Y.; Watkins, M.; Heralde, F.M., III; Stevenson, B.; Chen, P.; Hansson, K.; Stenflo, J.; Ownby, J.P.; Bouchet, P.; et al. Venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): Molecular phylogeny, foregut anatomy and comparative toxinology. J. Exp. Zool. B Mol. Dev. Evol. 2007, 308, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Grigoryan, A.; Bhuiyan, M.H.; Ueberheide, B.; Russell, V.; Quinoñez, J.; Moy, P.; Chait, B.T.; Poget, S.F.; Holford, M.; et al. Sample Limited Characterization of a Novel Disulfide-Rich Venom Peptide Toxin from Terebrid Marine Snail Terebra variegata. PLoS ONE 2014, 9, e94122. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Olenzek, A.M.; Duda, T.F., Jr. Effects of geographical heterogeneity in species interactions on the evolution of venom genes. Proc. Biol. Sci. 2015, 282, 201419874. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.D.; Miller, J.A. A new type of gastropod proboscis: The foregut of Hastula bacillus (Gastropoda: Terebridae). J. Zool. 1990, 220, 603–617. [Google Scholar] [CrossRef]
- Kantor, Y.I.; Fedosov, A.E.; Marin, I.N. An Unusually High Abundance and Diversity of the Terebridae (Gastropoda: Conoidea) in Nha Trang Bay, Vietnam. Zool. Stud. 2012, 51, 663–670. [Google Scholar]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 2009, 276, 2443–2449. [Google Scholar]
- Fry, B.G.; Grigoryan, A.; Bhuiyan, M.H.; Ueberheide, B.; Russell, V.; Quinoñez, J.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef] [Green Version]
- Kordis, D.; Gubensek, F. Adaptive evolution of animal toxin multigene families. Gene 2000, 261, 43–52. [Google Scholar] [CrossRef]
- Phuong, M.A.; Mahardika, G.N. Targeted sequencing of venom genes from cone snail genomes reveals coupling between dietary breadth and conotoxin diversity. Mol. Biol. Evol. 2018, 35, 1210–1224. [Google Scholar] [CrossRef] [Green Version]
- Van Valen, L. Morphological Variation and Width of Ecological Niche. Am. Nat. 1965, 99, 377–390. [Google Scholar] [CrossRef]
- Mans, B.J.; Louw, A.I.; Neitz AW, H. The major tick salivary gland proteins and toxins from the soft tick, Ornithodoros savignyi, are part of the tick Lipocalin family: Implications for the origins of tick toxicoses. Mol. Biol. Evol. 2003, 20, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Biggs, J.S.; Olivera, B.M.; Kantor, Y.I. Alpha-conopeptides specifically expressed in the salivary gland of Conus pulicarius. Toxicon 2008, 52, 101–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, G.-S.; Sarkar, I.N.; Siddall, M.E. Salivary transcriptome of the North American medicinal leech, Macrobdella decora. J. Parasitol. 2010, 96, 1211–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stafford-Banks, C.A.; Rotenberg, D.; Johnson, B.R.; Whitfield, A.E.; Ullman, D.E. Analysis of the Salivary Gland Transcriptome of Frankliniella occidentalis. PLoS ONE 2014, 9, e94447. [Google Scholar] [CrossRef]
- Bose, U.; Wang, T.; Zhao, M.; Motti, C.A.; Hall, M.R.; Cummins, S.F. Multiomics analysis of the giant triton snail salivary gland, a crown-of-thorns starfish predator. Sci. Rep. 2017, 7, 6000. [Google Scholar] [CrossRef] [Green Version]
- Duda, T.F., Jr.; Chang, D.; Lewis, B.D.; Lee, T. Geographic Variation in Venom Allelic Composition and Diets of the Widespread Predatory Marine Gastropod Conus ebraeus. PLoS ONE 2009, 4, e6245. [Google Scholar] [CrossRef]
- Illumina Nextera (R) XT DNA Library Preparation Guide. 2014.
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Lab, H. FASTX Toolkit; Cold Spring Harbor Laboratory: New York, NY, USA, 2014. [Google Scholar]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.-I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gatew. Comput. Environ. Workshop 2010. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 29 January 2021).
- Eriksson, A.; Anand, P.; Gorson, J.; Grijuc, C.; Hadelia, E.; Stewart, J.C.; Holford, M.; Claridge-Chang, A. Using Drosophila behavioral assays to characterize terebrid venom-peptide bioactivity. Sci. Rep. 2018, 8, 15276. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorson, J.; Fassio, G.; Lau, E.S.; Holford, M. Diet Diversity in Carnivorous Terebrid Snails Is Tied to the Presence and Absence of a Venom Gland. Toxins 2021, 13, 108. https://doi.org/10.3390/toxins13020108
Gorson J, Fassio G, Lau ES, Holford M. Diet Diversity in Carnivorous Terebrid Snails Is Tied to the Presence and Absence of a Venom Gland. Toxins. 2021; 13(2):108. https://doi.org/10.3390/toxins13020108
Chicago/Turabian StyleGorson, Juliette, Giulia Fassio, Emily S. Lau, and Mandë Holford. 2021. "Diet Diversity in Carnivorous Terebrid Snails Is Tied to the Presence and Absence of a Venom Gland" Toxins 13, no. 2: 108. https://doi.org/10.3390/toxins13020108
APA StyleGorson, J., Fassio, G., Lau, E. S., & Holford, M. (2021). Diet Diversity in Carnivorous Terebrid Snails Is Tied to the Presence and Absence of a Venom Gland. Toxins, 13(2), 108. https://doi.org/10.3390/toxins13020108